Abstract
Both ammonia-oxidizing bacteria (AOB) and archaea (AOA) might be the key microorganisms in ammonia conversion in ecosystems. However, the depth-related change of AOA and AOB in sediment ecosystem is still not clear. The relative contribution of AOA and AOB to nitrification in wetland sediment remains also unclear. Moreover, information about ammonia-oxidizing microorganisms in high-altitude freshwater wetland is still lacking. The present study investigated the relative abundances and community structures of AOA and AOB in sediments of a high-altitude freshwater wetland in Yunnan Province (China). Variations of the relative abundances and community structures of AOA and AOB were found in the wetland sediments, dependent on both sampling site and sediment depth. The relative abundances of AOA and AOB (0.04–3.84 and 0.01–0.52 %) and the AOA/AOB ratio (0.12–4.65) showed different depth-related change patterns. AOB community size was usually larger than AOA community size. AOB diversity was usually higher than AOA diversity. AOA diversity decreased with the increase of sediment depth, while AOB diversity showed no obvious link with the sediment depth. Pearson’s correlation analysis showed that AOA diversity had a positive significant correlation with available phosphorus. Nitrosospira-like sequences, with different compositions, predominated in the wetland sediment AOB communities. This work could provide some new insights toward nitrification in freshwater sediment ecosystems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrification, the biological conversion of ammonia to nitrate via nitrite, has been regarded as one important process for global nitrogen cycling. Ammonia oxidation, as the first and rate-limiting step in nitrification process, was usually considered to be exclusively carried out by ammonia-oxidizing bacteria (AOB), mainly belonging to genera Nitrosomonas, Nitrosospira, and Nitrosococcus (Li et al. 2012; Wang et al. 2014). These microorganisms harbor the ammonia monooxygenase (amoA) gene, which is responsible for ammonia oxidation. However, after the identification of the archaeal amoA gene and isolation of ammonia-oxidizing archaea (AOA) (Könneke et al. 2005; Venter et al. 2004), the abundance of AOA has been found in various terrestrial and aquatic ecosystems, suggesting the importance of AOA in ammonia oxidation (Erguder et al. 2009; Feng et al. 2012; Sims et al. 2012; Wang et al. 2014). Both AOA and AOB might be the key microorganisms in ammonia oxidation in the environment (He et al. 2007; Wang et al. 2014).
Aquatic sediment harbors a complex microbial ecosystem consisting of huge amounts of viable microorganisms, which might be involved in a variety of biogeochemical processes (Cheng et al. 2014). AOA could usually outnumber AOB in estuary sediment (Caffrey et al. 2007; Cao et al. 2011a), salt marsh sediment (Moin et al. 2009), and marine sediment (Cao et al. 2011a; Park et al. 2008; Sakami 2012). Few investigations regarding the abundances of AOA and AOB in freshwater sediment ecosystems have also been carried out. Wang et al. (2014) reported the higher abundance of AOA than AOB in freshwater reservoir sediment. Zhao et al. (2013) found that AOA abundance was higher than AOB abundance in sediments of Lake Taihu (China). Hou et al. (2013) also indicated that AOA usually predominated over AOB in sediments of Lake Taihu and Lake Chaohu (China). These previous studies suggested that AOA could usually make more contribution to nitrification than AOB in both saline and freshwater sediment ecosystems. However, the lower abundance of AOA than AOB was found in the subsurface sediments of the northern South China Sea (Cao et al. 2012). Liu et al. (2013) showed that, although AOA outnumber AOB in the sediment of the upstream reaches of the Qiantang River, AOB dominate over AOA in downstream sediment. Therefore, the relative contribution of AOA and AOB to nitrification in sediment ecosystems remains still unresolved.
Sediment is a stratified habitat. The sediment depth-related chemical features can provide niches for metabolically diverse microorganisms (Zhao et al. 2008). Previous studies have shown the depth-related changes of archaeal and bacterial community compositions in lake sediments (Liu et al. 2010; Shivaji et al. 2011; Zhao et al. 2008, 2013). The abundances and community structures of AOA and AOB can be influenced by habitat type and many environmental factors (Chen et al. 2010; Wang et al. 2014). Long et al. (2012) revealed that niche properties could have a significant impact on genotype distribution of the two ammonia-oxidizing groups. Unfortunately, there has been very limited information on the depth-related changes of the abundances and community structures of AOA and AOB (Li et al. 2011; Zhao et al. 2013). The links between sediment ammonia-oxidizing community and environmental conditions remain unclear.
Wetlands play important roles in ecosystems and environmental sustainability, such as storm and flood control, sediment retention, offering habitats, supporting abundant life, possessing high diversity, and nutrient control (Sims et al. 2012; Wang and Gu 2013). Ammonia-oxidizing organisms can be key microorganisms in wetland water purification and nitrogen cycling (Sims et al. 2012). However, little is known about ammonia-oxidizing microorganisms in wetland ecosystems. The Luoshijiang Wetland is a freshwater wetland in the Yunnan-Kweichow Plateau (China). The high-altitude wetland ecosystem is adjacent to Rrhai Lake (Daili City, Yunnan Province) and is annually inundated. It covers an area of about 1 km2 with an elevation of about 2,000 m (Zhang et al. 2013). The input water of the wetland consisted of runoff from adjacent agricultural field and hills mixed with wastewater from nearby villages. To date, information about ammonia-oxidizing microorganisms in high-altitude freshwater wetland is still lacking. Therefore, the objective of the present study was to investigate the depth-related change of the abundance and community structure of AOA and AOB in sediments of the Luoshijiang Wetland.
Materials and methods
Description of sites and sampling
Sediment samples in triplicate were collected using a core sampler at four different sites of the Luoshijiang Wetland (Fig. S1): A (25° 57′ 25″ N–100° 06′ 06″ E; nonvegetated zone), B (25° 57′ 12″ N–100° 05′ 59″ E; reed-vegetated zone), C (25° 57′ 4″ N–100° 06′ 00″ E; densely water lily-vegetated zone), and D (25° 56′ 55″ N–100° 05′ 59″ E; sparsely water lily-vegetated zone). Sediment samples were sliced into layers. During this study, the upper layer (0–5 cm) and the lower layer (10–15 cm) were used for further analyses. These sediment samples were immediately transported back to the laboratory after collection. The chemical parameters of the sediment samples are shown in Fig. 1.
Molecular analyses
Wetland sediment DNA was extracted using the Powersoil DNA extraction kit (Mobio Laboratories, USA). DNA concentration was quantified using Nanodrop® ND-1000 UV-vis spectrophotometry (USA). Each replicate sediment DNA sample was individually subjected to quantitative PCR assays. For quantitative PCR assays, the specific primers for amplification of the archaeal and bacterial amoA genes were as previously described (Wang et al. 20134): Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′), Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′), AmoA-1F (5′-GGGGTTTCTACTGGTGGT-3′), and AmoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′). The primers for amplification of the archaeal and bacterial 16S ribosomal RNA (rRNA) genes were selected according to the literature (Jung et al. 2011): Arch 349F (5′-GYGCASCAGKCGMGAAW-3′), Arch 806R (5′-GGACTACVSGGGTATCTAAT-3′), 341F (5′-CCTACGGGAGGCAGCAG-3′), and 534R (5′-ATTACCGCGGCTGCTGGCA-3′). Standard curves ranging from 104 to 108 gene copies/mL were obtained using serial dilutions of linearized plasmids (pGEM-T, Promega) containing cloned bacterial amoA gene, archaeal amoA gene, bacterial 16S rRNA gene, and archaeal 16S rRNA gene amplified from sediment. The amplification efficiency and coefficient (r 2) for the archaeal and bacterial amoA genes and the archaeal and bacterial 16S rRNA genes were 95 % and 0.998, 92 % and 0.997, 92 % and 0.998, and 101 % and 0.995, respectively.
For clone library analysis of community structures of AOA and AOB, the PCR amplification was conducted using the above-mentioned primer pairs Arch-amoAF/Arch-amoAR and amoA1F/amoA2R (Wang et al. 2014). Triplicate PCR products for each sediment sample were pooled and purified with a QIAquick PCR purification kit (Qiagen Inc.). The purified fragments were cloned into pMD19-T vector (Takara Corp, Japan) and the clones containing the correct size were sequenced. Chimera-free sequences with ≥98 % similarity were assigned as the same operational taxonomic units (OTUs) using the DOTUR program (Schloss and Handelsman 2005). OTU-based community diversity indices and rarefaction curves were also obtained using the DOTUR program (Schloss and Handelsman 2005). Phylogenetic analyses of AOA and AOB communities in all the sediment samples were performed using MEGA software version 4.0 (Tamura et al. 2007). Pearson’s correlation analysis of the relative abundances and community structures of AOA and AOB with the sediment chemical parameters (ammonia nitrogen, total nitrogen, total phosphorous, available phosphorus, and total organic carbon) was carried out using SPSS 20.0 software. The amoA gene sequences obtained in this study were submitted to GenBank under accession numbers KF618627–KF618889 for AOA and KF618890–KF619241 for AOB, respectively.
Results
Relative abundances of AOA and AOB
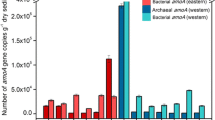
In this study, the densities of archaeal and bacterial amoA and 16S rRNA genes were determined using real-time PCR. Ratios of archaeal amoA gene to archaeal 16S rRNA gene and bacterial amoA gene to bacterial 16S rRNA gene were used to estimate the relative abundances of AOA and AOB in each sediment sample, respectively. Figure 2 shows large variations in the relative abundances of both AOA and AOB in the eight sediment samples. The relative abundance of AOA ranged between 0.04 and 3.84 %. The AOA proportion in sampling site A was much higher than that in other sampling sites. In sampling site A, the AOA proportion in the upper layer sediment was much lower than that in the lower layer. Conversely, in sampling sites B and C, the upper layer had a much higher AOA proportion. In addition, in sampling site D, the upper and lower layers had a similar AOA proportion.
Relative abundance of archaeal and bacterial amoA genes in the different samples. Samples AU and AL, BU and BL, CU and CL, and DU and DL represent the upper layer and lower layer sediments in sampling sites A–D, respectively. Each replicate sediment sample was individually subjected to quantitative PCR assays. Error bars represent standard deviation of mean (n = 3)
The relative abundance of AOB ranged between 0.01 and 0.52 %. The AOB proportion in sampling site D was much higher than that in other sampling sites. In sampling sites A and B, the upper layer had a higher AOB proportion than the lower layer. However, an opposite trend was found in the other two sampling sites. Moreover, the AOA proportion was much higher than the AOB proportion in sites A, B, and C, but lower in sampling site D.
Figure 3 shows a large variation in AOA to AOB ratio in the eight sediment samples. The highest value of AOA/AOB ratio was found in sample CU (4.65) followed by sample AL (1.71). However, for the other samples, the values of AOA/AOB ratio ranged between 0.12 and 0.54. In addition, the upper layer had a lower AOA/AOB ratio than the lower layer in sampling sites A and B, but a higher one in sampling sites C and D. In this study, Pearson’s correlation analysis was applied to investigate the relationships between the determined chemical properties and the AOA and AOB proportions, or the AOA/AOB ratio (Table 1). The results indicated that the AOA and AOB proportions or the AOA/AOB ratio did not show significant correlations with the measured sediment chemical parameters (p > 0.05).
Diversity of AOA and AOB communities
In this study, a total of 263 archaeal and 352 bacterial amoA sequences were retrieved from AOA and AOB clone libraries, respectively (Table 2). The number of OTUs and OTU-based community Shannon diversity index and rarefaction curves were obtained using the DOTUR program at a 2 % difference level (Schloss and Handelsman 2005). Table 2 shows that the number of AOA OTUs in each sediment sample ranged between 4 and 14, while the AOB clone libraries were composed of 9–26 OTUs. The rarefaction curves for AOA and AOB clone libraries nearly leveled off (Fig. 4a, b), suggesting that these communities were well sampled.
A large difference in AOA community diversity was observed in the different sampling sites. The values of AOA Shannon index ranged between 0.82 and 2.23. Samples AU and BU have much a greater AOA diversity than the other samples. Moreover, at each sampling site, the upper layer had a greater AOA diversity than the lower layer. A marked variation in AOB community diversity (Shannon index = 1.52–2.63) was also found. The highest and lowest AOB diversities occurred in the upper layer and the lower layer in sampling B, respectively. In sampling site C, the upper layer had a much lower AOB diversity, compared with the lower layer. However, in sampling sites A and D, the two sediment layers had a similar AOB diversity. In addition, the Shannon diversity index value of AOA was higher than that of AOB in sample AU, but lower in the other seven sediment samples. AOA Shannon diversity was positively correlated with available phosphorus (AP; p < 0.05) (Table 1). No significant correlation was found between AOB Shannon diversity and the determined chemical parameters.
Phylogeny of AOA and AOB communities
In this study, the representative AOA and AOB sequences used for phylogenetic analysis were selected from the OTUs that had two or more sequence members. Figure 5 shows that all of the archaeal amoA sequences of the eight AOA clone libraries could be grouped into five clusters. There was no sediment sample that could share all of the five AOA clusters. The archaeal amoA gene sequences recovered from sample AU were mainly distributed in clusters I and III and became much less abundant in clusters II and V. The AOA sequences recovered from sample AL were mainly present in cluster V, with several ones in clusters I, III, and IV. The AOA sequences retrieved from samples BU, BL, and CU were affiliated with clusters I, IV, and V. Members of the AOA clone libraries constructed with samples BU and BL were mainly found in clusters IV and V, while most of the AOA sequences from sample CU existed in cluster V. The AOA sequences from samples DU and DL belonged to clusters II, IV, and V. Members of AOA clone library constructed with sample DU were more evenly distributed in these three clusters. However, the archaeal amoA gene sequences from sample CL were mainly detected in cluster IV, but became much less abundant in cluster V. These results illustrated a large variation in the AOA community structure in the different samples. AOA community structure could be affected by both sampling site and sediment depth.
Phylogenetic tree of representative archaeal amoA sequences and reference sequences from GenBank. The obtained archaeal sequences beginning with AU and AL, BU and BL, CU and CL, and DU and DL were referred to the sequences retrieved from the upper layer and lower layer sediments in sampling sites A–D, respectively. The bold number in parentheses represent the numbers of the sequences in the same OTU in a given clone library. Numbers at the nodes indicate the levels of bootstrap support based on neighbor-joining analysis of 1,000 resampled datasets. The bar represents 5 % sequence divergence
Figure 5 also indicates that most of the obtained AOA sequences were distributed in clusters IV and V. Cluster I had 31 AOA sequences that were retrieved from sampling sites A, B, and C. These archaeal gene sequences were grouped with the uncultured ones from West Pacific deep-sea sediment (Dang et al. 2009), Qiantang River sediment (Liu et al. 2013), Yangtze Estuary sediment, Chaohu Lake sediment, stream biofilm, agricultural soil, wastewater bioreactor, and Chongming eastern intertidal sediment (Zheng et al. 2013). Cluster II was the smallest cluster and only had 11 members retrieved from sampling sites A and D. The sequences in cluster II were related to two isolated soil AOA strains, Nitrososphaera sp. JG1 and EN76 (Kim et al. 2012; Tourna et al. 2011). The AOA sequences in cluster III were only detected in sampling site A, and they were grouped with those from Yangtze Estuary sediment and Inner Mongolia grassland soil. Moreover, cluster IV was the second largest AOA group containing 74 sequences. These AOA sequences were retrieved from all of the four sampling sites, and they were affiliated with those from Icelandic grassland soil, Qiantang River sediment (Liu et al. 2013), granular activated carbon in advanced water purification plant, land-water ecotone sediment, and King Highway landfill (Im et al. 2011). In addition, cluster V was the largest group with 100 AOA members which were recovered from all of the four sampling sites. These sequences were related to the uncultured ones from a variety of ecosystems such as grassland soil, land-water ecotone sediment, water in drinking water distribution system (van der Wielen et al. 2009), river water, wastewater treatment plant, estuary sediment, and river and lake sediment.
Figure 6 indicates that bacterial amoA sequences of the eight AOB clone libraries could be grouped into three AOB clusters, namely clusters a, b, and c. Cluster a could be further divided into subclusters a1, a2, a3, and a4. The bacterial amoA gene sequences recovered from sample AU were mainly detected in clusters a1 and a4, while most of the AOB sequences from sample AL existed in cluster a4. The AOB sequences from sample BU were distributed in clusters a1, a2, a3, a4, and c. Members of AOB clone library constructed with sample BL were mainly present in cluster c, but became much less abundant in clusters a4 and b. The AOB sequences from sample CU mainly belonged to cluster c, while those from sample CL mainly to clusters a1 and a4. The bacterial amoA gene sequences retrieved from sample DU existed in clusters a1, a2, and a3, while those from sample DL were only detected in cluster a1. Therefore, a large shift in AOB community structure occurred in the different samples. AOB community structure could be affected by both sampling site and sediment depth.
Phylogenetic tree of representative bacterial amoA sequences and reference sequences from GenBank. The obtained bacterial sequences beginning with AU and AL, BU and BL, CU and CL, and DU and DL were referred to the sequences retrieved from the upper layer and lower layer sediments in sampling sites A–D, respectively. The bold numbers in parentheses represent the numbers of the sequences in the same OTU in a given clone library. Numbers at the nodes indicate the levels of bootstrap support based on neighbor-joining analysis of 1,000 resampled datasets. The bar represents 5 % sequence divergence
Figure 6 also illustrates that most of the obtained bacterial amoA gene sequences were distributed in clusters a1, a4, and c. Cluster a1 was the largest AOB group, composed of 102 bacterial amoA gene sequences. The sequences in cluster a1 were recovered from all of the four sampling sites, and they were related to some cultivated Nitrosospira species (TCH716, PJA1, and 9SS1). Cluster a2 was a nine-member group. Its members were from sampling sites B, C, and D, grouped with the uncultured sequences from sandy loam soil. Cluster a3 only had six AOB sequences. These sequences were recovered from sampling sites A, B, and D and were related to an isolated AOB strain (Nitrosospira sp. Nsp62) (Purkhold et al. 2003). Cluster a4 was the second largest group and contained 47 AOB sequences. The AOB sequences in cluster a4 were recovered from sampling sites A, B, and C. They were grouped with the uncultured sequences from a variety of soil ecosystems. Cluster b was the smallest group and only included three AOB sequences from sample BL. These sequences were related to an activated sludge AOB isolate (Suwa et al. 1997). In addition, the 49 AOB sequences in cluster c were retrieved from sampling sites A, B, and C. They were affiliated with the bacterial amoA gene sequences from Yangtze Estuary sediment and Chongming eastern intertidal sediment (Zheng et al. 2013).
Discussion
Depth-related change of the relative abundances of AOA and AOB
To date, there have few previous studies related to ammonia-oxidizing microorganisms in wetland ecosystems. Sims et al. (2012) found that AOA could outnumber AOB by at least an order of magnitude in soils and waters of freshwater wetlands. Cao et al. (2011b) reported higher abundance of AOA than AOB in mangrove sediment, while Li et al. (2011) found the dominance of AOB in coastal mangrove sediment. The abundance of bacterial amoA gene was greater than that of archaeal amoA gene in wetland sediments of Chongming eastern tidal flat (Zheng et al. 2013). Wang and Gu (2013) revealed the different distributions of AOA and AOB in sediments of coastal marine and freshwater wetlands. To the authors’ knowledge, this was the first report on ammonia-oxidizing microorganisms in sediment of high-altitude freshwater wetland. In this study, six out of eight sediment samples had the AOA/AOB ratio of less than 1. These results indicated that the dominance of AOB over AOA usually occurred in the Luoshijiang Wetland. Therefore, AOB might play a more important role than AOA in the nitrification process in the high-altitude freshwater wetland.
The spatial variations in the relative abundances of AOA and AOB and AOA/AOB ratios were found in sediment samples of the Luoshijiang Wetland. Previous researches have shown the spatial shift in the relative abundances (or abundances) of AOA and AOB or AOA/AOB ratio in various sediment ecosystems (Cao et al. 2011b; Hou et al. 2013; Moin et al. 2009; Li et al. 2011; Park et al. 2008; Sakami 2012; Zheng et al. 2013). Wang and Gu (2013) also found that the abundances of AOA and AOB and AOA/AOB ratio varied significantly among the different sites in the freshwater wetland. However, limited information exists on the depth-related change of the abundances of AOA and AOB. Zhao et al. (2013) revealed that the abundances of AOA and AOB in lake sediment exhibited negative correlations with the increased depths, while Li et al. (2011) found no relationship between the abundances of AOA and AOB and the sediment depth in mangrove sediment. To the authors’ knowledge, this was the first report on the depth-related change of the AOA and AOB abundances and AOA/AOB ratio in freshwater wetland sediment. In this study, a rise of the relative abundance of AOA with the decreased sediment depth was found in sampling sites B and C, while the AOA proportion in the upper layer sediment was much lower than that in the lower layer in sampling site A. Moreover, the relative abundance of AOB decreased with the increased depth in sampling sites A and B, but increased in sampling sites C and D. Conversely, the AOA/AOB ratio increased with the increased depth in sampling sites A and B, but declined in sampling sites C and D. These results were in agreement with those of others’ previous study (Li et al. 2011). In addition, Wang et al. (2013) suggested that sediment type could affect the abundances of ammonia-oxidizing organisms. Therefore, the depth-related changes of the AOA and AOB abundances could be dependent on sampling site and sediment type.
AOA is known for higher specific affinity for ammonia and preference for living in low ammonium nitrogen environments (Erguder et al. 2009). However, high ammonium nitrogen environments can favor the AOB abundance (Di et al. 2009). It has been commonly accepted that ammonia concentration is one of the important factors contributing to the definition of distinct ecological niches of AOA and AOB in the environment (Verhamme et al. 2011; Wang et al. 2014). The abundances of AOA and AOB can be influenced by ammonia availability (Hu et al. 2012). However, the impact of ammonia concentration on the abundances of AOA and AOB in sediment ecosystems remains unclear. Li et al. (2011) showed that the abundance of archaeal amoA sequences was significantly correlated with the ammonia concentration in mangrove sediments. Cao et al. (2011b) reported that the AOA/AOB ratio was correlated with the concentration of ammonium in mangrove sediments. Wu et al. (2010) found that the AOA abundance and the AOA/AOB ratio were negatively related to ammonia concentration in sediment of Lake Taihu. Hou et al. (2013) also indicated the AOB abundance was positively correlated with the ammonia concentration in lake sediment, while the AOA abundance was negatively correlated with the ammonia concentration. However, in this study, the AOA and AOB proportions or the AOA/AOB ratio did not show significant correlations with the ammonia concentration (p > 0.05). Zheng et al. (2013) also indicated that no significant correlations were observed between the abundances of AOA and AOB and the ammonia concentration in Chongming eastern intertidal sediments (Zheng et al. 2013). In addition, no significant relationship was observed between the amoA abundance and the ammonium concentration in the surface sediments of Matsushima Bay (Sakami 2012). In this study, Pearson’s correlation analysis indicated that the AOA and AOB proportions or the AOA/AOB ratio did not show significant correlations with the other measured sediment chemical parameters (p > 0.05). This suggest that the factors regulating AOA and AOB abundances in sediments of the Luoshijiang Wetland were quite complex. Multiple factors might be involved in shaping the dynamics of AOA and AOB in sediment ecosystem (Cao et al. 2011b).
Depth-related change of AOA and AOB community diversities
There have been numerous reports on the AOA and AOB community diversities in sediment ecosystems. AOA community diversity was higher than AOB diversity in salt marsh sediment (Moin et al. 2009), intertidal wetland sediment (Zheng et al. 2013), beach sediment (Santoro et al. 2008), and estuary sediment (Cao et al. 2011a), but lower in mangrove sediment (Cao et al. 2011b; Li et al. 2011) and reservoir sediment (Wang et al. 2014). Moreover, Wang and Gu (2013) revealed that AOA diversity was much higher than AOB diversity in sediment of freshwater wetland, but AOB diversity was higher in sediment of coastal marine wetland. Wang et al. (2013) suggested that sediment type could affect community structures of ammonia-oxidizing organisms. These previous studies also showed the spatial shift in the AOA and AOB community diversities in various sediment ecosystems. To date, information about the sediment depth-related change of the AOA and AOB community diversities is still very limited. Zhao et al. (2013) indicated that diversities of both the AOA and AOB amoA genes increased with the elevated depth in lake sediments. Li et al. (2011) showed the rise of AOB diversity with the increased depth in mangrove sediments, while they found no relationship between the AOA community diversity and the sediment depth. However, there has been no report on the depth-related change of the AOA and AOB community diversities in freshwater wetland sediment. In this study, large differences in the AOA and AOB community diversities were observed in the different sampling sites in the Luoshijiang Wetland. The AOB diversity was higher than the AOA diversity in most of the sediment samples (seven out of eight). This result was different from that of others’ previous study on the AOA and AOB community diversities in sediment of freshwater wetland (Wang and Gu 2013). In addition, in the Luoshijiang Wetland, the AOA diversity always decreased with the elevated depth, while no relationship was found between the AOB diversity and the sediment depth. These results were not in agreement with others’ previous studies on the AOA and AOB community diversities in lake and mangrove sediments (Li et al. 2011; Zhao et al. 2013). Therefore, the depth-related change of the AOA and AOB community diversities could be dependent on sediment type.
A few previous studies using Pearson’s correlation analysis have investigated the links between the AOA and AOB community diversities and the sediment environmental factors. The reported factors shaping the sediment AOA and AOB community diversities include pH (Cao et al. 2011a), temperature (Cao et al. 2011a), total phosphorous (Wu et al. 2010), and ammonia nitrogen (Li et al. 2011). In this study, no significant correlations were observed between the AOA and AOB community diversities and the levels of ammonia and total phosphorous. However, the AOA diversity was founded to be positively correlated with available phosphorus (p < 0.05). To the authors’ knowledge, this was the first report on the links between the AOA and AOB community diversities and available phosphorus. Further work is necessary in order to elucidate the links between the sediment depth and the abundances and diversities of AOA and AOB communities in sediment ecosystems.
AOA and AOB community structures
In this study, the obtained AOA sequences could be affiliated with those from various ecosystems, such as marine, river, estuary, lake, intertidal and wetland sediments, agricultural and grassland soils, stream biofilm, drinking and wastewater treatment, drinking water, and river water. This suggested that AOA species detected in different ecosystems could survive in the high-altitude freshwater wetland. Wang and Gu (2013) suggested that AOA might adapt to a variety of habitats and the AOA species detected in one type of habitat might occur in other types of habitats. Moreover, although most of the obtained archaeal amoA gene sequences were not related to any known AOA strains, a few AOA sequences recovered from sampling sites A and D could be affiliated with two soil Nitrososphaera strains (Kim et al. 2012; Tourna et al. 2011). Members of Nitrososphaera have been only isolated from hot spring (Hatzenpichler et al. 2008) and soil (Tourna et al. 2011). The Nitrososphaera-like sequences have been found in mangrove sediment (Cao et al. 2011b), intertidal wetland sediment (Zheng et al. 2013), and freshwater wetland sediment (Wang and Gu 2013). Therefore, Nitrososphaera might exist in various wetland sediment ecosystems. In addition, Zhao et al. (2013) reported marked differences for the AOA community compositions with the increase of sediment depth in Lake Taihu. AOA community compositions were also affected by the change of sediment depth in the Luoshijiang Wetland.
Nitrosospira favor low ammonia environments, while Nitrosomonas favor polluted environments of high ammonia (Wang and Gu 2013). Nitrosospira and Nitrosomonas are usually the predominant AOB in soil and sediment ecosystems, respectively (Wang et al. 2014). In this study, only several Nitrosomonas-like sequences were detected, suggesting their minor role in nitrification in the high-altitude freshwater wetland. In contrast, Nitrosospira-like sequences predominated in the wetland sediment AOB community, indicating their important contribution to ammonia oxidation. These Nitrosospira-like sequences were closely related to the AOB sequences recovered from soil ecosystems. This suggested that Nitrosospira could adapt to a wider range of habitats. Previous studies have confirmed the high abundance or predominance of Nitrosospira-like sequences in estuary and marine sediments (Cao et al. 2011a), intertidal wetland sediments (Zheng et al. 2013), sediments of freshwater wetland, and coastal marine wetland (Wang and Gu 2013). In addition, although Nitrosospira-like sequences dominated the high-altitude freshwater wetland, their compositions were quite different. They could be divided into two groups (clusters a1and a3). Cluster a1 had much more members than cluster a3. The AOB sequences in cluster a1 were recovered from seven sediment samples, while those in clusters a3 only from three samples. Moreover, a marked variation of AOB community structures with the increase of sediment depth was observed in the Luoshijiang Wetland. However, there was no significant variation of AOB community structures among the different sediment depths in Lake Taihu (Zhao et al. 2013). Therefore, the depth-related change of AOA and AOB community structures could differ in different sediment ecosystems.
In conclusion, depth-related variations of AOA and AOB community sizes and structures occurred in sediments of the Luoshijiang Wetland. AOB usually had higher abundance and diversity than AOA. A few Nitrososphaera-like AOA species were observed. Nitrosospira-like AOB species predominated in the wetland sediments.
References
Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT (2007) Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J 1:660–662
Cao HL, Hong YG, Li M, Gu JD (2011a) Diversity and abundance of ammonia-oxidizing prokaryotes in sediments from the coastal Pearl River estuary to the South China Sea. Antonie Van Leeuwenhoek 100:545–556
Cao HL, Li M, Hong YG, Gu JD (2011b) Diversity and abundance of ammonia-oxidizing archaea and bacteria in polluted mangrove sediment. Syst Appl Microbiol 34:513–523
Cao HL, Hong YG, Li M, Gu JD (2012) Lower abundance of ammonia oxidizing archaea than ammonia-oxidizing bacteria detected in the subsurface sediments of the Northern South China Sea. Geomicrobiol J 29:332–339
Chen X, Zhang LM, Shen JP, Xu ZH, He JZ (2010) Soil type determines the abundance and community structure of ammonia-oxidizing bacteria and archaea in flooded paddy soils. J Soils Sediments 10:1510–1516
Cheng W, Zhang JX, Wang Z, Wang M, Xie SG (2014) Bacterial communities in sediments of a drinking water reservoir. Ann Microbiol. doi:10.1007/s13213-013-0712-z
Dang H, Li J, Zhang X, Li T, Tian F, Jin W (2009) Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J Appl Microbiol 106:1482–1493
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624
Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W (2009) Environmental actors shaping the ecological niches of ammonia oxidizing archaea. FEMS Microbiol Rev 33:855–869
Feng S, Zhang XJ, Wang QF, Wan R, Chen C, Xie SG (2012) Heterogeneity of ammonia-oxidizing community structures in a pilot-scale drinking water biofilter. Int Biodeterior Biodegrad 70:148–152
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139
He J, Shen J, Zhang L, Zhu Y, Zheng Y, Xu M, Di HJ (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374
Hou J, Song CL, Cao XY, Zhou YY (2013) Shifts between ammonia-oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu). Water Res 47:2285–2296
Hu BL, Liu SA, Shen LD, Zheng P, Xu XY, Lou LP (2012) Effect of different ammonia concentrations on community succession of ammonia-oxidizing microorganisms in a simulated paddy soil column. PLOS One. doi:10.1371/journalpone0044122
Im J, Lee SW, Bodrossy L, Barcelona MJ, Semrau JD (2011) Field application of nitrogen and phenylacetylene to mitigate greenhouse gas emissions from landfill cover soils: effects on microbial community structure. Appl Microbiol Biotechnol 89:189–200
Jung J, Yeom J, Kim J, Han J, Lim HS, Park H, Hyun S, Park W (2011) Change in gene abundance in the nitrogen biogeochemical cycle with temperature and nitrogen addition in Antarctic soils. Res Microbiol 162:1018–1026
Kim JG, Jung MY, Park SJ, Rijpstra WIC, Damste JSS, Madsen EL, Min D, Kim JS, Kim GJ, Rhee SK (2012) Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I1b from an agricultural soil. Environ Microbiol 14:1528–1543
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Li M, Cao HL, Hong YG, Gu JD (2011) Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia oxidizing bacteria (AOB) in mangrove sediments. Appl Microbiol Biotechnol 89:1243–1254
Li XR, Xiao YP, Ren WW, Liu ZF, Shi JH, Quan ZX (2012) Abundance and composition of ammonia-oxidizing bacteria and archaea in different types of soil in the Yangtze River estuary. J Zhejiang Univ-SCI B 13:769–782
Liu L, Peng Y, Zheng XH, Xiao L, Yang LY (2010) Vertical structure of bacterial and archaeal communities within the sediment of a eutrophic lake as revealed by culture-independent methods. J Freshw Ecol 25:565–573
Liu S, Shen L, Lou L, Tian G, Zheng P, Hu B (2013) Spatial distribution and factors shaping the niche segregation of ammonia-oxidizing microorganisms in the Qiantang River, China. Appl Environ Microbiol 9:4065–4071
Long X, Chen CR, Xu ZH, Oren R, He JZ (2012) Abundance and community structure of ammonia-oxidizing bacteria and archaea in a temperate forest ecosystem under ten-years elevated CO2. Soil Biol Biochem 46:163–171
Moin NS, Nelson KA, Bush A, Bernhard AE (2009) Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl Environ Microbiol 75:7461–7468
Park SJ, Park BJ, Rhee SK (2008) Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12:605–615
Purkhold U, Wagner M, Timmermann G, Pommerening-Roser A, Koops HP (2003) 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int J Syst Evol Microbiol 53:1485–1494
Sakami T (2012) Distribution of ammonia-oxidizing archaea and bacteria in the surface sediments of Matsushima Bay in relation to environmental variables. Microbes Environ 27:61–66
Santoro AE, Francis CA, de Sieyes NR, Boehm AB (2008) Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ Microbiol 10:1068–1079
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Shivaji S, Kumari K, Kishore KH, Pindi PK, Rao PS, Srinivas TNR, Asthana R, Ravindra R (2011) Vertical distribution of bacteria in a lake sediment from Antarctica by culture-independent and culture-dependent approaches. Res Microbiol 162:191–203
Sims A, Horton J, Gajaraj S, McIntosh S, Miles RJ, Mueller R, Reed R, Hu ZQ (2012) Temporal and spatial distributions of ammonia-oxidizing archaea and bacteria and their ratio as an indicator of oligotrophic conditions in natural wetlands. Water Res 46:4121–4129
Suwa Y, Sumino T, Noto K (1997) Phylogenetic relationships of activated sludge isolates of ammonia oxidizers with different sensitivities to ammonium sulfate. J Gen Appl Microbiol 43:373–379
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4, molecular evolutionary genetics analysis, MEGA software version 4.0. Mol Biol Evol 24:1596–1599
Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425
van der Wielen PWJJ, Voost S, van der Kooij D (2009) Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl Environ Microbiol 75:4687–4695
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Verhamme DT, Prosser JI, Nicol GW (2011) Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5:1067–1071
Wang YF, Gu JD (2013) Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl Microbiol Biotechnol 97:7015–7033
Wang YF, Feng YY, Ma XJ, Gu JD (2013) Seasonal dynamics of ammonia/ammonium-oxidizing prokaryotes in oxic and anoxic wetland sediments of subtropical coastal mangrove. Appl Microbiol Biotechnol 97:7919–7934
Wang XY, Wang C, Bao LL, Xie SG (2014) Abundance and community structure of ammonia-oxidizing microorganisms in reservoir sediment and adjacent soils. Appl Microbiol Biotechnol 98:1883–1892
Wu YC, Xiang Y, Wang JJ, Zhong JC, He JZ, Wu QLL (2010) Heterogeneity of archaeal and bacterial ammonia oxidizing communities in Lake Taihu, China. Environ Microbiol Rep 2:569–576
Zhang JX, Zhang XL, Liu Y, Xie SG, Liu YG (2013) Bacterioplankton communities in a high-altitude freshwater wetland. Ann Microbiol. doi:10.1007/s13213-013-0785-8
Zhao XQ, Yang LY, Yu ZY, Peng NY, Xiao L, Yin DQ, Qin BQ (2008) Characterization of depth-related microbial communities in lake sediment by denaturing gradient gel electrophoresis of amplified 16S rRNA fragments. J Environ Sci 20:224–230
Zhao DY, Zeng J, Wan WH, Liang HD, Huang R, Wu QLL (2013) Vertical distribution of ammonia-oxidizing archaea and bacteria in sediments of a eutrophic lake. Curr Microbiol 67:327–332
Zheng YL, Hou LJ, Liu M, Lu M, Zhao H, Yin GY, Zhou JL (2013) Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments. Appl Microbiol Biotechnol 97:8351–8363
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Nos. 51279001 and 41222002).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 106 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Zhang, J., Zhang, X. et al. Depth-related changes of sediment ammonia-oxidizing microorganisms in a high-altitude freshwater wetland. Appl Microbiol Biotechnol 98, 5697–5707 (2014). https://doi.org/10.1007/s00253-014-5651-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5651-5