Abstract
Both ammonia-oxidizing archaea (AOA) and bacteria (AOB) can contribute to ammonia biotransformation in freshwater lake ecosystems. However, the factors shaping the distribution of sediment AOA and AOB in plateau freshwater lake remains unclear. The present study investigated sediment AOA and AOB communities in two freshwater lakes (hypertrophic Dianchi Lake and mesotrophic Erhai Lake) on the Yunnan Plateau (China). A remarkable difference in the abundance, diversity, and composition of sediment AOA and AOB communities was observed between Dianchi Lake and Erhai Lake. AOB usually outnumbered AOA in Dianchi Lake, but AOA showed the dominance in Erhai Lake. Organic matter (OM), total nitrogen (TN), and total phosphorus (TP) might be the key determinants of AOB abundance, while AOA abundance was likely influenced by the ration of OM to TN (C/N). AOA or AOB community structure was found to be relatively similar in the same lake. TN and TP might play important roles in shaping sediment AOA and AOB compositions in Dianchi Lake and Erhai Lake. Moreover, Nitrososphaera-like AOA were detected in Dianchi Lake. Nitrosospira- and Nitrosomonas-like AOB were dominant in Dianchi Lake and Erhai Lake, respectively. Sediment AOA and AOB communities in Dianchi Lake and Erhai Lake were generally regulated by trophic state.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Freshwater sediment ecosystems harbor a complex microbial community that participates in a variety of biogeochemical processes [2, 10, 29]. Transformation of ammonia to nitrite plays a central role in nitrogen cycle in sediment ecosystems as it is the first and rate-limiting step in nitrification process. This step is mainly catalyzed by ammonia monooxygenase (amoA) gene-carrying ammonia-oxidizing archaea (AOA) and bacteria (AOB) [14, 27]. The amoA gene has been widely used as a functional biomarker to investigate the distribution of sediment AOA and AOB in a variety of freshwater ecosystems, such as wetland [11, 27], reservoir [24], lake [5, 24, 14], and river [12, 20]. However, the links between ammonia-oxidizing microorganisms in freshwater lake sediment and environmental factors still remain unclear, although several previous studies suggested that trophic status might regulate the abundance and structure of AOA and AOB communities in freshwater lake sediment [1, 5, 6, 25, 31]. Compared to plain lakes, plateau lakes are usually more sensitive to anthropogenic disturbance due to their poor water exchange [29]. Information on sediment AOA and AOB in plateau freshwater lake is still very limited [14, 26], yet Yang et al. suggested that plateau lakes may harbor a unique ammonia-oxidizing microbial community of different evolutionary origin from those in other lakes worldwide [26]. In addition, the effect of trophic status on the distribution of sediment AOA and AOB in plateau freshwater lake is still not understood.
The Yunnan Plateau (southwestern China), located in the subtropical or temperate climate zone, has about 40 freshwater lakes. Dianchi Lake is the largest lake (309 km2), and its average water depth and altitude are 4.4 and 1886 m, respectively [33]. Erhai Lake is the second largest lake (251 km2), with an average water depth and elevation of 10.5 and 1974 m, respectively [9]. Dianchi Lake and Erhai Lake are characterized as highly eutrotrophic and mesotrophic, respectively. Therefore, the objective of the present study was to investigate the effect of trophic level on the distribution of sediment AOA and AOB in freshwater lakes.
Materials and Methods
Site Description and Sample Collection
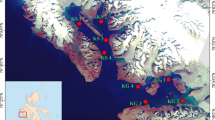
A total of 11 surface sediment samples (0–5 cm) in triplicate were collected using a core sampler from Dianchi Lake (102.6128–102.6825° E, 24.6960–24.9541° N) and Erhai Lake (100.1497–100.2213° E, 25.6858–25.9083° N) in April 2014 (Fig. 1). These sediment samples were placed in sterile plastic bottles and were kept on ice and immediately transported to the laboratory. These samples were homogenized and dried with a freeze dryer (Alpha 1–2 LD plus, Martin Christ, German).
Physicochemical Analysis
Sediment organic matter (OM) was determined according to the literature [28]. Total nitrogen (TN), nitrate nitrogen (NO3 −-N), ammonium nitrogen (NH4 +-N), and total phosphorus (TP) were determined according to the literature [19]. Sediment pH was determined using an IQ150 pH meter.
Molecular Analyses
Lake sediment DNA was extracted using the Powersoil DNA extraction kit (Mobio Laboratories, USA) according to the manufacturer’s instructions. The primer sets Arch-amoAF/Arch-amoAR and AmoA-1F/AmoA-2R were used for quantitative PCR (qPCR) assay of archaeal and bacterial amoA genes and for construction of AOA and AOB clone libraries, following the previously reported conditions [11, 27]. One-way analysis of variance (ANOVA) followed by Student-Newman-Keuls test was used to determine the quantitative differences (P < 0.05) in the density of amoA gene in different sediment samples. Chimera-free amoA gene sequences with similarity equal to or greater than 97 % were grouped into the same operational taxonomic units (OTUs), and OTU-based Shannon diversity index was further calculated using the MOTHUR program [18]. In order to compare the AOA or AOB community dissimilarity in lake sediments, phylogeny-based weighted UniFrac environmental clustering was also performed using the online UniFrac program [15]. Moreover, phylogenetic analysis of the retrieved archaeal and bacterial amoA genes from lake sediments were performed using MEGA 6.0 software [22].
The correlation between microbial communities and sediment physicochemical properties was discriminated using Pearson’s correlation analysis with the SPSS 20.0 software. In addition, detrended correspondence analysis (DCA) was also applied to choose the appropriate ordination analysis method. Since the longest DCA eigenvalue of AOA (3.497) or AOB (3.085) was between 3 and 4, either linear model or unimodal model would be suitable. In this study, the relationships between microbial OTU composition and environmental factors were identified with redundancy analysis (RDA) using CANOCO 4.5 software [8]. The number of amoA gene sequence in each OTU was used as species input but the determined environmental factors as environment input. The significance test of Monte Carlo permutations was conducted to construct the appropriate models of the microbe-environment relationships.
Nucleotide Sequence Accession Number
The obtained amoA gene sequences in this study were deposited in the GenBank database under accession numbers KP197191–KP197396 and KP197397–KP197611 for AOA and AOB in Dianchi Lake, respectively, while KM116520–KM116727 and KM250454–KM250522 for AOA and AOB in Erhai Lake, respectively.
Results
Sediment Physicochemical Properties
Sediment physicochemical properties (OM, TN, ration of OM to TN (C/N), NO3 −-N, NH4 +-N, TP, and pH) are listed in Table S1. They were 24.3–158.7 g/kg, 1.0–8.1 g/kg, 7.3–39, 0–118 mg/kg, 3.13–36 mg/kg, 0.41–4.28 g/kg, and 7.01–7.34, respectively.
Abundance of AOA and AOB Communities
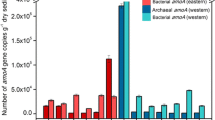
The density of archaeal amoA gene ranged from 4.23 × 104 to 7.07 × 105 copies per gram dry sediment in the sediment samples (D1–D6) from Dianchi Lake (Fig. 2). Sediment D2 had significantly higher AOA abundance than other five sediments in Dianchi Lake (P < 0.05). Sediment AOA was found to be much more abundant in Erhai Lake than in Dianchi Lake. Significant difference in AOA abundance was also observed in the sediment samples (E1–E5) from Erhai Lake (P < 0.05), ranging from 9.88 × 105 to 7.64 × 106 amoA gene copies per gram dry sediment, and sediment E5 had the highest AOA community size followed by sediment E4.
AOB community size ranged from 1.31 × 105 to 5.98 × 105 amoA gene copies per gram dry sediment in the sediment samples (D1–D6) from Dianchi Lake, and sediment D1 had the highest AOB community size followed by sediment D2. Compared with sediments D1–D6, sediments E1 and E5 had relatively high AOB abundance (1.69 × 106 or 1.36 × 106 amoA gene copies per gram dry sediment). However, the density of AOB amoA gene in sediments E2–E4 was found to be below PCR detection limit. In addition, AOB community size was usually much larger than AOA in the sediment samples from Dianchi Lake, but lower in those from Erhai Lake.
Diversity of AOA and AOB Communities
In this study, sediments E2–E4 were not successfully amplified for the construction of AOB clone libraries, due to the very low abundance of bacterial amoA gene (below PCR detection). A total of 414 archaeal and 284 bacterial sequences were obtained from lake sediments. The AOA libraries with the sediment samples from Dianchi Lake consisted of 8–11 OTUs, while only 1–7 OTUs were found in AOA communities in sediments E1–E5 (Table 1). A remarkable variation in AOA community diversity was found in sediments D1–D6, with Shannon index of 1.4–1.95. Sediments E1–E5 had much lower AOA community diversity (Shannon index = 0–0.88) than sediments D1–D6 (Table 1).
The AOB libraries with sediments D1–D6 were composed of 5–26 OTUs, while 10 and 11 OTUs were observed in AOB communities in sediments E1 and E5, respectively. Sediments E1 and E5 (Shannon index = 1.38 or 1.79) had higher AOB community diversity than sediment D2 (0.87), but lower than sediments D1 and D3–D6 (2.57–3.14). In addition, for the sediment samples from Dianchi Lake, AOB usually showed higher community diversity than AOA. For sediments E1 and E5, AOB Shannon diversity was also higher than AOA.
Comparison of AOA and AOB Communities
Phylogeny-based weighted UniFrac clustering analysis showed the existence of two distinctive AOA clades in plateau lake sediments (Fig. 3a). The sediment samples from Dianchi Lake were grouped together. Except for sediment E4, the sediment samples from Erhai Lake were also clustered. These results suggested that sediments in the same lake tended to have relatively similar AOA community structure.
Two distinctive AOB clades were found in plateau lake sediments (Fig. 3b). Except for sediment D2, the sediment samples from Dianchi Lake were grouped together. Moreover, the two sediments from Erhai Lake fell into another clade. These results suggested that sediments in the same lake also tended to have relatively similar AOB community structure.
Phylogeny of AOA and AOB Communities
In this study, the representative sediment amoA gene sequences used for phylogenetic analysis were selected only from the OTUs that had at least two sequence members. The sediment AOA sequences retrieved from Dianchi Lake and Erhai Lake could be grouped into three clusters (Fig. 4). The archaeal amoA gene sequences from Dianchi Lake were mainly distributed in clusters I and II, but those from Erhai Lake mainly in cluster III. This further confirmed the distinctive difference of AOA community structure between sediments in Dianchi Lake and Erhai Lake. Cluster I was the second largest AOA group containing 146 archaeal sequences. The AOA sequences in this cluster could be affiliated with the uncultured ones from paddy and purple soils, tidal, lake, river, and marine sediments, stream biofilm, and wastewater treatment plant [3, 4, 32]. Cluster II was the smallest AOA group containing 39 sequences that could be related to two cultivated soil Nitrososphaera strains (JG1 and EN76) [7, 23]. Moreover, cluster III was a 203-member AOA group. The archaeal amoA gene sequences in this cluster could be grouped with those from various soil and sediment ecosystems [6, 32].
Phylogenetic tree of representative archaeal amoA sequences and reference sequences from GenBank. The obtained archaeal sequences beginning with “D1”–“D6” and “E1”–“E5” were referred to the sequences retrieved from sediments D1–D6 and E1–E5, respectively. The bold number in parentheses represents the numbers of the sequences in the same OTU in a given clone library. The amoA sequences from Dianchi Lake are highlighted in green, while the ones from Erhai Lake are in blue. Numbers at the nodes indicate the levels of bootstrap support based on neighbor-joining analysis of 1000 resampled datasets. The bar represents 5 % sequence divergence (color figure online)
The bacterial amoA gene sequences retrieved from sediments in Dianchi Lake and Erhai Lake could be grouped into two clusters (Fig. 5). The AOB sequences from Dianchi Lake mainly existed in cluster a, but those from Erhai Lake mainly in cluster b, suggesting the distinctive difference of AOB community structure between sediments in Dianchi Lake and Erhai Lake. However, all the bacterial sequences from sediment D2 were only detected in cluster b, while 12 sequences from sediment E5 were found in cluster a. This indicated that sediment AOB community structure could be much different even in the same lake. Cluster a included a total of 120 bacterial amoA gene sequences that were related to numerous cultivated Nitrosospira species (Nsp65, CT2F, Nsp62, LT1FMf, Enl299, NRS527, GS832, TYM9, 9SS1, PJA1, KAN8, TCH711, and TCH716 [16, 17]. In addition, cluster b was a 77-member AOB group. The bacterial amoA gene sequences in this cluster could be affiliated with a cultivated Nitrosomonas species (AL212) [21].
Phylogenetic tree of representative bacterial amoA sequences and reference sequences from GenBank. The obtained bacterial sequences beginning with “D1”–“D6,” “E1,” and “E5” were referred to the sequences retrieved from sediments D1–D6, E1, and E5, respectively. The bold number in parentheses represents the numbers of the sequences in the same OTU in a given clone library. The amoA sequences from Dianchi Lake are highlighted in green, while the ones from Erhai Lake are in blue. Numbers at the nodes indicate the levels of bootstrap support based on neighbor-joining analysis of 1000 resampled datasets. The bar represents 2 % sequence divergence (color figure online)
Influential Factors Regulating AOA and AOB Communities
Pearson’s correlation analysis indicated that AOA abundance was positively correlated with sediment C/N (P < 0.05), while AOB abundance showed negative correlations with OM (P < 0.05), TN and TP (P < 0.01) (Table 2). The AOB/AOA ratio showed a positive correlation with NO3-N (P < 0.05). In addition, AOA Shannon diversity showed highly significant positive correlations with TN and TP (P < 0.01), while AOB Shannon diversity was negatively correlated with pH (P < 0.01).
The environmental factors in the first two RDA axes respectively accounted for 80.9 and 6.6 % of the total variance for lake sediment AOA OTU composition (Fig. 6a).TN (P = 0.0010, F = 22.134, 999 Monte Carlo permutations) and TP (P = 0.0270, F = 4.265, 999 Monte Carlo permutations) significantly contributed to the AOA community–environment relationship. For AOB OTU composition, the environmental factors in the first two RDA axes respectively explained 46.8 and 34.1 % of the total variance (Fig. 6b). Environmental factors including TP (P = 0.0320, F = 2.917, 999 Monte Carlo permutations), TN (P = 0.0250, F = 4.412, 999 Monte Carlo permutations), and C/N (P = 0.0380, F = 4.502, 999 Monte Carlo permutations) significantly contributed to the AOB assemblage–environment relationship.
Discussion
AOA and AOB Abundance in Freshwater Lake Sediment
So far, information on sediment AOA and AOB abundance in freshwater lake is still very limited. AOA were usually found to be more abundant than AOB in Danish softwater lakes [5], Chaohu Lake [6], and Taihu Lake [6, 24, 31]. However, our previous study indicated that AOB outnumbered AOA in sediments of a number of freshwater lakes on the Yunnan Plateau [14]. Hence, the advantage of sediment AOA or AOB could vary in different freshwater lakes, which made it difficult to determine the relative contribution of AOA and AOB to nitrification in freshwater lake sediment. In addition, for the Laurentian Great Lakes system, Bollmann et al. indicated that AOB dominated in sediments of mesotrophic Lake Erie, while AOA outnumbered AOB in sediments of oligotrophic Lake Superior [1]. In this study, AOB was also usually much more abundant than AOA in the sediment samples from highly eutrophic Dianchi Lake, but lower in those from mesotrophic Erhai Lake. The results of these two studies suggested that AOA and AOB abundance could be affected by lake trophic status.
AOA and AOB abundance in freshwater lake sediment can be regulated by multiple environmental factors. AOA abundance in freshwater lake sediment was found to be affected by pH [24], NH4 +-N [13, 25, 31], NO3 −-N [1, 13], TN [25, 31], TP [13], and total organic carbon (TOC) [25], while AOB abundance by NH4 +-N [1], pH [31], and NO3 −-N [31]. In contrast, in this study, AOB abundance was found to be likely shaped by OM, TN, and TP, which was not in agreement with the results reported in the previous studies [1, 31]. In addition, to the authors’ knowledge, this was the first report to show that, in freshwater lake sediment, AOA abundance could be likely shaped by C/N, and the AOB/AOA ratio by NO3 −-N. However, in our previous research on the distribution of sediment AOA and AOB across many freshwater lakes on the Yunnan Plateau, no obvious link was found between sediment properties and AOA and AOB abundance or the AOB/AOA ratio [14]. Therefore, further work is still required in order to elucidate the distribution of AOA and AOB abundance in freshwater lake sediment and the influential factors.
AOA and AOB Community Diversity in Freshwater Lake Sediment
There have been few reports on comparing AOA and AOB community diversity in freshwater lake sediment. Zhao et al. indicated higher AOA diversity than AOB in unvegetated sediment and rhizosphere sediment of Potamogeton crispus in Taihu Lake [31]. Our previous study also found that AOA diversity was usually higher than AOB in sediments of freshwater lakes on the Yunnan Plateau [14]. However, in this study, AOB generally showed higher community diversity than AOA in sediments of both Dianchi Lake and Erhai Lake. Higher sediment AOB community diversity than AOA was also found in Jinshan Lake [13] and Taihu Lake [30]. Hence, it remains unclear whether AOA or AOB have relatively high community diversity in freshwater lake sediment.
Herrmann et al. found an increase in the diversity of ammonia-oxidizing prokaryotes from oligotrophic lakes to mesotrophic ones [5]. Bollmann et al. showed higher AOA diversity in mesotrophic Lake Erie than in oligotrophic Lake Superior [1]. In this study, Dianchi Lake generally showed higher AOA or AOB diversity than Lake Erhai. Therefore, the diversity of sediment ammonia-oxidizing microbial populations might increase with increasing lake eutrophic level. The result of Pearson’s correlation analysis also suggested that sediment AOA community diversity was positively influenced by TN and TP. Liu et al. suggested that AOB community diversity in freshwater lake sediment could be affected by nitrogen content [13]. However, in this study, no significant correlation was found between nutrient level and AOB community diversity. In addition, Herrmann et al. suggested that pH could regulate sediment AOA and AOB richness in freshwater lake [5], while the present study showed that pH might be a key determinant of AOB community diversity in freshwater lake sediment.
AOA and AOB Community Structure in Freshwater Lake Sediment
Based on comparison of AOA community composition using weighted UniFrac clustering, Bollmann et al. pointed out that sediment samples from oligotrophic Lake Superior and sediment samples from mesotrophic Lake Erie, respectively, could be grouped together [1]. In addition, Herrmann et al. also indicated that freshwater lake sediment AOA communities clustered together according to lake trophic status and pH [5]. In this study, the results of both weighted UniFrac clustering and phylogenetic analysis indicated that sediment samples in the same lake tended to have relatively similar community structure of either AOA or AOB. These studies suggested that lake trophic status played a crucial role in shaping the community structure of sediment ammonia-oxidizing populations. The result of RDA also confirmed that TN and TP might be the key determinants of sediment AOA and AOB assemblages in Dianchi Lake and Erhai Lake.
Yang et al. suggested that the lakes on the northeastern Qinghai-Tibetan Plateau may harbor a unique AOA population of different evolutionary origin from those in other lakes worldwide [26]. However, in the present study, the retrieved sediment archaeal amoA gene sequences from Dianchi Lake and Erhai Lake could be affiliated with the uncultured ones from sediments in several plain freshwater lakes in China (e.g., Chaohu Lake, Donghu Lake, and Taihu Lake). Our previous study also indicated that the sediment AOA sequences from freshwater lakes on the Yunnan Plateau could be related to those from plain freshwater lakes [14]. Moreover, Nitrososphaera-like AOA were usually present in freshwater lake sediments [5, 13, 31]. So far, the links between Nitrososphaera-like AOA and environmental factors remains unknown. However, Nitrososphaera-like AOA were found to be dominant in mesotrophic Lake Erie but were almost not detected in oligotrophic Lake Superior [1]. In the current study, the Nitrososphaera-like microorganisms were detected in all the sediment samples from Dianchi Lake, but not detected in those from Erhai Lake. Therefore, it could be assumed that that lake trophic status might lead to a niche separation for Nitrososphaera-like AOA species.
Nitrosospira and Nitrosomonas are two AOB groups that are commonly present in freshwater ecosystems [13, 31]. However, their relative advantage in sediment AOB community in freshwater lake remains unclear. The proportion of Nitrosomonas and Nitrosospira can vary in lakes and sampling sites [14]. Several previous researches showed the advantage of sediment Nitrosospira-like AOB in Taihu Lake [25, 30], and in Danish freshwater lakes [5]. Our previous studies also showed the dominance of Nitrosospira-like AOB in sediment from most of the freshwater lakes (8 out of 13) [14]. However, Liu et al. reported the dominance of Nitrosomonas-like AOB in sediments from Jinshan Lake [13]. Moreover, both Nitrosomonas- and Nitrosospira-like AOB were found to dominate in sediments of Taihu Lake and Chaohu Lake [6, 31]. In the present study, Nitrosospira-like organisms were the predominant AOB in most of sediments from Dianchi Lake, while Nitrosomonas-like AOB dominated in two sediments from Erhai Lake and one from Dianchi Lake (sediment D2). This suggested that lake trophic status might lead to a niche separation for AOB species. A previous study also revealed the existence of spatial heterogeneity of AOB populations across the lake trophic state [6].
Conclusions
The abundance, diversity, and composition of AOA and AOB communities in sediments of Dianchi Lake and Erhai Lake showed a remarkable difference. AOB and AOA dominated in Dianchi Lake and Erhai Lake, respectively. AOB usually showed higher community diversity than AOA in these two lakes. AOA or AOB community structure tended to be relatively similar in the same lake. Nitrososphaera-like AOA were present in Dianchi Lake, but absent in Erhai Lake. Nitrosospira and Nitrosomonas were the dominant AOB species in Dianchi Lake and Erhai Lake, respectively. In addition, lake trophic status might play an important role in shaping sediment AOA and AOB communities in Dianchi Lake and Erhai Lake.
References
Bollmann A, Bullerjahn GS, Mckay RM (2014) Abundance and diversity of ammonia-oxidizing archaea and bacteria in sediments of trophic end members of the Laurentian Great Lakes, Erie and Superior. PLoS One 9, e97068
Cheng W, Zhang JX, Wang Z, Wang M, Xie SG (2014) Bacterial communities in sediments of a drinking water reservoir. Ann Microbiol 64:875–878
Dang H, Li J, Zhang X, Li T, Tian F, Jin W (2009) Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical west pacific continental margin. J Appl Microbiol 106:1482–1493
Gao JF, Luo X, Wu GX, Li T, Peng YZ (2014) Abundance and diversity based on amoA genes of ammonia-oxidizing archaea and bacteria in ten wastewater treatment systems. Appl Microbiol Biotechnol 98:3339–3354
Herrmann M, Saunders AM, Schramm A (2009) Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl Environ Microbiol 75:3127–3136
Hou J, Song CL, Cao XY, Zhou YY (2013) Shifts between ammonia oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu). Water Res 47:2285–2296
Kim J, Jung M, Park S, Rijpstra WIC, Damste JSS, Madsen EL, Min D, Kim J, Kim G, Rhee S (2012) Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group i.1b from an agricultural soil. Environ Microbiol 14:1528–1543
Lepš J, Šmilauer P (2003) Multivariate analysis of ecological data using CANOCO. Cambridge University Press, New York
Li YP, Wang SR, Zhang L, Zhao HC, Jiao LX, Zhao YL, He XS (2014) Composition and spectroscopic characteristics of dissolved organic matter extracted from the sediment of Erhai Lake in China. J Soils Sediments 14:1599–1611
Liu Y, Zhang JX, Zhao L, Zhang XL, Xie SG (2014) Spatial distribution of bacterial communities in high-altitude freshwater wetland sediment. Limnology 15:249–256
Liu Y, Zhang JX, Zhang XL, Xie SG (2014) Depth-related changes of sediment ammonia-oxidizing microorganisms in a high-altitude freshwater wetland. Appl Microbiol Biotechnol 98:5697–5707
Liu B, Wu CD, Zhou XH (2014) Community structure of ammonia-oxidizing microorganisms in the Grand Canal, Zhenjiang, of Jiangsu Province, China. Water Sci Technol 70:990–995
Liu B, Li YM, Zhang JP, Zhou XH, Wu CD (2014) Abundance and diversity of ammonia-oxidizing microorganisms in the sediments of Jinshan Lake. Curr Microbiol 69:751–757
Liu Y, Zhang JX, Zhao L, Li YZ, Dai Y, Xie SG (2015) Distribution of sediment ammonia-oxidizing microorganisms in plateau freshwater lakes. Appl Microbiol Biotechnol 99:4435–4444
Lozupone C, Hamady M, Knight R (2006) UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371
Mintie AT, Heichen RS, Cromack K, Myrold DD, Bottomley PJ (2003) Ammonia-oxidizing bacteria along meadow-to-forest transects in the Oregon Cascade Mountains. Appl Environ Microbiol 69:3129–3136
Purkhold U, Wagner M, Timmermann G, Pommerening-Roser A, Koops HP (2003) 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the Nitrosomonads. Int J Syst Evol Microbiol 53:1485–1494
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur, open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–754
Song H, Li Z, Du B, Wang G, Ding Y (2012) Bacterial communities in sediments of the shallow Lake Dongping in China. J Appl Microbiol 112:79–89
Sun W, Xia CY, Xu MY, Guo J, Wang AJ, Sun GP (2013) Distribution and abundance of archaeal and bacterial ammonia oxidizers in the sediments of the Dongjiang River, a drinking water supply for Hong Kong. Microbes Environ 28:457–465
Suwa Y, Sumino T, Noto K (1997) Phylogenetic relationships of activated sludge isolates of ammonia oxidizers with different sensitivities to ammonium sulfate. J Gen Appl Microbiol 43:373–379
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6, Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425
Wang XY, Wang C, Bao LL, Xie SG (2014) Abundance and community structure of ammonia-oxidizing microorganisms in reservoir sediment and adjacent soils. Appl Microbiol Biotechnol 98:1883–1892
Wu YC, Xiang Y, Wang JJ, Zhong JC, He JZ, Wu QLL (2010) Heterogeneity of archaeal and bacterial ammonia oxidizing communities in Lake Taihu, China. Environ Microbiol Rep 2:569–576
Yang J, Jiang HC, Dong HL, Wang HY, Wu G, Hou WG, Liu WG, Zhang CL, Sun YJ, Lai ZP (2013) amoA-encoding archaea and thaumarchaeol in the lakes on the northeastern Qinghai-Tibetan Plateau, China. Front Microbiol 4:329
Yang YY, Shan JW, Zhang JX, Zhang XL, Xie SG, Liu Y (2014) Ammonia- and methane-oxidizing microorganisms in high-altitude wetland sediments and adjacent agricultural soils. Appl Microbiol Biotechnol 98:10197–10209
Ye WJ, Liu XL, Lin SQ, Tan J, Pan JL, Li DT, Yang H (2009) The vertical distribution of bacterial and archaeal communities in the water and sediment of Lake Taihu. FEMS Microbiol Ecol 70:263–276
Zhang JX, Yang YY, Zhao L, Li YZ, Xie SG, Liu Y (2015) Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes. Appl Microbiol Biotechnol 99:3291–3302
Zhao DY, Zeng J, Wan WH, Liang HD, Huang R, Wu QLL (2013) Vertical distribution of ammonia-oxidizing archaea and bacteria in sediments of a eutrophic lake. Curr Microbiol 67:327–332
Zhao DY, Luo J, Zeng J, Wang M, Yan WM, Huang R, Wu QLL (2014) Effects of submerged macrophytes on the abundance and community composition of ammonia-oxidizing prokaryotes in a eutrophic lake. Environ Sci Pollut Res 21:389–398
Zheng YL, Hou LJ, Liu M, Lu M, Zhao H, Yin GY, Zhou JL (2013) Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments. Appl Microbiol Biotechnol 7:8351–8363
Zhou J, Liu Y, Guo HC, He D (2014) Combining the SWAT model with sequential uncertainty fitting algorithm for streamflow prediction and uncertainty analysis for the lake Dianchi Basin, China. Hydrol Process 28:521–533
Acknowledgments
This work was financially supported by the National Basic Research Program of China (2015CB458900), National Natural Science Foundation of China (41222002), and special fund of State Key Joint Laboratory of Environment Simulation and Pollution Control (14Y02ESPCP).
Ethical Statement
I would like to declare on behalf of my co-authors that the work described was original research that has not been published previously, and not under consideration for publication elsewhere, in whole or in part.
Conflict of Interest
No conflict of interest exits in the submission of this manuscript, and the manuscript is approved by all authors for publication.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yuyin Yang and Jingxu Zhang contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 78 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Zhang, J., Zhao, Q. et al. Sediment Ammonia-Oxidizing Microorganisms in Two Plateau Freshwater Lakes at Different Trophic States. Microb Ecol 71, 257–265 (2016). https://doi.org/10.1007/s00248-015-0642-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0642-3