Abstract
Both ammonia-oxidizing archaea (AOA) and bacteria (AOB) can play important roles in the microbial oxidation of ammonia nitrogen in freshwater lake, but information on spatiotemporal variation in water column and sediment community structure is still limited. Additionally, the drivers of the differences between sediment and water assemblages are still unclear. The present study investigated the variation of AOA and AOB communities in both water columns and sediments of eutrophic freshwater Dianchi Lake. The abundance, diversity, and structure of both planktonic and sediment ammonia-oxidizing microorganisms in Dianchi Lake showed the evident changes with sampling site and time. In both water columns and sediments, AOB amoA gene generally outnumbered AOA, and the AOB/AOA ratio was much higher in summer than in autumn. The total AOA amoA abundance was relatively great in autumn, while sediment AOB was relatively abundant in summer. Sediment AOA amoA abundance was likely correlated with ammonia nitrogen (rs = 0.963). The AOB/AOA ratio in lake sediment was positively correlated with total phosphorus (rs = 0.835), while pH, dissolved organic carbon, and ammonia nitrogen might be the key driving forces for the AOB/AOA ratio in lake water. Sediment AOA and AOB diversity was correlated with nitrate nitrogen (rs = -0.786) and total organic carbon (rs = 0.769), respectively, while planktonic AOB diversity was correlated with ammonia nitrogen (rs = 0.854). Surface water and sediment in the same location had a distinctively different microbial community structure. In addition, sediment AOB community structure was influenced by total phosphorus, while total phosphorus might be a key determinant of planktonic AOB community structure.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microbial ammonia oxidation plays a crucial role in nitrogen cycling in aquatic ecosystems. Ammonia conversion can be achieved by both ammonia-oxidizing archaea (AOA, affiliated with archaeal phylum Thaumarchaeota) and bacteria (AOB, within classes Beta- and Gamma-proteobacteria) (Hayden and Beman 2014). Both use ammonia monoxygenase (AMO) to catalyze the oxidation of ammonia nitrogen. The amoA gene has become a well-established functional biomarker to investigate the distribution of ammonia-oxidizing archaeal and bacterial populations in marine and estuarine ecosystems. Salinity has been known as a key factor shaping the abundance of ammonia-oxidizing microbial community (Zhang et al. 2015a). AOA are typically more abundant than AOB in marine environment (Bouskill et al. 2012; Lipsewers et al. 2014; Nakagawa et al. 2007; Newell et al. 2013), suggesting that AOA might play a more important role in ammonia oxidation. However, the numerical advantage of AOA versus AOB in estuarine ecosystem remains ambiguous (Magalhaes et al. 2009; Mosier and Francis 2008; Puthiya Veettil et al. 2015; Santoro et al. 2008; Veettil et al. 2015; Zhang et al. 2015a,b).
The distribution of sediment ammonia-oxidizing archaeal and bacterial populations has been investigated in a variety of freshwater ecosystems, such as river (Reis et al. 2015; Sonthiphand et al. 2013; Sun et al. 2013; Xie et al. 2014), reservoir (Wang et al. 2014), lake (Bollmann et al. 2014; Hou et al. 2013; Liu et al. 2015; Mukherjee et al. 2016; Zhao et al. 2013, 2014), wetland (Liu et al. 2014a; Wang et al. 2013; Yang et al. 2014), and pond (Lu et al. 2015), yet the relative importance of AOA and AOB to nitrification process in freshwater sediments remains under debate. Several previous studies indicated that AOA amoA gene abundance was usually greater than AOB in lake sediments (Herrmann et al. 2009; Hou et al. 2013; Zhao et al. 2014), while the numerical dominance of AOB over AOA amoA gene abundance was observed in sediments of many freshwater lakes on the Yunnan Plateau (Liu et al. 2015). AOA and AOB can have comparable contribution to nitrification in lake sediment (Liu et al. 2014b). So far, the links between lake sediment ammonia-oxidizing microbial populations and environmental factors still remain largely unclear. A number of factors (etc., pH, nitrogen compounds, phosphorus, and organic matter) might mutually govern the abundance, diversity and structure of lake sediment AOA and AOB (Liu et al. 2014b, 2015; Yang et al. 2016). Moreover, although the spatial variation of lake sediment AOA and AOB communities has been well-documented, information on their temporal change is still very limited. In addition, plateau lakes may harbor ammonia-oxidizing assemblages of different evolutionary origin from those in other lakes worldwide (Yang et al. 2013). However, little is known about the distribution of sediment AOA and AOB in plateau freshwater lake (Liu et al. 2015; Yang et al. 2016). Several studies have shown that planktonic AOA and AOB communities in oligotrophic high-altitude lakes can vary with sampling site and time (Auguet et al. 2011; Auguet and Casamayor 2013; Hayden and Beman 2014; Hu et al. 2010; Vissers et al. 2013; Yang et al. 2013). To date, the difference of the diversity and structure of ammonia-oxidizing microorganisms between in water column and in sediment has not been addressed, although Auguet et al. (2012) implied that AOA assemblages in water column were distantly related to their sediment counterparts.
Dianchi Lake (309 km2), located on the Yunnan Plateau, is the sixth largest freshwater lake in China, with average water depth and altitude of 4.4 and 1886 m, respectively (Yang et al. 2016). Dianchi Lake is regarded as one of the three most eutrophic lakes in China. Because it provides the substrate for nitrogen removal through denitrification and/or anammox, ammonia oxidation can be of great importance to diminish the external nitrogen pollutants (Hou et al. 2013). Therefore, detailed knowledge of AOA and AOB communities in Dianchi Lake can aid in the possible solutions for its eutrophication. Therefore, the main objective of the present study was to investigate the variation of AOA and AOB communities in both the water columns and sediments of highly eutrophic Dianchi Lake.
Materials and methods
Study sites and sampling
Four sampling locations were located in the northern part of Dianchi Lake (Fig. S1). Triplicate water and sediment samples were collected in July (summer) and October (autumn) in 2014. Water samples (1-m depth below water surface) were collected using plexiglass water sampler, while sediment samples (0–5 cm) were collected using core sampler. In this study, samples SW1, SW2, SW3, and SW4, respectively, represented the water samples collected from the sampling sites 1–4 in summer, while samples AW1, AW2, AW3, and AW4 correspondingly represented the water samples from these four sites in autumn. Moreover, samples SS1, SS2, SS3, and SS4, respectively, were referred to the sediment samples from the sampling sites 1–4 in summer, while samples AS1, AS2, AS3, and AS4 correspondingly were referred to the four sediment samples in autumn. After collection, these water and sediment samples were immediately transported to the laboratory. For water samples, pH, dissolved oxygen (DO), ammonium nitrogen (NH4 +-N), nitrate nitrogen (NO3 --N), total nitrogen (TN), and total phosphorus (TP) were determined according to the standard protocols (China Environmental Protection Agency 2002). The level of dissolved organic carbon (DOC) in water was measured using a Shimadzu 5000A TOC analyzer. The levels of sediment total organic carbon (TOC), NH4 +-N, NO3 --N, TN, and TP were determined according to the literatures (Song et al. 2012; Yang et al. 2016). The properties of water and sediment samples are listed in Table S1 and Table S2, respectively.
Molecular analyses
Microbial cells in each water sample (300 mL) were retained using 0.22-μm pore-size membrane (diameter 50 mm; Millipore), and the total genomic DNA was extracted independently from each replicate using E.Z.N.A. Water DNA kit (Omega, USA). Sediment DNA was extracted independently from each replicate using Powersoil DNA extraction kit (Mobio Laboratories, USA). DNA quality was checked by 1.0 % agarose gel electrophoresis and DNA concentration was quantified using a biophotometer (Eppendorf, Hamburg, Germany). Each replicate lake water or sediment DNA sample was individually subjected to real-time quantitative PCR (q-PCR) assays. The number of archaeal and bacterial amoA genes were quantified using the primer sets Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′)/Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′), and AmoA-1 F (5′-GGGGTTTCTACTGGTGGT-3′)/AmoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′), respectively, following the same PCR reactions as previously described (Francis et al. 2005; Rotthauwe et al. 1997; Wang et al. 2014). SYBR Green q-PCR assays was performed using an ABI 7500 FAST (Applied Biosystems). Standard curves ranging from 103 to 108 gene copies/mL were generated using serial dilutions of linearized plasmids (pGEM-T, Promega) containing cloned amoA gene amplified from environmental DNA. The amplification efficiency and coefficient (r 2) for the amplification of AOA and AOB amoA genes were 93 and 99 %, 0.994 and 0.998, respectively. One-way analysis of variance (ANOVA) followed by the Student–Newman–Keuls test were applied to assess the significant differences (P < 0.05) in the copy number of amoA gene among different samples.
The primer sets Arch-amoAF/Arch-amoAR and AmoA-1 F/AmoA-2R were also adopted for construction of AOA and AOB clone libraries, following the same conditions as previously described in the literature (Wang et al. 2014). The PCR products from triplicate samples were mixed in equal amounts and cloned into pMD19-T vector (Takara Corp, Japan). The clones containing correct size were sequenced at Beijing Genomics Research Center Co., Ltd. The obtained valid amoA gene sequences were assigned into the same operational taxonomic units (OTUs) with a maximum distance of 3 % using the MOTHUR program (Schloss et al. 2009), and OTU-based Shannon diversity index was further calculated. Phylogenetic analysis of the obtained amoA gene sequences was performed using the MEGA 6.0 software (Tamura et al. 2013). Moreover, the links between ammonia-oxidizing assemblages and water or sediment properties were identified using Pearson’s correlation analysis with the SPSS 20.0 software. Redundancy analysis (RDA) using the CANOCO 4.5 software was also applied to discriminate the links between microbial community composition and environmental factors. The fraction of amoA gene sequence in each OTU was assigned as species input and the measured water/sediment physicochemical properties as input for the environmental variables. The significance test of Monte Carlo permutations was performed to choose a suitable model of the microorganism-environment relationships. The obtained sequences in this study were deposited in the GenBank database under accession number KT317786–KT318082 and KT274817–KT275131 for AOA and KT323361–KT323795, KP902850–KP902954, and KP903025–KP903057 for AOB, respectively.
Results
Abundance of archaeal and bacterial amoA genes in summer and autumn
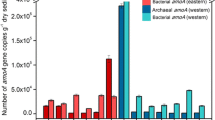
In this study, AOB amoA abundance was significantly greater than AOA in water samples (P < 0.05) except for sample AW3 (Fig. 1). The number of archaeal amoA gene ranged from 3.72 to 1.16 × 105 copies per mL water in the water samples from Dianchi Lake. Sample AW3 (1.16 × 105 copies per mL water) and sample AW1 (6.86 × 104 copies per mL water) had much greater AOA amoA gene abundance than other water samples (less than 7 × 103 copies per mL water) (P < 0.05). At each sampling site, the archaeal amoA gene was more abundant in the water column in autumn than in summer (P < 0.05). Moreover, the significant difference of the bacterial amoA gene abundance was observed in the water columns in either summer or autumn (P < 0.05), ranging from 8.02 × 102 to 1.42 × 105 bacterial amoA gene copies per mL water. Sample AW1 had the greatest AOB amoA abundance followed by sample SW1. These two water samples (more than 6.6 × 104 bacterial amoA gene copies per mL water) had significantly greater AOB amoA abundance than the rest (less than 3.5 × 104 bacterial amoA gene copies per mL water) (P < 0.05). At both sampling sites 1 and 4, AOB amoA was more abundant in autumn than summer, but the reverse was observed at sites 2 and 3. In addition, the ratio of AOB to AOA amoA genes ranged between 533 and 905.6 in summer water samples, much greater than that in autumn water samples (0.04–2.07).
For each sediment sample, AOB showed greater amoA gene abundance than AOA (P < 0.05) (Fig. 2). The number of archaeal amoA gene varied from 2.54 × 103 to 3.7 × 104 copies per gram dry sediment in summer and autumn sediments in Dianchi Lake. Samples AS1, AS3, and AS4 (more than 1.3 × 104 gene copies per gram dry sediment) had greater AOA amoA abundance than other sediment samples (less than 7.3 × 103 gene copies per gram dry sediment) (P < 0.05). At each sampling site, the archaeal amoA gene in autumn sediment sample numerically outnumbered the corresponding summer one (P < 0.05). Moreover, sediment samples from Dianchi Lake showed a significant difference in AOB amoA gene abundance (P < 0.05), varying from 4.75 × 104 to 3.84 × 105 bacterial amoA gene copies per gram dry sediment. Sample AS2 had much lower AOB amoA gene abundance than other sediment samples (P < 0.05). At each sampling site, the summer sediment sample showed greater AOB amoA gene abundance than the corresponding autumn one (P < 0.05). In addition, the ratio of AOB to AOA in summer sediment samples (71.6–151) was much greater than that in autumn sediment samples (5.7–2.07).
Diversity of archaeal and bacterial amoA genes in summer and autumn
In the present study, a total of 612 archaeal and 573 bacterial amoA gene sequences were retrieved from water and sediment samples from Dianchi Lake. AOA libraries were composed of 5–15 archaeal OTUs (Table 1). A wide range of AOA community diversity was found in both sediment samples (with Shannon index of 0.68–2.1) and water samples (with Shannon index of 1.14–1.96). At sampling sites 1, 2, and 4, summer water samples had lower AOA community diversity than the corresponding autumn ones.
AOB libraries consisted of 1–15 bacterial OTUs (Table 2). A wide range of AOB community diversity also occurred in either sediment samples (with Shannon index of 0–2.08) or water samples (with Shannon index of 0.64–2.28). Summer sediment samples had greater AOB community diversity than the corresponding autumn ones at three sampling sites (sites 1, 3, and 4). However, at each sampling site, summer water samples showed greater AOB diversity than the corresponding autumn ones. Moreover, in both summer and autumn seasons, sediment samples showed lower AOB diversity than the corresponding surface water samples in most of the sampling sites (3 out of 4). In addition, autumn water and sediment samples always showed lower AOB diversity than AOA diversity. In contrast, summer water and sediment samples usually had relatively high AOB diversity.
Phylogeny of AOA and AOB communities in summer and autumn
In the present study, the representative sequences used for further phylogenetic analysis were selected only from the major OTUs (containing at least two amoA gene sequences). The archaeal amoA gene sequences from the major AOA OTUs could be assigned into six distinctive AOA groups (clusters A–F) (Fig. 3). Both sediment samples and water samples from Dianchi Lake differed greatly in the proportion of each AOA amoA cluster (Fig. S2a). In the same sampling location, a distinctive structure difference of AOA communities between in surface water and in sediment was also observed. Cluster-A was the second largest AOA group in both waters and sediments. The proportion of AOA sequences affiliated with cluster-A in water samples AW1, SW3, SW4, AW2, AW3, and AW4 (25–45.5 %) was higher than SW1 (11.4 %) and SW2 (19.5 %). The AOA sequences affiliated with cluster-A comprised a greater proportion in sediment samples SS1, SS4, AS2, and AS3 (30.6–41.9 %) than in other sediment samples (0–19.4 %). The AOA sequences in this cluster were related to ≥97 % similarity to several cultured ones from wastewater treatment plant, flooding paddy soil, lake sediment. Moreover, cluster-C was the largest AOA group in both waters and sediments. They comprised a greater fraction of the total sequence library in summer water samples (SW1 65.7 %, SW2 65.6 %, SW3 50 %, and SW4 64.1 %) than autumn ones (AW1 36.4 %, AW2 30.8 %, AW3 42.4 %, and AW4 41.7 %). In addition, AOA sequences affiliated with cluster-C consisted of a greater proportion in sediment samples SS2, SS3, AS1, and AS4 (64.5–87.2 %) than other sediments (38.7–54.3 %). The sequences in cluster-C were related with ≥95 % similarity to several cultured ones from a variety of soil and sediment ecosystems, including river side, wetland, and grassland soils, and wetland, river estuary, and lake estuary sediments. Cluster-F was a minor AOA group. The sequences in this group could be related to ≥89 % similarity to the AOA amoA gene from two cultivated AOA strains (Nitrososphaera JG1 and EN76) (Kim et al. 2012; Tourna et al. 2011). They were only observed in water samples SW1 (8.6 %), AW1 (6.1 %), and AW4 (11.1 %) and sediment samples SS3 (7.7 %), AS1 (6.5 %), and AS3 (20.6 %).

The sequences from the major AOB amoA OTUs could be grouped into five distinctive AOB clades (clusters I–VI) (Fig. 4). Both sediments and waters showed an evident difference in the proportion of each AOB amoA gene cluster (Fig. S2b). Surface water and sediment samples from the same site also showed distinct community structures. AOB sequences affiliated with cluster I were mainly observed in summer waters SW1, SW2 and SW3 (31–67.6 %) and summer sediments SS1, SS3 and SS4 (55.6–85.7 %). AOB sequences affiliated with cluster I were not detected in all the autumn waters and most of the autumn sediments (3 out of 4). These sequences could be related with ≥90 % similarity to the uncultured ones from aquaculture pond sediment, rice soil, plateau soil and riparian sediment. Cluster IV was the second largest AOB group. AOB sequences affiliated with cluster IV were mainly found in autumn waters AW1, AW2, AW3 and AW4 (57.6–79.4 %) and autumn sediments AS2 (51.4 %) and AS3 (80 %). Most of AOB sequences in cluster IV showed ≥93 % similarity to the uncultured ones from lake sediment. In addition, cluster V was the largest AOB amoA group and the members in this group could be distantly related (less than 82 % similarity) to the amoA gene from one cultivated Nitrosomonas strain. AOB sequences affiliated with cluster V predominated in water sample SW4 (71.4 %) and sediment samples SS2, AS1 and AS4 (89.3–100 %), and also comprised a considerable proportion in water samples AW1, AW2, AW3, AW4 and SW2 (13.8–42.4 %) and sediment samples AS2 (48.6 %) and AS3 (20 %). However, AOB sequences affiliated with cluster V were not detected in other water or sediment samples.
Phylogenetic tree of representative bacterial amoA sequences and reference sequences from Genbank. The obtained bacterial sequences beginning with “SW1”– “SW4”, “AW1”– “AW4”, “SS1” –“SS4” and “AS1” –“AS4” were referred to the sequences retrieved from samples SW1– SW4, AW1– AW4, SS1 –SS4 and AS1 –AS4, respectively. The number in parentheses represents the numbers of the sequences in the same OTU in a given clone library. Numbers at the nodes indicate the levels of bootstrap support based on neighbor-joining analysis of 1000 resampled datasets. The bar represents 5 % sequence divergence
Influential factors regulating AOA and AOB communities
Pearson’s correlation analysis indicated that the AOB/AOA ratio in lake water was positively correlated with the levels of pH, DOC, and NH4 +-N (P < 0.05), while no significant links were found between the determined water parameters and AOA or AOB amoA gene abundance (P > 0.05) (Table 3). Planktonic AOB diversity was found to show a positive correlation with NH4 +-N (P < 0.05). Moreover, sediment AOA amoA gene abundance was positively correlated with sediment NH4 +-N (P < 0.05), while the sediment AOB/AOA ratio showed a positive correlation with TP (P < 0.05) (Table 4). In addition, sediment AOA diversity was negatively correlated with NO3 --N (P < 0.05), while sediment AOB diversity showed a positive correlation with TOC (P < 0.05).
Water environmental factors accounted for 74.9 % (the first two RDA axes, respectively, 30.5 % and 21.6 %) of the total variance for planktonic AOA OTU composition (Fig. 5a). In this study, no water parameter was found to significantly contribute to the planktonic AOA community–environment relationship. For planktonic AOB OTU composition, the water environmental factors accounted for 90.7 % (the first two RDA axes, respectively, 32 % and 27.7 %) of the total variance (Fig. 5b). TP (P = 0.0340, F = 2.821, 999 Monte Carlo permutations) significantly contributed to the planktonic AOB assemblage–environment relationship. In addition, sediment environmental factors explained 88.7 % (the first two RDA axes, respectively, 35.9 % and 25.3 %) of the total variance for sediment AOA OTU composition (Fig. 6a). No sediment parameter significantly contributed to the sediment AOA–environment relationship. The sediment environmental factors accounted for 88.2 % (the first two RDA axes, respectively, 41.5 % and 20.2 %) of the total variance for lake sediment AOB OTU composition (Fig. 6b). Only TP (P = 0.012, F = 4.259, 999 Monte Carlo permutations) showed significant contribution to the sediment AOB–environment relationship.
Discussion
AOA and AOB amoA gene abundance in freshwater lake
It remains unclear whether AOA or AOB amoA gene is dominant in lake sediment ecosystem. Previous studies indicated that AOA amoA gene outnumbered AOB in the sediments of freshwater lakes at different trophic states, such as eutrophic freshwater lakes (Chaohu Lake and Taihu Lake) (Hou et al. 2013; Zhao et al. 2014), mesotrophic Erhai Lake (Yang et al. 2016), and oligotrophic Lake Superior (Bollmann et al. 2014). AOA amoA gene also showed numerical dominance over AOB in the sediments of eight freshwater lakes in Jiangsu (China) (Sun et al. 2014). However, bacterial amoA gene was more abundant than archaeal amoA gene in the sediments of several small freshwater lakes on the Yunnan Plateau (Liu et al. 2015) and eutrophic freshwater Lake Erie (Bollmann et al. 2014). AOA amoA gene was also found to be less abundant than AOB in the sediments of saline Qinghai Lake (Jiang et al. 2009). In this study, sediment AOB always displayed greater amoA gene abundance than AOA, suggesting that AOB might have a greater contribution to nitrification in lake sediment of eutrophic freshwater Dianchi Lake. This was in agreement with the result reported in our recent study (Yang et al. 2016).
So far, although the spatial variation of AOA and AOB amoA gene abundance in freshwater lake sediment has been well-documented, there is a paucity of information on the temporal change. The present study showed the time-related change of both AOA and AOB amoA gene abundance in lake sediment. At a given sampling site, sediment AOA abundance was greater in autumn than summer, while sediment AOB amoA gene abundance was greater in summer. In addition, the sediment AOB/AOA ratio was also subject to temporal change, and it was much greater in summer than in autumn.
It remains in debate whether AOA or AOB amoA gene is more abundant in lake water column. AOA amoA gene were more abundant than AOB in the waters of saline Qinghai Lake (Jiang et al. 2009). Vissers et al. (2013) indicated that AOA amoA gene generally outnumbered AOB in the water column of oligotrophic freshwater Lake Lucerne, while Hayden and Beman (2014) showed that AOB amoA gene was more abundant than AOA in all the water samples from nine oligotrophic high-altitude freshwater lakes in California. Mukherjee et al. (2016) revealed that AOA amoA gene dominated compared to AOB in oligotrophic freshwater Lake Superior, whereas AOB amoA gene were more abundant than AOA in eutrophic freshwater Lake Erie. In the present study, both AOA and AOB amoA genes in water columns of eutrophic freshwater Dianchi Lake showed the evident time- and site-related changes. This was consistent with the results reported in oligotrophic freshwater lake (Vissers et al. 2013). In Dianchi Lake, similar to sediment AOA amoA gene, at a given sampling site, planktonic AOA amoA gene was also found to be more abundant in autumn than in summer. In addition, AOB amoA gene generally showed greater abundance than AOA in lake water, suggesting that AOB might play a more important role in nitrification in lake water. The temporal shift in the AOB/AOA ratio in water columns was also observed, and the AOB/AOA ratio was much greater in summer than in autumn.
AOA and AOB community diversity in freshwater lake
Several previous studies indicated that AOA generally had higher community diversity than AOB in sediments of Taihu Lake (Zhao et al. 2014) and small freshwater lakes on the Yunnan Plateau (Liu et al. 2015), while other studies showed higher AOB diversity than AOA in sediments of Jinshan Lake (Liu et al. 2014b) and Dianchi Lake and Erhai Lake (Yang et al. 2016). In the present study, both AOA and AOB diversity in sediments of Dianchi Lake illustrated a temporal shift. In addition, autumn sediment samples always showed lower AOB diversity than AOA, while summer sediment samples usually had relatively high AOB diversity. This suggested that the diversity difference between lake sediment AOA and AOB could vary with time.
Auguet et al. (2011) reported the seasonal changes of AOA diversity in waters in oligotrophic alpine lakes. Hu et al. (2010) revealed higher planktonic AOA diversity than AOB in two high-altitude oligotrophic freshwater lakes. In this study, both AOA and AOB diversity in water columns of eutrophic Dianchi Lake illustrated the evident time- and site-related changes. At a given sampling site, planktonic AOA generally had lower diversity in summer than in autumn, while planktonic AOB tended to have relatively high diversity in summer. In addition, planktonic AOB diversity was generally higher than planktonic AOA in summer, but lower in autumn, which also suggested the temporal change of diversity difference between lake planktonic AOA and AOB. Moreover, at a given sampling site, sediment usually tended to have lower AOB diversity than surface water.
AOA and AOB community structure in freshwater lake
The spatial heterogeneity of the structure of either sediment AOA or AOB community has been found in a single freshwater lake or across different freshwater lakes (Bollmann et al. 2014; Hou et al. 2013; Liu et al. 2014b; Sun et al. 2014; Yang et al. 2016). In this study, the results of phylogenetic analysis indicated the site-related difference in AOA or AOB community structure in sediments of Dianchi Lake. Moreover, the present study also provided the evidence that lake sediment AOA or AOB community structure could be subject to temporal change. In addition, the waters from eutrophic Dianchi Lake showed the evident time- and site-related variation in either AOA or AOB community structure. This was consistent with the results in other previous studies (Auguet et al. 2011; Auguet and Casamayor 2013; Hu et al. 2010). So far, to the authors’ knowledge, there has been no report available on comparing the structures of lake ammonia-oxidizing microbial populations between in sediment and in water column. In the current study, surface water and sediment in the same sampling location showed a difference in both AOA and AOB community structure.
In this study, only a small proportion of archaeal amoA gene sequences from Dianchi Lake could be related (with ≥89 % similarity) to the AOA amoA gene from two cultivated AOA strains (Nitrososphaera JG1 and EN76) (Kim et al. 2012; Tourna et al. 2011). Most of the obtained archaeal amoA gene sequences from Dianchi Lake were distributed in AOA cluster C, and they could be grouped together with the uncultured ones from a variety of soil and sediment ecosystems. Moreover, only a small proportion of AOB sequences from Dianchi Lake could be affiliated with several cultivated Nitrosospira strains (NIJS18, PJA1, and TCH711). A majority of the obtained AOB sequences from Dianchi Lake showed no close relation to those from any known isolated AOB sequences. They were affiliated with the uncultured bacterial sequences from various soil and sediment ecosystems and wastewater bioreactor.
Environmental factors influencing AOA and AOB community
A number of environmental factors can regulate AOA and AOB amoA gene abundance in freshwater lake sediment (Yang et al. 2016). In this study, sediment AOA amoA gene abundance in Dianchi Lake was found to be likely shaped by the level of sediment ammonia nitrogen, which was consistent with the results reported in other freshwater lakes (Liu et al. 2014b; Zhao et al. 2014). Little is known about the environmental factors governing the AOB/AOA ratio in freshwater lake sediment. Our previous study indicated that the AOB/AOA ratio in freshwater lakes could be influenced by sediment NO3 --N (Yang et al. 2016). In contrast, the current study revealed that TP might positively influence the AOB/AOA ratio in lake sediment. TP can be a key determinant of AOA or AOB amoA gene abundance in freshwater lake sediment (Liu et al. 2014b; Yang et al. 2016). The present study further suggested the potential role of TP in determining the AOB/AOA ratio in lake sediment. In addition, previous studies indicated that lake sediment AOA diversity was likely influenced by lake trophic status (Bollmann et al. 2014; Herrmann et al. 2009; Yang et al. 2016), TN and TP (Yang et al. 2016), and temperature (Zeng et al. 2014), while AOB diversity by lake trophic status (Hou et al. 2013; Yang et al. 2016), pH (Yang et al. 2016), NO3 --N and TN (Liu et al. 2014b), and temperature (Zeng et al. 2014). However, in this study, lake sediment AOA and AOB diversity was found to be likely affected by NO3 --N and TOC, respectively, which was not in agreement with the results reported in these previous studies. Moreover, it has been widely accepted that lake trophic status is a key determinant of AOA and AOB community structure (Bollmann et al. 2014; Herrmann et al. 2009; Yang et al. 2016). The result of RDA in the current study also showed that TP might play an important role in shaping sediment AOB community structure in Dianchi Lake, while no obvious links were found between sediment properties and AOA community structure.
Previous work has reported that planktonic AOA amoA gene abundance in freshwater lake could be influenced by lake elevation (Hayden and Beman 2014), NH4 +-N and nitrite nitrogen (NO2 --N) (Auguet et al. 2011, 2012), and temperature and conductivity (Vissers et al. 2013), while planktonic AOB amoA gene abundance by phosphate (Hayden and Beman 2014). In this study, the links between environmental factors and planktonic AOA or AOB amoA gene abundance was not clear. However, the AOB/AOA ratio in waters of Dianchi Lake was found to be likely influenced by pH, DOC, and NH4 +-N. Moreover, the present study showed that AOB diversity in waters of Dianchi Lake might be positively affected by the level of NH4 +-N. In addition, several previous studies suggested that planktonic AOA community structure in freshwater lake was shaped by pH (Auguet and Casamayor 2013), temperature (Auguet et al. 2011), and water depth (Auguet et al. 2012). Hu et al. (2010) revealed the salinity-related differentiation of AOA community composition in high-altitude lakes on the Tibetan Plateau. To date, the report on the driving force for planktonic AOB community structure has been yet unavailable. In the current study, the result of RDA suggested that TP was likely a key determinant of planktonic AOB community structure in Dianchi Lake, while the driving force for planktonic AOA community structure was not identified.
Conclusions
The abundance, diversity and structure of planktonic and sediment AOA and AOB communities in eutrophic Dianchi Lake were subject to the evident time- and site-related changes. AOB amoA gene was generally more abundant than AOA in both water columns and sediments. At a giving sampling location, surface water and sediment showed a difference in either AOA or AOB community structure. TP might play an important role in shaping sediment AOB community structure, while planktonic AOB community structure was likely determined by TP as well as TN.
References
Auguet JC, Casamayor EO (2013) Partitioning of Thaumarchaeota populations along environmental gradients in high mountain lakes. FEMS Microbiol Ecol 84:154–164
Auguet JC, Nomokonova N, Camarero L, Casamayor EO (2011) Seasonal changes of freshwater ammonia-oxidizing archaeal assemblages and nitrogen species in oligotrophic alpine lakes. Appl Environ Microbiol 77:1937–1945
Auguet JC, Triado-Margarit X, Nomokonova N, Camarero L, Casamayor EO (2012) Vertical segregation and phylogenetic characterization of ammonia-oxidizing Archaea in a deep oligotrophic lake. ISME J 6:1786–1797
Bollmann A, Bullerjahn GS, Mckay RM (2014) Abundance and diversity of ammonia-oxidizing archaea and bacteria in sediments of trophic end members of the Laurentian Great Lakes, Erie and Superior. PLoS One 9:e97068
Bouskill NJ, Eveillard D, Chien D, Jayakumar A, Ward BB (2012) Environmental factors determining ammonia-oxidizing organism distribution and diversity in marine environments. Environ Microbiol 14:714–729
China Environmental Protection Agency (2002) Methods for Water and Wastewater Determination. China Environmental Science Press, Beijing
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688
Hayden CJ, Beman JM (2014) High abundances of potentially active ammonia-oxidizing bacteria and archaea in oligotrophic, high-altitude lakes of the Sierra Nevada, California, USA. PLoS One 9:e111560
Herrmann M, Saunders AM, Schramm A (2009) Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl Environ Microbiol 75:3127–3136
Hou J, Song CL, Cao XY, Zhou YY (2013) Shifts between ammonia oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu). Water Res 47:2285–2296
Hu AY, Yao TD, Jiao NZ, Liu YQ, Yang Z, Liu XB (2010) Community structures of ammonia-oxidising archaea and bacteria in high-altitude lakes on the Tibetan Plateau. Freshw Biol 55:2375–2390
Jiang HC, Dong HL, Yu BS, Lv G, Deng SC, Berzins N, Dai MH (2009) Diversity and abundance of ammonia-oxidizing archaea and bacteria in Qinghai Lake, northwestern China. Geomicrobiol J 26:199–211
Kim JG, Jung MY, Park SJ, Rijpstra WIC, Damste JSS, Madsen EL, Min D, Kim JS, Kim GJ, Rhee SK (2012) Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I1b from an agricultural soil. Environ Microbiol 14:1528–1543
Lipsewers YA, Bale NJ, Hopmans EC, Schouten S, Damste JSS, Villanueva L (2014) Seasonality and depth distribution of the abundance and activity of ammonia oxidizing microorganisms in marine coastal sediments (North Sea). Front Microbiol 5:472
Liu Y, Zhang JX, Zhang XL, Xie SG (2014a) Depth-related changes of sediment ammonia-oxidizing microorganisms in a high-altitude freshwater wetland. Appl Microbiol Biotechnol 98:5697–5707
Liu B, Li YM, Zhang JP, Zhou XH, Wu CD (2014b) Abundance and diversity of ammonia-oxidizing microorganisms in the sediments of Jinshan Lake. Curr Microbiol 69:751–757
Liu Y, Zhang JX, Zhao L, Li YZ, Dai Y, Xie SG (2015) Distribution of sediment ammonia-oxidizing microorganisms in plateau freshwater lakes. Appl Microbiol Biotechnol 99:4435–4444
Lu SM, Liao MJ, Xie CX, He XG, Li DP, He LL, Chen J (2015) Seasonal dynamics of ammonia-oxidizing microorganisms in freshwater aquaculture ponds. Ann Microbiol 65:651–657
Magalhaes CM, Machado A, Bordalo AA (2009) Temporal variability in the abundance of ammonia-oxidizing bacteria vs. archaea in sandy sediments of the Douro River estuary, Portugal. Aquat Microb Ecol 56:13–23
Mosier AC, Francis CA (2008) Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol 10:3002–3016
Mukherjee M, Ray A, Post AF, McKay RM, Bullerjahn GS (2016) Identification, enumeration and diversity of nitrifying planktonic archaea and bacteria in trophic end members of the Laurentian Great Lakes. J Gt Lakes Res 42:39–49
Nakagawa T, Mori K, Kato C, Takahashi R, Tokuyama T (2007) Distribution of cold-adapted ammonia-oxidizing microorganisms in the deep-ocean of the northeastern Japan Sea. Microbes Environ 22:365–372
Newell SE, Fawcett SE, Ward BB (2013) Depth distribution of ammonia oxidation rates and ammonia-oxidizer community composition in the Sargasso Sea. Limnol Oceanogr 58:1491–1500
Puthiya Veettil V, Abdulaziz A, Chekidhenkuzhiyil J, Kalanthingal Ramkollath L, Karayadi Hamza F, Kizhakkepat Kalam B, Kallungal Ravunnikutty M, Nair S (2015) Bacterial domination over archaea in ammonia oxidation in a monsoon-driven tropical estuary. Microbial Ecol 69:544–553
Reis MP, Avila MP, Keijzer RM, Barbosa FAR, Chartone-Souza E, Nascimento AMA, Laanbroek HJ (2015) The effect of human settlement on the abundance and community structure of ammonia oxidizers in tropical stream sediments. Front Microbiol 6:898
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Santoro AE, Francis CA, de Sieyes NR, Boehm AB (2008) Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ Microbiol 10:1068–1079
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur, open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–75
Song H, Li Z, Du B, Wang G, Ding Y (2012) Bacterial communities in sediments of the shallow Lake Dongping in China. J Appl Microbiol 112:79–89
Sonthiphand P, Cejudo E, Schiff SL, Neufeld JD (2013) Wastewater effluent impacts ammonia-oxidizing prokaryotes of the Grand River, Canada. Appl Environ Microbiol 79:7454–7465
Sun W, Xia CY, Xu MY, Guo J, Wang AJ, Sun GP (2013) Distribution and abundance of archaeal and bacterial ammonia oxidizers in the sediments of the Dongjiang River, a drinking water supply for Hong Kong. Microbes Environ 28:457–465
Sun X, Wang AL, Yang LY, Guo LY, Chen QK, Hu ZX, Jiang LJ, Xiao L (2014) Spatial distribution of ammonia-oxidizing archaea and bacteria across eight freshwater lakes in sediments from Jiangsu of China. J Limnol 73:312–324
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6, molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425
Veettil VP, Abdulaziz A, Chekidhenkuzhiyil J, Ramkollath LK, Hamza FK, Kalam BK, Ravunnikutty MK, Nair S (2015) Bacterial domination over Archaea in ammonia oxidation in a monsoon-driven tropical estuary. Microb Ecol 69:544–553
Vissers EW, Anselmetti FS, Bodelier PLE, Muyzer G, Schleper C, Tourna M, Laanbroek HJ (2013) Temporal and spatial coexistence of archaeal and bacterial amoA genes and gene transcripts in Lake Lucerne. Archaea 2013:289478
Wang YF, Feng YY, Ma XJ, Gu JD (2013) Seasonal dynamics of ammonia/ammonium-oxidizing prokaryotes in oxic and anoxic wetland sediments of subtropical coastal mangrove. Appl Microbiol Biotechnol 97:7919–7934
Wang XY, Wang C, Bao LL, Xie SG (2014) Abundance and community structure of ammonia-oxidizing microorganisms in reservoir sediment and adjacent soils. Appl Microbiol Biotechnol 98:1883–1892
Xie W, Zhang CL, Zhou XD, Wang P (2014) Salinity-dominated change in community structure and ecological function of Archaea from the lower Pearl River to coastal South China Sea. Appl Microbiol Biotechnol 98:7971–7982
Yang J, Jiang HC, Dong HL, Wang HY, Wu G, Hou WG, Liu WG, Zhang CL, Sun YJ, Lai ZP (2013) amoA-encoding archaea and thaumarchaeol in the lakes on the northeastern Qinghai-Tibetan Plateau, China. Front Microbiol 4:329
Yang YY, Shan JW, Zhang JX, Zhang XL, Xie SG, Liu Y (2014) Ammonia- and methane-oxidizing microorganisms in high-altitude wetland sediments and adjacent agricultural soils. Appl Microbiol Biotechnol 98:10197–10209
Yang YY, Zhang JX, Zhao Q, Zhou QH, Li NN, Wang YL, Xie SG, Liu Y (2016) Sediment ammonia-oxidizing microorganisms in two plateau freshwater lakes at different trophic states. Microb Ecol 71:257–265
Zeng J, Zhao DY, Yu ZB, Huang R, Wu QLL (2014) Temperature responses of ammonia-oxidizing prokaryotes in freshwater sediment microcosms. PLoS One 9:e100653
Zhang Y, Chen LJ, Dai TJ, Tian JP, Wen DH (2015a) The influence of salinity on the abundance, transcriptional activity, and diversity of AOA and AOB in an estuarine sediment: a microcosm study. Appl Microbiol Biotechnol 99:9825–9833
Zhang QF, Tang FY, Zhou YJ, Xu JR, Chen HP, Wang MK, Laanbroek HJ (2015b) Shifts in the pelagic ammonia-oxidizing microbial communities along the eutrophic estuary of Yong River in Ningbo City, China. Front Microbiol 6:1180
Zhao DY, Zeng J, Wan WH, Liang HD, Huang R, Wu QLL (2013) Vertical distribution of ammonia-oxidizing archaea and bacteria in sediments of a eutrophic lake. Curr Microbiol 67:327–332
Zhao DY, Luo J, Zeng J, Wang M, Yan WM, Huang R, Wu QLL (2014) Effects of submerged macrophytes on the abundance and community composition of ammonia-oxidizing prokaryotes in a eutrophic lake. Environ Sci Pollut Res 21:389–398
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (No. 41571444), National Basic Research Program of China (2015CB458900), and special fund of State Key Joint Laboratory of Environment Simulation and Pollution Control (15L02ESPC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
The work has not been published previously and not under consideration for publication elsewhere.
Additional information
Responsible editor: Robert Duran
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 229 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Li, N., Zhao, Q. et al. Ammonia-oxidizing archaea and bacteria in water columns and sediments of a highly eutrophic plateau freshwater lake. Environ Sci Pollut Res 23, 15358–15369 (2016). https://doi.org/10.1007/s11356-016-6707-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-6707-0