Abstract
Objectives
Accurate and efficient knee cartilage and bone segmentation are necessary for basic science, clinical trial, and clinical applications. This work tested a multi-stage convolutional neural network framework for MRI image segmentation.
Materials and methods
Stage 1 of the framework coarsely segments images outputting probabilities of each voxel belonging to the classes of interest: 4 cartilage tissues, 3 bones, 1 background. Stage 2 segments overlapping sub-volumes that include Stage 1 probability maps concatenated to raw image data. Using six fold cross-validation, this framework was tested on two datasets comprising 176 images [88 individuals in the Osteoarthritis Initiative (OAI)] and 60 images (15 healthy young men), respectively.
Results
On the OAI segmentation dataset, the framework produces cartilage segmentation accuracies (Dice similarity coefficient) of 0.907 (femoral), 0.876 (medial tibial), 0.913 (lateral tibial), and 0.840 (patellar). Healthy cartilage accuracies are excellent (femoral = 0.938, medial tibial = 0.911, lateral tibial = 0.930, patellar = 0.955). Average surface distances are less than in-plane resolution. Segmentations take 91 ± 11 s per knee.
Discussion
The framework learns to automatically segment knee cartilage tissues and bones from MR images acquired with two sequences, producing efficient, accurate quantifications at varying disease severities.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Knee osteoarthritis (OA) is a chronic joint disease estimated to affect more than 7% of the United Stated population [1]. While OA damages all joint tissues, cartilage degeneration is the hallmark [2, 3]. As such, MRI is recommended for assessing structural OA by quantifying cartilage outcomes [4,5,6]. Medial joint cartilage is of particular interest due to a higher prevalence of medial knee osteoarthritis [7]. Cartilage localization must be highly accurate to enable measurements specific and sensitive enough to characterize its structure for identifying disease-modifying drugs and clinical use.
A major hurdle to quantifying cartilage outcomes from MRI scans is the resources necessary for tissue segmentation. Manual segmentation is the gold standard; however, it takes hours to perform, requires highly specialized knowledge, and suffers from inter- and intra-rater errors [8,9,10]. As a result, manual segmentation is not a realistic strategy to manage large volumes of data necessary to understand knee OA progression or for clinical trials. For example, the Osteoarthritis Initiative (OAI), a private–public collaboration by the National Institutes of Health (NIH), acquired serial MRI scans of thousands [11]. Not surprisingly, quantitative cartilage outcomes are unavailable for most OAI scans.
Automated extraction of cartilage outcomes is necessary to enable efficient analysis. Conventional methods include statistical shape modelling (SSM), active appearance models, and traditional machine learning (e.g., support vector machines, k-nearest neighbours). These methodologies require 10 min to 48 h [12,13,14,15] and fall short of human accuracies [12,13,14,15,16,17,18,19,20]. Convolutional neural networks (CNN) have become state-of-the-art for many tasks in computer vision [21] and have shown promise in cartilage segmentation [22,23,24,25].
Segmenting 3D medical data using deep learning is limited by memory available on graphics processing units (GPUs) and small samples. A 3D CNN trained on a 12 GB GPU typically handles images shaped 128 × 128 × 64 [26] with a batch size of only one. Images from the OAI commonly used for cartilage quantification (384 × 384 × 160) would thus require downsampling to less than 5% of their original size. Deep learning methods have drastically reduced segmentation times, with the fastest algorithms taking < 15 s but still fall short of traditional automated methods on accuracy [23, 25]. Meanwhile, Ambellan et al. [22] showed that a combination of SSMs with deep learning matched or exceeded traditional methodologies in accuracy, with a segmentation time of 10 min per knee. Nevertheless, heterogeneity introduced by osteophytes required manual definitions in this SSM, making transfer to new tissues more challenging. We require methods that better balance speed and accuracy to enable wide-spread use of cartilage analyses.
Multiple methods have been proposed to overcome the memory constraints. Original methods used sub-volume or patch-based methods [27]. Zeng and Zheng [28] used 3D tiled convolutions with periodic down-shuffling to reduce memory use, enabling larger patches, and outperforming conventional U-Net and V-Net architectures. Multi-stage methods have also been proposed, using stage one to localize and stage two to produce a higher-resolution segmentation, or to vote using machine learning methods other than deep learning. For example, the SSM plus deep learning method by Ambellan et al. [22] leverages stage one to localize the bone surface using a CNN, refines the CNN segmentation using an SSM and then does sub-volume segmentation using a smaller 3D-CNN over the bone surface. In applications to segment abdominal tissues, Zhu et al. [29] use a patch-based method. First, to identify a bounding box of the tissue of interest, the whole image was segmented in large patches (643 pixels), then smaller patches re-segmented this smaller region. Roth et al. [30] use a similar approach but without a bounding box; instead, this approach identified a new region of interest of arbitrary shape from stage one and then re-segmented this smaller region using a second network in stage two. Separately, Roth et al. [31] use holistically nested CNNs to segment all three orthogonal planes of an image in 2D and then aggregate the 2D predictions using a random forest classifier. Similarly, Pang et al. [32] used a CNN at stage one and then refined the segmentation in stage two using a custom conditional random field [53] also used a final conditional random field to refine a segmentation produced by a CNN; however, their initial CNN used a patch-based method that included two paths [53]. The different paths processed patches of the input image centered on the same image location at different scales, providing local and global contexts.
The purpose of this study is to define a general multi-stage fully CNN framework for segmenting knee cartilage and bone from MRI data that overcomes memory constraints, while balancing computation time and accuracy. Our method builds upon previous segmentation literature. In our multi-stage framework, Stage 1 uses multiple CNNs to segment the image coarsely and Stage 2 conducts a fine segmentation, where the CNN takes inputs of both raw image data as well as the probabilities outputted from Stage 1. Providing probabilities gives global context to the second stage that segments image sub-volumes in 3D; this global context has not been used in previously proposed methods. We describe the segmentation algorithm, then train and test the algorithm on images acquired from two MRI vendors using different pulse sequences, and in samples with and without OA.
Materials and methods
Segmentation framework
The framework (Fig. 1) uses two stages of CNNs. Stage 1 performs two coarse segmentations (one 2D, one 3D CNN) of the entire MRI volume. Each outputs a 4D probability map, where the indices along the last (4th) dimension contain the probabilities for each voxel belonging to the eight classes of interest. Classes are the different categories that each voxel can be categorized to; here 7 anatomical tissues and the background. Stage 2 segments overlapping sub-volumes of the image, taking inputs of full-resolution image data and priors (probability maps) outputted from Stage 1. The segmented sub-volumes are then combined to create a full-resolution segmentation.
Visualization of the algorithm. Stage 1 coarsely segments the input image using two convolutional neural networks (CNNs). Probability outputs of Stage 1 are combined with the raw image to perform full-resolution segmentation of sub-volumes using a third CNN in Stage 2. Stage 2 segmentations from the main tissues, or classes, of interest are combined with coarse segmentations at the periphery to produce the final segmentation
Stage 1
Two networks that share the same U-Net style architecture segmented the background and 7 tissues of interest: femur, tibia, patella, femoral cartilage, medial tibial cartilage, lateral tibial cartilage, and patellar cartilage [33]. The first had an input shape of 128 × 128 × 64 and segmented an entire image in a single pass. The second network segmented individual slices at an image size of 384 × 384. The 2D network was applied to each slice, then the slices were combined to create the full segmentation. Results of the 2D and 3D analyses were resampled to the original image shape. For both networks, prior to segmentation, the whole 3D image was normalized to have mean zero and unit variance.
The coarse network used for Stage 1 is described in Fig. 2. Like U-Net, it included long residual connections from the compression to decompression branches and short residual connections at each level of the compression and decompression branches [26, 27]. Short residual connections used a summation methodology and long residual connections used concatenation [34]. The compression branch iteratively reduced the image dimension by 1/2 using a stride of 2 with a regular convolution. The decompression branch iteratively doubled image dimensions using a stride of 2 with a transpose convolution. The number of filters for each convolution is included in Fig. 2. Convolutions throughout the network comprised 5 × 5 (2D) or 3 × 3 × 3 (3D) filters, followed by batch normalization, dropout (probability = 0.2), and a parametric rectified linear unit (PReLU) [35]. In addition, we included a form of deep supervision inspired by previous work [34]. Deep supervision directly passed data from the deep layers to the final output, using the same number of filters as classes. However, we used a PReLU activation instead of the logistic or Softmax functions [34]. The final convolution used a Softmax activation which gives probabilities of each voxel belonging to the 8 classes (4 cartilage, 3 bone, 1 background).
Visual depiction of the Stage 1 (coarse) and Stage 2 (fine) segmentation network architectures. Yellow cubes represent inputs, white are regular convolutions with parametric rectified linear unit (PReLU) activations, red are down convolutions (2 × 2 × 2 stride) with PReLU activations, green are transpose (up) convolutions (2 × 2 × 2) with PReLU activations, orange are transpose convolutions (2 × 2 × 2) with Softmax activations, blue are regular convolutions with a 1 × 1 × 1 filter and Softmax activations. Orange circles represent addition and blue circles represent concatenation. The number of filters in a convolution is printed on the face of each cube
During training, the 3D network used batch sizes of 1 and the 2D network used batch sizes of 16. Image augmentation included 50% probability of flipping along the slice-axis (3D network only), random height and width shift (± 20%), and random in-plane rotation (± 6°). The Adam optimizer [36] with a learning rate of 10–4 and early stopping was implemented. Early stopping was invoked after 10 epochs with < 0.02 improvement in the loss, when evaluated on validation data. Learning rate and early stopping criteria were determined using a grid search in a previous study [37]. The loss function was a custom generalized Dice similarity coefficient (gen-DSC). In the following, (1) is the traditional DSC, and (2), which utilizes (1) in its definition, is the gen-DSC:
where N is the number of classes, Xn is the predicted probability map for class n, and Yn is the ground truth (manual) segmentation for class n. Under this paradigm, the best possible gen-DSC score was − 8 (the negative of the number of classes).
Stage 2
This single 3D CNN (Fig. 2) segmented sub-volumes, sized 32 × 32 × 32, of the original image. This network took a 4-dimensional input, where the first 3 dimensions were the physical dimensions of the sub-volume, and the 4th dimension was length 17; 1 for original image data, 8 for probabilities from the coarse 3D segmentation, and 8 for probabilities from the coarse 2D segmentation. The whole image was normalized before sub-volume extraction. For efficiency, a region 30% bigger than the region containing all cartilage, determined using results from Stage 1, was extracted and segmented (Fig. 3). Sub-volumes that overlap by 50% in each of the three dimensions were extracted resulting in 8 predictions for all but the most peripheral voxels.
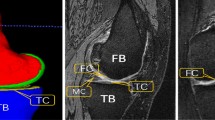
Example of the region that was extracted (orange) then segmented in Stage 2. The region is 30% larger than the cartilage segmentations and includes all cartilage as well as the primary bone areas of interest, e.g., locations of osteophyte formation. Osteophytes are bony growths at the joint margin characteristic of osteoarthritis (OA). The example image is a participant from the OA dataset
The fine network (Fig. 2) used 3 × 3 × 3 convolutions. The same as the Stage 1 network, the final filter used a Softmax activation. Compared to the Stage 1 network, the fine network included 3 down convolutions (versus 4), a greater number of filters, and used addition for long and short residual connections. More filters enabled high-level feature extraction not possible in the coarse network due to memory constraints. Shallower depth enabled bigger batch sizes. Finally, deep supervision used a Softmax activation as originally designed [34].
During training of Stage 2, every 33 epochs 1000 sub-volumes were selected from each image using stratified random sampling. Stratified random sampling ensured that, at a minimum, each tissue was included in the following percentages of sub-volumes: femur 9%, tibia 9%, patella 7%, femoral cartilage 22%, medial tibial cartilage 15%, lateral tibial cartilage 15%, patellar cartilage 15%, and background 8%. A batch size of 8 and the Adam optimizer with a learning rate of 10–3 with early stopping was implemented. Early stopping was invoked after 10 epochs with < 0.00025 improvement when tested on the validation data. These parameters were determined using a grid search.
To train the class-imbalanced tissues, a custom-weighted generalized DSC (weighted-DSC) was utilized. Class-imbalance is the unequal occurrences of the different classes (that is, tissues). The weighted-DSC multiplied a DSC per class by a weighting factor. The background class weighting factor was 1. Weighting factors for the non-background classes summed to 7 (number of non-background classes). For each sub-volume, weightings for non-background classes were calculated to distribute the available weighting. Weighting for each sub-volume was distributed based on the percentage of non-background voxels (from both prediction and ground truth), which each class occupied. For example, if the sub-volume only contained one class in both the prediction and ground truth segmentations, then that class received a weighting of 7 (1.0 × 7) and all others 0 (0.0 × 7); if there were two classes in the sub-volume, one in 25% of the voxels and the other in 75%, then they would have weightings of 1.75 (0.25 × 7) and 5.25 (0.75 × 7), respectively, with all other classes being weighted at 0 (0.0 × 7). The primary goal of this implementation was to weight classes that are not present to 0, and to distribute the total available metric over the tissues that were present. In preliminary testing, on only cartilage tissues [37], implementing this loss function produced better results than the Gen-DSC. Performance of the final Stage 2 predictions when trained using the same Gen-DSC as the Stage 1 networks is provided in Supplemental 1. Because predictions were not binary, the number of voxels for any class was calculated by summing the probabilities of each voxel belonging to that class. In the following, (3) describes the calculation for the number of non-background-voxels (NBV), (4) describes the proportion of NBVs for a given class (pNBV), and (5) describes the full weighted-DSC calculation:
where N is the number of classes, and 1 is the background class; I is image dimension 1, J image dimension 2, and K image dimension 3; X is the predicted probability map, and Y the ground truth probability map; the first 3 dimensions of X and Y are the image dimensions, and the 4th dimension is the segmentation class. Weighted-DSC efficiently learned to segment the tissues and outperformed the generalized DSC used in previous implementations of this algorithm [37].
Compiling final segmentations
To create full-resolution segmentation probability maps, first the full-sized 2D and 3D segmentation probabilities from Stage 1 were averaged, yielding the average-coarse-segmentation. From Stage 2, the segmentation probabilities for each class from overlapping sub-volumes were averaged, yielding the average-fine-segmentation. The final segmentation consisted of the average-fine-segmentation in the region it was extracted from (Fig. 3), and the average-coarse-segmentation in the area outside this region. This scheme efficiently segmented the full image, with high resolution for cartilage and bone approximating the articular surfaces.
The final segmentation was one-hot-encoded by classifying each voxel according to the class it had the highest probability of belonging to. To remove spurious segmentations, any regions of a class that were < 500 connected voxels were labelled as background. This threshold was based on qualitative interpretation of interim results [37]. Supplemental 2 includes the segmentation accuracies if the post-processing (removing regions < 500 voxels) was not performed. The secondary analysis showed that this post-processing did not consistently affect cartilage segmentation accuracies compared to the unprocessed results. Bone segmentations were consistently improved by post-processing. Thus, post-processing is not recommended for cartilage segmentation as it may omit true cartilage in the severe OA knee. However, we suggest that only the single largest connected bone region is included, as done previously [38].
Testing accuracy and segmentation times
To test accuracy and segmentation times, we conducted a sixfold cross-validation. Data from two samples were used: (1) 176 knee MRI scans from 88 individuals enrolled in the OAI, and (2) 60 MRI scans from 15 healthy young men.
Osteoarthritis sample
Data were from individuals [45 Male, 43 Female; mean (SD); 61.2 (10.0) years, 31.1 (4.6) kg/m2] in the OAI with doubtful (grade 1; n = 2), minimal (grade 2; n = 31), moderate (grade 3, n = 52), and severe (grade 4; n = 3) OA severity defined on the gold standard Kellgren and Lawrence (KL) classification system [39]. Images were acquired using a 3T Siemens MRI scanner and quadrature transmit—receive knee coil (USA Instruments, Aurora, OH) at one of four sites using a 3D sagittal water excited T1-weighted (TE 5 ms, TR 16 ms, flip angle 25º) Dual Echo in the Steady State (DESS) sequence with in-plane resolution of 0.365 × 0.365 mm, matrix size 384 × 384, and slice thickness of 0.7 mm [11, 40]. Manual segmentations of cartilage were performed for the OAI by an industrial partner (iMorphics) [41]. Manual segmentations of the 3 bones (femur, tibia, patella) were conducted by one researcher (AAG); manual segmentations were completed once. Quality control ensured that every image was carefully reviewed and updated where necessary. This quality control was completed > 6 months after initial segmentation.
Healthy sample
In a previous study [42], healthy male participants [mean (SD); 25.8 (4.2) years, 23.71 (2.62) kg/m2] completed 2 MRI visits, obtaining 2 sets of knee MRI scans per visit (4 image sets per participant). Images were acquired using a 3T GE Discovery MR750 (GE Healthcare, Milwaukee WI) and 16-channel receive knee coil array (Invivo Corp) using a 3D sagittal fat saturated T1-weighted (TE 5.83 ms, TR 17.39 ms, flip angle 18º) fast spoiled gradient recalled (FSPGR) sequence, in-plane resolution of 0.3125 × 0.3125 mm, matrix size 512 × 512, and slice thickness of 1.0 mm [42]. Manual cartilage and bone segmentations were conducted by one researcher (AAG); manual segmentations were completed once. Quality control ensured that every image was carefully reviewed and updated where necessary. This quality control was completed > 6 months after initial segmentation.
Experiments
For sixfold cross-validation, 236 MRI volumes from 103 participants were split into 6 partitions by participant. Partitions are described in Table 1. Partitions included data from both the OA and healthy samples and thus had a broader range of disease states and a broader array of MR sequences than previous investigations. These additional data, along with differences in data splits in the OAI sample, limit definitive identification of the optimal method when compared to the literature. Using all data were chosen so that one pipeline could be used for future predictions, instead of requiring creation of a new pipeline for every new data source. Six cross-validation folds balance the trade-offs between bias and variance [43]. During each fold, one partition was held out for final testing. Of the remaining five partitions, one was used for validation (to assess early stopping) and the remaining four were used to train all stages. Assessments of segmentation quality were performed for five cartilage classes (femoral, medial tibial, lateral tibial, all tibial, patellar) and three bones (femur, tibia, patella).
Assessments were run on the entire image volume, as well as on cropped image volumes that were 30% bigger than the region containing all cartilage, with cropping performed based on the reference segmentations. Assessments were performed and reported for all folds of the final testing data, and separately for all folds of the validation data.
Assessment of segmentation accuracies was performed using the DSC (Eq. 1), the volume difference [VD; (6)], and the average surface distance [ASD; (7)] [16, 22, 23, 44]. The DSC and ASD are symmetric; VD is a percent difference relative to the reference volume [22]. Time to complete all segmentation steps was recorded. VD and ASD were defined as follows:
where Xn is the predicted segmentation for tissue n, and Yn is the ground truth, ∂X and ∂Y are the boundary voxels of segmentations X and Y, n∂X and n∂Y are the number of boundary voxels for ∂X and ∂Y, \(\left|\bullet \right|\) is the volume, and \({\Vert \bullet \Vert }_{2}\) the Euclidean distance.
To explore the role of disease severity and morphometry on segmentation accuracies, two analyses were performed independently for each cartilage tissue of interest. In the OA sample, we explored whether cartilage DSC was dependent on radiographic disease severity (KL grade), using one-way analysis of variance (ANOVA). In all participants, we determined whether the DSC was dependent on tissue volume using linear regression; separate regression models were run for the OA and healthy samples.
Finally, an ablation study was performed to determine how Stage 2 (fine) added to prediction accuracy. Using only the testing data, we report the segmentation accuracies (DSC, VD, and ASD) separately for the two Stage 1 segmentations, as well as a simple average of the two Stage 1 segmentations. All post-processing was implemented the same as the full model; e.g., the segmentation was one-hot-encoded, and islands less than 500 connected voxels were removed.
All experiments were conducted using a virtual machine with 12 CPU threads, 78 GB of RAM, and an NVIDIA Tesla V100 GPU on the Google Cloud Platform. Keras with a Tensorflow backend in Python was used.
Results
Summary statistics of segmentation accuracies, by sample, are presented in Table 2 and graphically displayed in Fig. 4. For the testing data of the OA sample, the mean (SD) DSCs were: femoral cartilage 0.907 (0.022); medial tibial cartilage 0.876 (0.042); lateral tibial cartilage 0.913 (0.026); all tibial cartilage 0.897 (0.026); and patellar cartilage 0.840 (0.128). DSC distributions by KL grade (Fig. 5, Table 4) show small decreases in accuracy with worsening disease severity. In the OA sample, mean VDs were systematically larger using the proposed methodology for the femoral and tibial cartilage, and smaller for the patellar cartilage, compared to the reference. The mean and median ASD for all OA segmentations were less than in-plane resolution (0.365 mm). OA sample segmentation times were on average 91.4 (9.6) s.
Visualizations of secondary analyses of dependence of Dice similarity coefficient (DSC) on Kellgren and Lawrence (KL) disease severity (top) and cartilage volume (bottom). Top: violin plots of DSC by KL disease severity for individuals with osteoarthritis (OA). One-way analyses of variance were run to compare DSCs across KL grades. Post-hoc significant p values, at Bonferroni corrected p < 0.0125, are included. Bottom: scatter plots and fitted regression lines predicting DSC from cartilage volume. Blue is for OA, and orange for healthy knees. The equation, its R2 and significance are presented for each fitted line in the respective legend
In the healthy sample, the mean (SD) DSC values were: femoral cartilage 0.938 (0.015); medial tibial cartilage 0.911 (0.015); lateral tibial cartilage 0.930 (0.011); all tibial cartilage 0.922 (0.010); and patellar cartilage 0.955 (0.013). Average ASD for all cartilage classes in the healthy sample were < 0.135 mm, which is less than one-half of in-plane resolution (0.3125 mm). Average ASD for bone segmentations had higher errors (femur: 0.240 mm, tibia: 0.317 mm, patella: 0.113 mm), however, when only analyzing the cropped region, the errors for the femur (0.108 mm) and tibia (0.086 mm) dropped to < 1/3 of the in-plane resolution. Healthy sample segmentation times were on average 91.4 (10.8) s.
Dependence of DSC on cartilage volume (Fig. 5) was observed in femoral cartilage in the healthy sample (R2 = 0.30) and patellar cartilage in the OA sample (R2 = 0.33). All other R2 for correlations between cartilage DSC and volume were < 0.14, indicating negligible volume dependence.
The ablation study showed that, other than for VD in the tibial cartilage of the healthy group, the full algorithm (Stage 2) outperformed all Stage 1 (2D, 3D, average) segmentations for all metrics (Table 3). In the exception, (healthy tibial cartilage regions), the Stage 1 2D model had a small overestimation of volume (medial tibial: 0.87, lateral tibial: 0.24) whereas the full model had a small underestimation for medial tibial cartilage (− 1.66) and lateral tibial cartilage (− 1.30). Under the current training paradigm, the Stage 1 2D model outperformed the Stage 1 3D model, and the average segmentation.
Comparing the Stage 1 2D model to the full model (Stage 2), in the OA sample, the full model outperformed in terms of DSC by 0.002–0.007 across all tissues (Table 3; p < 0.001 for all tissues). Similarly, in the bones of the healthy knees, the full model outperformed in terms of DSC by 0.001–0.002 (Table 3; p < 0.001 for all tissues). In the healthy cartilage, there was a bigger difference with the full model outperforming the Stage 1 2D model by 0.016–0.022 (Table 3; p < 0.001 for all tissues). In terms of ASD, the full model performed considerably better than the Stage 1 2D model producing ASDs between 3.4% (Healthy tibia, p = 0.003) and 42.6% (OA patella, p < 0.001) better. In cartilage regions, the full model outperformed the Stage 1 2D model by 6.9% (medial tibial cartilage, p < 0.001) to 10.5% (lateral tibial cartilage, p < 0.001) for the OA knees and by 14.2% (lateral tibial cartilage, p < 0.001) to 28.4% (patellar cartilage, p < 0.001) in the healthy knees. Looking at improvements of the full model (Stage 2) compared to the Stage 1 2D model by KL grade (Table 4), the improvements of the full model are consistent across the range of disease severities.
The Stage 1 (coarse) 3D model was the worst in every region and outcome for both healthy and OA knees. The average segmentation outperformed the Stage 1 2D model for ASD of all OA bones (femur 0.251 mm versus 0.258 mm; tibia 0.308 mm versus 0.322 mm; patella 0.132 mm vs 0.162 mm), and effectively tied the Stage 1 2D model for DSC in those regions. With the exception of these OA bones, the average segmentation had outcomes in between the Stage 1 2D and 3D models.
Discussion
We present a novel framework for segmenting cartilage and bone from knee MRI scans using only CNNs. When benchmarked on the OAI dataset, results demonstrate excellent DSC and ASD for femoral and medial and lateral tibial cartilage (Table 5) [22, 44,45,46]. The results on the healthy dataset outperformed the OAI dataset in all cartilage regions. The full model outperformed the Stage 1 predictions in overlap (DSC) and particularly surface distance (ASD) metrics. The segmentations produced were among the fastest times reported [22, 25, 46, 47]. The proposed algorithm demonstrates an ability to learn, without human intervention, how to automatically segment cartilage and bone from both sagittal FSPGR and DESS MRI sequences on individuals with and without OA. This work enables an efficient quantification of cartilage outcomes for research (basic science, clinical trials) and clinical usage.
The proposed framework failed to match the best cartilage segmentation accuracies for patellar cartilage, when benchmarked on the OAI dataset, achieving mean DSC of 0.840 and mean ASD of 0.354 mm. Three algorithms beat the current implementation (Table 5) [44,45,46]. All of these models tested their results on small samples using < 1/2 the sample of this study (Table 5). As seen in Fig. 4, there is a long tail of patellar cartilage DSCs on the OAI dataset, indicating a few poor performances reduced accuracy. Figure 5 shows these poor results occurred in knees with low patellar cartilage volume. It is possible that previous algorithms were tested on sub-samples [44,45,46] that did not include these knees with low patellar cartilage volume (Fig. 5). There are a few reasons for this volume dependence. First, as thin structures like cartilage decrease in volume, a greater proportion of voxels are on the boundaries, where the majority of errors occur. Second, in low-volume segmentations, the cartilage itself is sparse, and at times disconnected, likely introducing greater error in manual segmentations (Fig. 6). Still, the framework proposed by Panfilov et al. produced excellent DSCs for patellar cartilage. The authors used a 2D approach with a deeper network and more filters than the current implementation. A bigger 2D coarse network may improve the current framework. It is also possible that the weighted-DSC, which was primarily intended to increase weighting of tissues that are present in a volume, may have had unintended consequences. This approach gave zero weighting to classes that were absent. Concurrently, for classes that were present, this approach gave a higher weighting to ones with greater volume. As such, it is possible that the low-volume patellar cartilage may have had a small weighting, particularly in severe knees. Finally, it should be noted that segmentation of patellar cartilage in the healthy sample produced the highest accuracy cartilage segmentations we are aware of. It is possible that this discrepancy could be attributed to differences in the FSPGR versus DESS sequences or between the samples.
Raw image of a participant in the osteoarthritis sample (left) and segmentations from the manual (middle) and predicted (right). Femur (light blue), femoral cartilage (blue), tibia (light green), tibial cartilage (green), patella (light red), and patellar cartilage (red) are overlaid on the raw image. Of note is the region on the trochlea where the framework correctly omitted cartilage (orange arrow). It is also interesting to note that the predicted regions of patellar cartilage (green arrows) are plausibly correct, and therefore potentially indicate an instance of outperforming the reference segmentation
The International Workshop on OA imaging (IWOAI) competition entries provide the most direct comparison of different cartilage segmentation algorithms performed on the OAI iMorphics dataset [44]. While the results of this study cannot be directly compared to those results because of differences in data splits, some comparisons are warranted. Results from the competition often outperformed our segmentation of patellar cartilage (ours: 0.84, IWOAI Teams 1–6: 0.83, 0.86, 0.85, 0.86, 0.81, 0.86). However, the current algorithm was either comparable or perhaps even outperformed implementations in terms of DSC for femoral (ours: 0.91, IWOAI Teams 1–6: 0.88, 0.90, 0.90, 0.90, 0.87, 0.90) and all tibial (ours: 0.90, IWOAI Teams 1–6: 0.87, 0.89, 0.89, 0.89, 0.85, 0.88) cartilage. The same trends hold for average ASD. The four best teams from the IWOAI (Teams 2, 3, 4, and 6) performed very similar to one another suggesting the field may be reaching an upper limit on segmentation accuracies when benchmarked against manual methods and with dataset sizes of this magnitude. Two of these algorithms used a single prediction (Teams 4 and 6) and two used an ensemble of predictions (Teams 2 and 3). In the current ablation study, we found that an ensemble of two networks did not improve upon the results of the best network, and essentially produced a DSC or average ASD equivalent to the average of the two networks. However, in our case, the 3D network did quite poorly, and thus using multiple 2D networks in orthogonal planes may have produced better results. The two ensemble methods tested in the IWOAI competition used a simple average as well as a learned linear combination of the constituent segmentations. It is unknown how well the learned weights approach performed compared to a simple average of the constituent predictions. In our case, the second-stage CNN improved cartilage segmentation accuracies, particularly surface distances. Future work could determine whether a second-stage CNN, such as that presented in the current study, broadly improves segmentation results by experimenting with multiple combinations of Stage 1 networks.
When benchmarked on the whole OAI segmentation dataset, the current implementation produced bone DSCs for the femur (0.989) and tibia (0.987) that were better than those reported by Ambellan et al. [22]. In terms of ASD, when considering the whole image, the current implementation was slightly worse than Ambellan (femur: 0.178 mm vs 0.17 mm; tibia: 0.247 mm vs. 0.18 mm); however, the current implementation only produced coarse segmentations for the periphery of the images. When considering only the high-resolution area, the ASD drops considerably to 0.102 mm and 0.096 mm for the femur and tibia, respectively. The DSC also improved considerably for the cropped regions, yielding DSCs of 0.993 and 0.994 for the femur and tibia. These cropped regions include osteophytes and all primary classes of interest (Fig. 3). The comparison to Ambellan et al. is noteworthy because they used multiple stages of segmentations as well as SSMs to control bone shape and minimize errors. The comparable results between the approaches indicate that a fully-learned CNN approach does not require the regularization imposed by a SSM.
Segmentation times (~ 1.5 min) fall between extremely fast algorithms (< 15 s) that produce lower-accuracy results [23, 25], and slower methods (10 + min) [22] that produce comparable results. One algorithm of note by Gaj et al. [46] reports segmentation times of less than 1 min with comparable segmentation accuracies to the current framework. The work presented by Gaj et al. was tested on MRI scans from 9 individuals and thus inherently has greater variability associated with the DSC point estimates.
In the current implementation, the relatively small OA and healthy datasets create potential shortcomings. In particular, probabilities outputted from Stage 1 act as priors in Stage 2 and we trained Stages 1 and 2 using the same images. With more data, Stages 1 and 2 would ideally be trained using different sets of data, letting Stage 2 better learn how to weight priors. In addition to more data, it is possible that different network architectures or loss functions may improve results. For example, a deeper network, such as that by Panfilov et al. [45] applied to the Stage 1 2D network. Or, since the Stage 1 3D network performed worst, it is possible that using two or three coarse 2D networks trained along orthogonal planes at Stage 1 may be better. Another alternative would be to train Stage 1 using multiple similar networks that have different loss functions. For example, one network could be trained on volume overlap, and another based on surface errors [38]. This scheme could provide better information for Stage 2 to predict the final segmentation.
A primary motivation for the proposed multi-stage algorithm was to overcome memory limitations. The two-stage approach improved segmentation outcomes from the Stage 1 2D, Stage 1 3D, and Stage 1 average segmentations. However, it is important to acknowledge that these methods still require considerable memory. First, the Stage 1 3D network was still bottlenecked by available GPU memory. On a 16 GB GPU we were limited to a batch size of 1. The practice of using a batch size of 1 is a limitation because it means that the gradient estimates as well as the mean and variance estimates from batch normalization are noisy (high variance) and may adversely affect training [48,49,50]. Second, our algorithm effectively overcomes limited parallel memory using memory in a serial nature which lengthened computation time, and when considering all three models, resulted in considerable memory use (Stage 1 3D weights: 633 MB, Stage 1 2D weights: 232 MB, Stage 2 3D weights: 213 MB, total model weights: 1.08 GB). However, this serial nature, particularly the Stage 2 patch-based segmentations, potentially had an additional benefit of increasing the number of training samples. Most MRI datasets, particularly annotated ones, have sample sizes (e.g., OAI iMorphics dataset includes 176 knees from 88 people) that are much smaller than those used to achieve state-of-the-art results using conventional CNN models [51, 52]. Continued improvements to cartilage segmentation will likely require methods that are more efficient in their use of memory and available annotated data.
The gold standard of cartilage segmentation is manual. The only work to test manual segmentations tested the inter-radiologist DSC of all tibial, femoral, and patellar cartilage as one segmented region with one label [10]. That work showed a mean DSC of 0.878 for 10 individuals from the OAI with primarily mild OA (KL0: 1, KL1: 3, KL2: 4, KL3: 2). The cartilage DSCs for 3 of 4 individual compartments (femur, medial tibial, lateral tibia) from the current framework, and from many published results using deep learning [22, 44,45,46], surpass this level [10]. The only class that falls short is the patellar cartilage with a DSC of 0.84–0.87 from the 4 best algorithms. When assessing the DSC of all femoral, tibial, and patellar cartilage as one label on predictions from the current framework, the mean (SD) DSC of the OAI dataset was 0.901 (0.023). By these standards, many currently published deep learning algorithms are more accurate than the current gold-standard, manual segmentation. This may also be why many algorithms are converging to similar accuracies (Table 5) as we may be approaching the highest possible metrics when the benchmark is manual segmentation; this is highlighted by the plausibly correct segmentations predicted in Fig. 6 where the manual segmentations said there was no cartilage. These performances are also achieved in timescales measured in seconds [23, 25] or minutes [22, 46] as opposed to hours [10]. Nonetheless, algorithms make mistakes [22, 46]. Spot-checking segmentations more likely to include errors (low cartilage volume, high KL) could enable efficient analysis of data while maintaining high fidelity. Checking segmentations for accuracy is recommended for individual analysis such as that required by clinical workflows. However, analyses of large datasets allow small errors to be overcome by group statistics.
Conclusion
The proposed multi-stage CNN segmentation framework provides excellent accuracies when segmenting knee cartilage from OAI DESS and healthy FSPGR images in an average of 1.5 min. The segmentation framework is flexible and fully learns from provided examples, therefore showing promise for segmenting other musculoskeletal tissues in future work. Furthermore, a single framework was trained to handle knees across the OA severity spectrum, and from different MRI vendors and sequences. Together, these results demonstrate an ability to efficiently analyze cartilage outcomes for basic science, clinical trials, and clinical usage.
References
Deshpande BR, Katz JN, Solomon DH, Yelin EH, Hunter DJ, Messier SP, Suter LG, Losina E (2016) Number of persons with symptomatic knee osteoarthritis in the US: impact of race and ethnicity, age, sex, and obesity: symptomatic knee OA in the US. Arthritis Care Res. https://doi.org/10.1002/acr.22897
Creamer P, Hochberg MC (1997) Osteoarthritis. Lancet 350:503–508
Kraus VB, Blanco FJ, Englund M, Karsdal MA, Lohmander LS (2015) Call for standardized definitions of osteoarthritis and risk stratification for clinical trials and clinical use. Osteoarthritis Cartilage 23:1233–1241
Hunter DJ, Altman RD, Cicuttini F, Crema MD, Duryea J, Eckstein F, Guermazi A, Kijowski R, Link TM, Martel-Pelletier J, Miller CG, Mosher TJ, Ochoa-Albíztegui RE, Pelletier J-P, Peterfy C, Raynauld J-P, Roemer FW, Totterman SM, Gold GE (2015) OARSI clinical trials recommendations: knee imaging in clinical trials in osteoarthritis. Osteoarthritis Cartilage 23:698–715
Conaghan PG, Hunter DJ, Maillefert JF, Reichmann WM, Losina E (2011) Summary and recommendations of the OARSI FDA osteoarthritis Assessment of Structural Change Working Group. Osteoarthritis Cartilage 19:606–610
Peterfy C, Woodworth T, Altman R (2006) Workshop for consensus on osteoarthritis imaging: MRI of the knee. Osteoarthritis Cartilage 14:44–45
Metcalfe AJ, Andersson ML, Goodfellow R, Thorstensson CA (2012) Is knee osteoarthritis a symmetrical disease? Analysis of a 12 year prospective cohort study. BMC Musculoskelet Disord. https://doi.org/10.1186/1471-2474-13-153
Pedoia V, Majumdar S, Link TM (2016) Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn Reson Mater Phys Biol Med 29:207–221
Duryea J, Neumann G, Brem MH, Koh W, Noorbakhsh F, Jackson RD, Yu J, Eaton CB, Lang P (2007) Novel fast semi-automated software to segment cartilage for knee MR acquisitions. Osteoarthritis Cartilage 15:487–492
Shim H, Chang S, Tao C, Wang J-H, Kwoh CK, Bae KT (2009) Knee cartilage: efficient and reproducible segmentation on high-spatial-resolution MR images with the semiautomated graph-cut algorithm method. Radiology 251:548–556
Peterfy CG, Schneider E, Nevitt M (2008) The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage 16:1433–1441
Shan L, Zach C, Charles C, Niethammer M (2014) Automatic atlas-based three-label cartilage segmentation from MR knee images. Med Image Anal 18:1233–1246
Ahn C, Bui TD, Lee Y, Shin J, Park H (2016) Fully automated, level set-based segmentation for knee MRIs using an adaptive force function and template: data from the osteoarthritis initiative. Biomed Eng Online. https://doi.org/10.1186/s12938-016-0225-7
Dodin P, Pelletier J, Martel-Pelletier J, Abram F (2010) Automatic human knee cartilage segmentation from 3-D magnetic resonance images. IEEE Trans Biomed Eng 57:2699–2711
Fripp J, Crozier S, Warfield SK, Ourselin S (2010) Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans Med Imaging 29:55–64
Dam EB, Lillholm M, Marques J, Nielsen M (2015) Automatic segmentation of high- and low-field knee MRIs using knee image quantification with data from the osteoarthritis initiative. J Med Imaging (Bellingham) 2:024001
Wang Q, Wu D, Lu L, Liu M, Boyer KL, Zhou SK (2014) Semantic Context Forests for Learning-Based Knee Cartilage Segmentation in 3D MR Images. In: Menze B, Langs G, Montillo A, Kelm M, Müller H, Tu Z (eds) Medical Computer Vision. Large Data in Medical Imaging. Springer International Publishing, Cham, pp 105–115
Prasoon A, Igel C, Loog M, Lauze F, Dam EB, Nielsen M (2013) Femoral cartilage segmentation in knee MRI scans using two stage voxel classification. In: Engineering in medicine and biology society (EMBC), 2013 35th annual international conference of the IEEE. IEEE, pp 5469–5472
Tamez-Pena JG, Farber J, Gonzalez PC, Schreyer E, Schneider E, Totterman S (2012) Unsupervised segmentation and quantification of anatomical knee features: data from the osteoarthritis initiative. IEEE Trans Biomed Eng 59:1177–1186
Yin Y, Zhang X, Williams R, Xiaodong Wu, Anderson DD, Sonka M (2010) LOGISMOS—layered optimal graph image segmentation of multiple objects and surfaces: cartilage segmentation in the knee joint. IEEE Trans Med Imaging 29:2023–2037
Shen D, Wu G, Suk H-I (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248
Ambellan F, Tack A, Ehlke M, Zachow S (2019) Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the osteoarthritis initiative. Med Image Anal 52:109–118
Liu F (2018) SUSAN: segment unannotated image structure using adversarial network. Magn Reson Med. https://doi.org/10.1002/mrm.27627
Zhou Z, Zhao G, Kijowski R, Liu F (2018) Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 80:2759–2770
Norman B, Pedoia V, Majumdar S (2018) Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 288:177–185
Milletari F, Navab N, Ahmadi S-A (2016) V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV). IEEE, Stanford, CA, USA, pp 565–571 http://arxiv.org/abs/1606.04797v1
Yu L, Yang X, Chen H, Qin J, Heng P-A (2017) Volumetric ConvNets with Mixed Residual Connections for Automated Prostate Segmentation from 3D MR Images. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI’17). AAAI Press, pp 66–72
Zeng G, Zheng G (2019) 3D tiled convolution for effective segmentation of volumetric medical images. In: Shen D, Liu T, Peters TM, Staib LH, Essert C, Zhou S, Yap P-T, Khan A (eds) Medical image computing and computer assisted intervention—MICCAI 2019. Springer International Publishing, Cham, pp 146–154
Zhu Z, Xia Y, Xie L, Fishman EK, Yuille AL (2019) Multi-scale coarse-to-fine segmentation for screening pancreatic ductal adenocarcinoma. Prepring at arXiv:1807.02941 [cs]
Roth HR, Oda H, Zhou X, Shimizu N, Yang Y, Hayashi Y, Oda M, Fujiwara M, Misawa K, Mori K (2018) An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput Med Imaging Graph 66:90–99
Roth HR, Lu L, Lay N, Harrison AP, Farag A, Sohn A, Summers RM (2018) Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med Image Anal 45:94–107
Pang S, Du A, He X, Díez J, Orgun MA (2019) Fast and accurate lung tumor spotting and segmentation for boundary delineation on CT slices in a coarse-to-fine framework. In: Gedeon T, Wong KW, Lee M (eds) Neural information processing. Springer International Publishing, Cham, pp 589–597
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Kayalibay B, Jensen G, van der Smagt P (2017) CNN-based segmentation of medical imaging data. Preprint at arXiv:1701.03056 [cs]
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. In: 2015 IEEE international conference on computer vision (ICCV). IEEE, Santiago, Chile, pp 1026–1034
Kingma DP, Ba J (2017) Adam: a method for stochastic optimization. Preprint at arXiv:1412.6980 [cs]
Gatti AA (2018) NEURALSEG: state-of-the-art cartilage segmentation using deep learning–analyses of data from the osteoarthritis initiative. Abstracts from the 2018 OARSI World Congress on Osteoarthritis. Osteoarthritis and Cartilage, pp 47–48
Caliva F, Iriondo C, Martinez AM, Majumdar S, Pedoia V (2019) Distance map loss penalty term for semantic segmentation. Preprint at arXiv:1908.03679 [cs, eess]
Kellgren JH, Lawrence JS (1957) Radiological assessment of osteo-arthrosis. Ann Rheum Dis 16:494–502
Schneider E, NessAiver M, White D, Purdy D, Martin L, Fanella L, Davis D, Vignone M, Wu G, Gullapalli R (2008) The osteoarthritis initiative (OAI) magnetic resonance imaging quality assurance methods and results. Osteoarthritis Cartilage 16:994–1004
Williams TG, Holmes AP, Bowes M, Vincent G, Hutchinson CE, Waterton JC, Maciewicz RA, Taylor CJ (2010) Measurement and visualisation of focal cartilage thickness change by MRI in a study of knee osteoarthritis using a novel image analysis tool. Br J Radiol 83:940–948
Gatti AA, Noseworthy MD, Stratford PW, Brenneman EC, Totterman S, Tamez-Peña J, Maly MR (2017) Acute changes in knee cartilage transverse relaxation time after running and bicycling. J Biomech 53:171–177
Hastie T, Tibshirani R, Friedman JH (2009) The elements of statistical learning: data mining, inference, and prediction, 2nd edn. Springer, New York
Desai AD, Caliva F, Iriondo C, Mortazi A, Jambawalikar S, Bagci U, Perslev M, Igel C, Dam EB, Gaj S, Yang M, Li X, Deniz CM, Juras V, Regatte R, Gold GE, Hargreaves BA, Pedoia V, Chaudhari AS (2021) The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset. Radiology: Artificial Intelligence 3:e200078
Panfilov E, Tiulpin A, Klein S, Nieminen MT, Saarakkala S (2019) Improving robustness of deep learning based knee MRI segmentation: mixup and adversarial domain adaptation. In: 2019 IEEE/CVF international conference on computer vision workshop (ICCVW). IEEE, Seoul, Korea (South), pp 450–459
Gaj S, Yang M, Nakamura K, Li X (2020) Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks. Magn Reson Med 84:437–449
Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R (2017) Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging: Deep Learning Approach for Segmenting MR Image. Magn Reson Med. https://doi.org/10.1002/mrm.26841
Ioffe S (2017) Batch renormalization: towards reducing minibatch dependence in batch-normalized models. Preprint at arXiv:1702.03275 [cs]
Masters D, Luschi C (2018) Revisiting small batch training for deep neural networks. Preprint at arXiv:1804.07612 [cs, stat]
Lian X, Liu J (2019) Revisit batch normalization: new understanding and refinement via composition optimization. In: Proceedings of machine learning research. pp 3254–3263
Deng J, Dong W, Socher R, Li L-J, Kai Li, Li Fei-Fei (2009) ImageNet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. IEEE, Miami, FL, pp 248–255
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25:1097–1105
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical Image Analysis 3661-78. https://doi.org/10.1016/j.media.2016.10.004
Acknowledgements
We would like to acknowledge Google for providing cloud compute credits used to conduct the experiments. The Osteoarthritis Initiative (OAI) is a public-private partnership funded by the National Institutes of Health (NIH) and private partners including Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. This manuscript was prepared using an OAI public use data set and does not reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
Funding
A. A. Gatti was supported by an Ontario Graduate Scholarship, The Arthritis Society, and a Mitacs Accelerate Entrepreneur award. M.R. Maly holds The Arthritis Society Stars Mid-Career Development Award funded by the Canadian Institutes of Health Research-Institute of Musculoskeletal Health and Arthritis and an NSERC Discovery grant that supported this work (MRM: 353715).
Author information
Authors and Affiliations
Contributions
AAG contributed to study conception and design, acquisition of data, analysis and interpretation of data, drafting of manuscript, and critical revision. MRM contributed to acquisition of data, interpretation of data, drafting of manuscript, and critical revision.
Corresponding author
Ethics declarations
Conflict of interest
A. A Gatti is the founder of NeuralSeg, Ltd. There are no other conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gatti, A.A., Maly, M.R. Automatic knee cartilage and bone segmentation using multi-stage convolutional neural networks: data from the osteoarthritis initiative. Magn Reson Mater Phy 34, 859–875 (2021). https://doi.org/10.1007/s10334-021-00934-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-021-00934-z