Abstract
Both ammonia-oxidizing bacteria (AOB) and archaea (AOA) can play important roles in ammonia biotransformation in ecosystems. However, the factors regulating the distribution of these microorganisms in lacustrine ecosystems remain essentially unclear. The present study investigated the effects of geographic location on the distribution of sediment AOA and AOB in 13 freshwater lakes on the Yunnan Plateau (China). The spatial dissimilarity in the abundance and structure of sediment AOA and AOB communities was observed in these plateau lakes. AOA abundance was usually less than AOB abundance, and the AOA/AOB ratio was positively correlated with water depth. Nitrososphaera-like AOA occurred in most of the studied lakes and were dominant in two lakes. Nitrosospira was the dominant AOB species in most of the lakes, while Nitrosomonas showed high abundance only in three lakes. In addition, geographic location was found to affect lake sediment AOB community structure.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chemolithotrophic oxidation of ammonia to nitrite, the first and rate-limiting step in nitrification process, has a critical significance to the global nitrogen cycle. Microbial ammonia oxidation is currently known to be performed by two groups of prokaryotes: ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB). Both of them harbor ammonia monooxygenase (amoA) gene, which has been widely used as a functional biomarker to investigate the distribution of ammonia-oxidizing microorganisms in a variety of natural ecosystems (Abell et al 2014; Han et al. 2013; Meyer et al. 2014; Wang and Gu 2013, 2014; Wang et al. 2014a). Both AOA and AOB can be key participators in ammonia biotransformation in natural environments (He et al. 2007; Liu et al. 2014a; Wang et al. 2014a). A few previous studies indicated that the abundance and community structure of AOA and AOB in lake sediments could be affected by trophy (Herrmann et al. 2009; Hou et al. 2013; Wu et al. 2010), pH (Herrmann et al. 2009), and sediment depth (Zhao et al. 2013). However, information on AOA and AOB communities in plateau freshwater lake sediment ecosystems is still very limited. So far, the inter-lake investigation on sediment AOA and AOB communities has been rarely addressed. The impact of geographic location on the distribution of lacustrine AOA and AOB communities remains unknown.
The Yunnan Plateau, located in the southwest of China, has about 40 freshwater lakes with an area of larger than 1 km2. The plateau is located in the subtropical or temperate climate zone. These plateau lakes are relatively isolated and distributed in different geographical regions (Liu et al. 2014b). However, the distribution of ammonia-oxidizing microorganisms in these lakes remains unclear. There has been no report available on the inter-lake distribution of ammonia-oxidizing microbial community on the Yunnan Plateau. Therefore, the objective of the present study was to investigate the effect of geographic location on the abundance and structure of sediment AOA and AOB communities in the lakes on the Yunnan Plateau.
Materials and methods
Study sites and sampling
Sediment cores were collected using a core sampler from the center of 13 freshwater lakes (99.2799–103.9863° E, 23.4325–28.6783° N) on the Yunnan Plateau. Bailonghai Lake, Puzhehei Lake, Changqiaohu Lake, Sanjiaohai Lake, Changhu Lake, and Qingshuihai Lake were located on the eastern Yunnan Plateau, while Jianhu Lake, Haixihai Lake, Cibihu Lake, Xihu Lake, Caohaishidi Lake, Lashihai Lake, and Tianchi Lake were located on the western Yunnan Plateau. These cores were sectioned at 5-cm intervals, and the surface sediment samples in triplicate (0–5 cm) were homogenized and subsampled for further analysis. The geographic and morphometric parameters of these freshwater lakes and the sediment properties were described in detail in the literature (Liu et al. 2014b).
Molecular analyses
Sediment DNA was extracted using the Powersoil DNA extraction kit (Mobio Laboratories, USA). The primer sets Arch-amoAF/Arch-amoAR and AmoA-1F/AmoA-2R were used for quantitative PCR (qPCR) assays of the archaeal and bacterial amoA genes, respectively, following the previously reported conditions (Wang et al. 2014a). A known copy number of linearized plasmid containing cloned amoA gene was used as a standard for archaeal or bacterial qPCR. The amplification efficiency and coefficient (r 2) for the archaeal and bacterial amoA genes were 94 % and 0.995 and 92 % and 0.997, respectively.
For construction of AOA and AOB clone libraries, the primer sets
Arch-amoAF/Arch-amoAR and AmoA-1F/AmoA-2R were also used (Wang et al. 2014a). Triplicate PCR products for each sediment sample were pooled and purified with a QIAquick PCR purification kit (Qiagen). The purified PCR products were cloned into pMD19-T vector (Takara Corp, Japan) following the manufacturer’s instructions. The clones containing the correct insert size were sequenced. Chimera-free sequences with similarity ≥ 97 % were grouped into the same operational taxonomic units (OTUs). OTU-based community Shannon diversity index was obtained using the MOTHUR program (Schloss et al. 2009). In order to perform AOA or AOB community comparisons, the phylogeny-based weighted UniFrac environmental clustering was also applied using the online UniFrac program (Lozupone et al. 2006). Phylogenetic analyses of archaeal and bacterial ammonia-oxidizing communities in all of the lake sediment samples were performed using MEGA software version 6.0 (Tamura et al. 2013). Pearson’s correlation analysis of the abundance and structure of AOA and AOB communities with lake water depth and sediment chemical properties was carried out using SPSS 20.0 software.
Nucleotide sequence accession number
The sequences obtained in the present study were deposited in the GenBank database under accession numbers KF898528–KF898872, KJ004926–KJ005122 for AOA, and KJ138225–KJ138266, KJ144199–KJ144248, KJ160448–KJ160493, and KJ640841–KJ641268 for AOB, respectively.
Results
Abundance of AOA and AOB
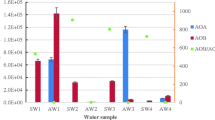
In this study, the sediment AOA and AOB abundances based on amoA genes were estimated using real-time PCR. Figure 1 illustrates a large variation in the densities of both AOA and AOB in the sediment samples from the 13 freshwater lakes on the Yunnan Plateau. The archaeal amoA gene copy numbers ranged from 1.35 × 103 to 2.21 × 105 copies per gram dry sediment. The largest AOA community size was found in Jianhu Lake (on the western Yunnan Plateau), followed by Qingshuihai Lake (on the eastern Yunnan Plateau) (1.12 × 105 copies per gram dry sediment). The other 11 plateau lakes had a much lower AOA community size (less than 2 × 104 copies per gram dry sediment). In addition, the bacterial amoA gene copy numbers ranged from 1.05 × 104 to 4.59 × 105 copies per gram dry sediment. Jianhu Lake also had the largest AOB community size. In contrast, the AOB abundance in the other plateau lakes was much lower (below 5 × 104 copies per gram dry sediment).
Abundance of archaeal and bacterial amoA genes in the different sediment samples. Samples BL, PZ, CQ, SJ, CH, QS, JH, HX, CB, XH, CS, LS, and TC represent the sediments collected from Bailonghai Lake, Puzhehei Lake, Changqiaohu Lake, Sanjiaohai Lake, Changhu Lake, Qingshuihai Lake, Jianhu Lake, Haixihai Lake, Cibihu Lake, Xihu Lake, Caohaishidi Lake, Lashihai Lake, and Tianchi Lake, respectively. Each replicate sediment sample was individually subjected to quantitative PCR assays. Error bars represent standard deviation of mean (n = 3)
Figure S1 shows a large variation in the AOA/AOB ratio in sediment samples from the 13 freshwater lakes on the Yunnan Plateau. The highest value of AOA/AOB ratio (3.23) was found in Qingshuihai Lake. However, in the other 12 lakes, the AOA/AOB ratio ranged between 0.03 and 0.78. Therefore, AOB outnumbered AOA in most of the studied lakes on the Yunnan Plateau. Moreover, Pearson’s correlation analysis indicates that the AOA/AOB ratio was positively correlated with lake water depth (P < 0.05) (Table 1). However, the AOA and AOB abundance or the AOA/AOB ratio did not show significant correlations with the measured lake sediment chemical parameters (P > 0.05).
Diversity of AOA and AOB
In this study, 542 archaeal and 566 bacterial amoA sequences were retrieved from the AOA and AOB clone libraries constructed with sediment samples from the 13 freshwater lakes on the Yunnan Plateau. Table 2 indicates that the AOA clone libraries were composed of 2– 20 OTUs, while the number of AOB OTUs in each sediment sample ranged between 1 and 25. A large variance in sediment AOA community diversity was found among different sediment samples, with the value of Shannon index of 0.11–2.59. On the western Yunnan Plateau, five lakes (Jianhu Lake, Haixihai Lake, Cibihu Lake, Lashihai Lake, and Tianchi Lake) had high AOA diversity (with Shannon diversity index value of 2.03–2.59), while the other two (Xihu Lake and Caohaishidi Lake) showed medium diversity (Shannon index = 0.94 or 1.14). On the eastern Yunnan Plateau, only two lakes (Sanjiaohai Lake and Bailonghai Lake) had high AOA Shannon diversity (2.05 or 1.61), while low AOA diversity was found in the other four lakes (0.36–0.85).
Table 2 also illustrates a marked difference in sediment AOB community diversity (Shannon index = 0–2.71). The lowest AOB diversity was found in the eastern plateau lakes (Changhu Lake and Changqiaohu Lake), while the highest was in the western plateau lakes (Jianhu Lake and Tianchi Lake). In addition, the Shannon index value of AOA was higher than that of AOB in most of sediment samples (8 out of 13). No significant correlation was found between amoA gene diversity and lake sediment chemical parameters or water depth (P > 0.05) (Table 1).
Comparison of AOA and AOB communities
Phylogeny-based weighted UniFrac clustering analysis was applied to identify the differences in amoA gene assemblages. Figure 2a shows a large difference in sediment archaeal amoA community structure. Except for the two samples from Puzhehei Lake and Qingshuihai Lake, samples from the eastern plateau lakes were distantly separated. Moreover, the samples from the western plateau lakes were also not grouped together. These results suggested that lake sediment AOA community structure could be much different even in the same geographic region.
Clustering of AOA (a) and AOB (b) clone libraries based on weighted UniFrac algorithm. Samples BL, PZ, CQ, SJ, CH, QS, JH, HX, CB, XH, CS, LS, and TC represent the sediments collected from Bailonghai Lake, Puzhehei Lake, Changqiaohu Lake, Sanjiaohai Lake, Changhu Lake, Qingshuihai Lake, Jianhu Lake, Haixihai Lake, Cibihu Lake, Xihu Lake, Caohaishidi Lake, Lashihai Lake and Tianchi Lake, respectively. The samples from the eastern plateau lakes are highlighted in red, while the samples from the western plateau lakes are in blue
The results of UniFrac clustering analysis of AOB clone libraries are illustrated in Fig. 2b. Except for the sample from Changhu Lake, the samples from the eastern plateau lakes were grouped together. Most of the samples (six out of seven) from the western plateau lakes were also clustered, but they were separated from most of the samples from the eastern plateau lakes. These results suggested that lake sediments in the same geographic region tended to have similar AOB community structure.
Phylogeny of AOA
In this study, the representative amoA gene sequences used for further phylogenetic analysis were selected only from the OTUs that had at least two members. Figure 3 shows that all of the AOA sequences of the 13 archaeal clone libraries could be grouped into four clusters. Cluster I contained 64 AOA sequences, including 33 from eastern plateau lakes (Bailonghai Lake, Sanjiaohai Lake, and Changqiaohu Lake ) and 31 from western plateau lakes (Jianhu Lake, Lashihai Lake, Tianchi Lake, Xihu Lake, Cibihu Lake, and Caohaishidi Lake). The archaeal amoA gene sequences from Sanjiaohai Lake and Xihu Lake were dominant in cluster I. The AOA sequences in this cluster were related to two cultivated soil AOA strains, Nitrososphaera sp. JG1 and EN76 (Kim et al. 2012; Tourna et al. 2011), and were also grouped with the uncultured ones from a variety of ecosystems such as wastewater bioreactors, river sediment, wetland sediment (Wang et al. 2011), estuary sediment (Zheng et al. 2013), agricultural soil, costal alkaline soil, and alpine meadow soil.
Phylogenetic tree of representative archaeal amoA sequences and reference sequences from GenBank. The obtained archaeal sequences beginning with BL, PZ, CQ, SJ, CH, QS, JH, HX, CB, XH, CS, LS, and TC were referred to the sequences retrieved from Bailonghai Lake, Puzhehei Lake, Changqiaohu Lake, Sanjiaohai Lake, Changhu Lake, Qingshuihai Lake, Jianhu Lake, Haixihai Lake, Cibihu Lake, Xihu Lake, Caohaishidi Lake, Lashihai Lake, and Tianchi Lake, respectively. The bold number in parentheses represents the numbers of the sequences in the same OTU in a given clone library. The amoA sequences from the eastern plateau lakes are highlighted in red, while the ones from the western plateau lakes are in blue. Numbers at the nodes indicate the levels of bootstrap support based on neighbor-joining analysis of 1000 resampled datasets. The bar represents 5 % sequence divergence
Cluster II was the second largest AOA group containing 125 archaeal amoA gene sequences. These AOA sequences were mainly retrieved from two eastern plateau lakes (Changhu Lake and Sanjiaohai Lake) and two western plateau lakes (Caohaishidi Lake Haixihai Lake). The AOA sequences in this cluster could be affiliated with those uncultured ones from meadow soil, grassland soil, agricultural soil (Glaser et al. 2010), and river sediment (Pereira E Silva et al. 2012).
Cluster III was the smallest AOA cluster and only had 28 members retrieved from four western plateau lakes (Lashihai Lake, Cibihu Lake, Jianhu Lake, and Tianchi Lake). The AOA sequences in cluster III were grouped with an uncultured one from Chaohu Lake. In contrast, cluster IV was the largest AOA cluster that had 269 archaeal amoA gene sequences. Most of the obtained archaeal amoA gene sequences from Bailonghai Lake, Puzhehei Lake, Changqiaohu Lake, and Qingshuihai Lake (on the eastern Yunnan Plateau) were distributed in this cluster. Moreover, the AOA sequences from the western plateau lakes (except Caohaishidi Lake) were also abundant in cluster IV. The members of cluster IV were related to the uncultured archaeal amoA gene sequences from river sediment (Liu et al. 2013), cave sediment, and lake water. Therefore, the results of phylogenetic analysis illustrated a large variation in the AOA community structure in the plateau lake sediment samples.
Phylogeny of AOB
Figure 4 shows that all of the AOB sequences of the 13 sediment bacterial amoA gene clone libraries could be grouped into five clusters. Cluster A was the largest AOB cluster and had 251 bacterial amoA gene sequences. A significant proportion of the obtained AOB sequences from Changhu Lake, Puzhehei Lake, Sanjiaohai Lake, Tianchi Lake, Caohaishidi Lake, Jianhu Lake, Lashihai Lake, and Xihu Lake were found in this cluster. The samples from Bailonghai Lake, Qingshuihai Lake, and Cibihu Lake had a much lower number of AOB sequences in cluster A. Moreover, the AOB sequences in this cluster were related to several of some cultivated Nitrosospira species (L115, En13, CT2F, APG3, PJA1, 9SS1, and TCH711) (Mintie et al. 2003; Purkhold et al. 2003). They were also grouped with the uncultured ones from various ecosystems such as meadow soil, protected land soil, grassland soil, plateau soil, agricultural soil (Wang et al. 2014b), lake water, prawn culture pond sediment, river sediment, wetland sediment, reservoir sediment (Wang et al. 2014a), and wastewater bioreactor.
Phylogenetic tree of representative bacterial amoA sequences and reference sequences from GenBank. The obtained bacterial sequences beginning with BL, PZ, CQ, SJ, CH, QS, JH, HX, CB, XH, CS, LS, and TC were referred to the sequences retrieved from Bailonghai Lake, Puzhehei Lake, Changqiaohu Lake, Sanjiaohai Lake, Changhu Lake, Qingshuihai Lake, Jianhu Lake, Haixihai Lake, Cibihu Lake, Xihu Lake, Caohaishidi Lake, Lashihai Lake, and Tianchi Lake, respectively. The bold number in parentheses represent the numbers of the sequences in the same OTU in a given clone library. The amoA sequences from the eastern plateau lakes are highlighted in red, while the ones from the western plateau lakes are in blue. Numbers at the nodes indicate the levels of bootstrap support based on neighbor-joining analysis of 1000 resampled datasets. The bar represents 2 % sequence divergence
Cluster B was the second largest AOB cluster and contained 110 bacterial amoA gene sequences. Most of the obtained AOB sequences from Bailonghai Lake, Qingshuihai Lake, and Haixihai Lake existed in this cluster. Several bacterial amoA gene sequences from Puzhehei Lake were also found. However, no AOB sequence from the other plateau lakes was detected in this AOB cluster. Moreover, the sequences in cluster B could be affiliated to the uncultured AOB sequences from river water, freshwater mining operation retention pond (Miazga-Rodriguez et al. 2012), and eutrophic urban lake sediment. In this study, cluster C was the smallest AOB group, composed of 12 bacterial amoA gene sequences retrieved from Cibihu Lake. They were closely related to an uncultured sequence from Yangtze Estuary sediment (Zheng et al. 2014). Cluster D was a 71-member group. This cluster mainly consisted of the AOB sequences from Changqiaohu Lake and Sanjiaohai Lake. Almost all of the bacterial sequences from Changqiaohu Lake were found in this AOB cluster. The member in cluster D could be grouped with an uncultured sequence from nitrogen-rich lake sediment (Wang et al. 2012).
Cluster E contained a total of 78 bacterial amoA gene sequences, including seven from two eastern plateau lakes and 71 from six western plateau lakes. This cluster was mainly composed of the AOB sequences from Cibihu Lake, Lashihai Lake, and Tianchi Lake. In addition, more than half of the total AOB sequences from Cibihu Lake were distributed in this cluster. The AOB sequences in this cluster were related to two cultivated Nitrosomonas species (NL7 and Nm84) (Park and Noguera 2007; Purkhold et al. 2003). They could be also grouped with the uncultured bacterial amoA gene sequences from eutrophic lake sediment and wastewater treatment bioreactor. Moreover, the results of phylogenetic analysis confirmed a large variation in the AOB community structure in the plateau lake sediment samples.
Discussion
AOA and AOB abundance in sediment
So far, information about the abundance of ammonia-oxidizing microorganisms in lake sediment is still very limited. In this study, for the sediments in the 13 lakes on the Yunnan Plateau, the archaeal and bacterial amoA gene copy numbers ranged from 1.35 × 103 to 2.21 × 105 copies per gram dry sediment and 1.05 × 104 to 4.59 × 105 copies per gram dry sediment, respectively. The sediment AOA and AOB abundance in the plateau lakes was usually relatively low compared to the other previously reported lakes (Hou et al. 2013; Wu et al. 2010; Yang et al. 2013; Zhao et al. 2013, 2014). Moreover, AOB was found to outnumber AOA in most of the studied lakes (12 out of 13) on the Yunnan Plateau. This result coincided with that of a previous study on sediment of high-altitude Qinghai Lake (Jiang et al. 2009) but was not in agreement with those of a number of previous studies on lake sediment (Herrmann et al. 2009; Hou et al. 2013; Wu et al. 2010; Zhao et al. 2013, 2014). Our recent study also found that the dominance of AOB over AOA usually occurred in sediment of Luoshijiang Wetland on the Yunnan Plateau (Liu et al. 2014a). Therefore, it seems that high elevation might favor the dominance of AOB over AOA in aquatic ecosystems. AOB might play a more important role than AOA in the nitrification process in high-altitude lake sediment.
AOA and AOB community diversity in sediment
Zhao et al. (2013) revealed that AOA community diversity was much lower than AOB diversity in sediments of Taihu Lake, while Zhao et al. (2014) found that, in Taihu Lake, AOA diversity was higher than AOB diversity in unvegetated sediment and rhizosphere sediment of Potamogeton crispus, but lower in rhizosphere sediments of Ceratophyllum demersum and Vallisneria spinulosa. In this study, AOA diversity was higher than AOB diversity in sediments of most of the plateau lakes (8 out of 13). In contrast, our previous study indicated that AOB community diversity was usually higher than AOA diversity in sediment of Luoshijiang Wetland on the Yunnan Plateau (Liu et al. 2014a). Therefore, sediment type could affect the difference between sediment AOA and AOB community diversity.
AOA and AOB community structures in sediment
AOA can adapt to a variety of habitats, and the AOA species found in one type of habitat can occur in other types of habitats (Wang and Gu 2013). In this study, the obtained archaeal amoA gene sequences could be related to those from various ecosystems such as river, lake, wetland and estuary sediments, grassland and agricultural soils, and wastewater bioreactors. Although Nitrososphaera strains have been only isolated from soil and hot spring (Hatzenpichler et al. 2008; Tourna et al. 2011), the Nitrososphaera-like microorganisms can usually exist in wetland sediment ecosystems (Liu et al. 2014a). Yang et al. (2013) found that the Nitrososphaera-like microorganisms were the major AOA components in sediments of the lakes on the northeastern Qinghai-Tibetan Plateau. However, Nitrososphaera-like microorganisms were in a low abundance in sediment AOA communities in Danish freshwater lakes (Herrmann et al. 2009) and Taihu Lake (Zhao et al. 2014). In this study, the Nitrososphaera-like sequences were detected in most of the lakes (9 out of 13) on the Yunnan Plateau but were only dominant in Sanjiaohai Lake and Xihu Lake. Therefore, Nitrososphaera might make different contributions to ammonia oxidation in sediments of various plateau lakes. In most of the plateau lakes, the AOA species not related to cultivated strains could play a more important role in nitrification than Nitrososphaera-related AOA.
Nitrosospira and Nitrosomonas are two commonly found AOB groups in freshwater ecosystems (Zhao et al. 2014). The compositions of sediment AOB communities in Taihu Lake have been extensively investigated. Wu et al. (2010) and Zhao et al. (2013) indicated that Nitrosospira was the predominant or even sole AOB. Zhao et al. (2014) revealed that Nitrosospira-like microorganisms were the sole AOB in unvegetated sediment and rhizosphere sediment of P. crispus, while Nitrosomonas-like microorganisms predominated in rhizosphere sediments of C. demersum and V. spinulosa. Hou et al. (2013) showed that both Nitrosomonas- and Nitrosospira-affiliated AOB dominated in sediments of Taihu Lake and Chaohu Lake. To date, information about the compositions of sediment AOB communities in the other lakes is still very limited. Jiang et al. (2009) found that the sediment AOB community in Qinghai Lake mainly consisted of both Nitrosomonas- and Nitrosospira-like microorganisms. Nitrosospira-related AOB was dominant in sediments of Danish freshwater lakes, with a much lower abundance of Nitrosomonas-related AOB (Herrmann et al. 2009). Therefore, the proportion of sediment Nitrosomonas- and Nitrosospira-affiliated AOB could vary in lakes and sampling sites. In this study, Nitrosospira-like microorganisms were the major or even predominant sediment AOB in most of the lakes (8 out of 13), suggesting their important contribution to ammonia oxidation in lakes on the Yunnan Plateau. Our recent study showed that Nitrosospira-like sequences also predominated in the wetland sediment AOB communities on the Yunnan Plateau (Liu et al. 2014a). However, high abundance of Nitrosomonas-like AOB was also found in three western plateau lakes (Cibihu Lake, Lashihai Lake, and Tianchi Lake). Moreover, the microorganisms related to uncultivated AOB species were dominant in four eastern plateau lakes (Bailonghai Lake, Qingshuihai Lake, Changqiaohu Lake, and Sanjiaohai Lake) and in two western plateau lakes (Haixihai Lake and Cibihu Lake). These results indicate that distinct AOB groups might have different functional significances in various plateau lakes. In addition, the obtained AOB sequences in this study were related to the microorganisms from a variety of soil and sediment ecosystems and wastewater bioreactors. This implied that AOB species of different evolutionary origins could survive in high-altitude lake sediment.
Regulating factors for AOA and AOB in sediment
Soil AOA and AOB are known to be influenced mainly by pH, temperature, and ammonium, while ammonium and oxygen concentrations play major roles in marine systems (Vissers et al. 2013). However, there is a paucity of information on ammonia-oxidizing microorganisms in lake sediment. Herrmann et al. (2009) indicated that AOA and AOB diversity increased from oligotrophic to mesotrophic sites in sediments of Danish lakes. They also found that the number of OTUs was positively correlated to ammonia availability and pH but negatively to sediment C/N ratios. Wu et al. (2010) reported that organic substances significantly influenced the sediment AOA abundance in Taihu Lake. Hou et al. (2013) indicated that the relative abundance of AOB was positively correlated to sediment ammonia and nitrite concentrations in Taihu Lake and Chaohu Lake, while the relative abundance of AOA showed negative correlation to nitrate level. They also revealed that the AOB diversity had a positive relationship with ammonia and organic matter. However, in this study, no significant correlation was found between the abundance and diversity of AOA and AOB and the determined sediment chemical parameters in lakes on the Yunnan Plateau. No significant correlation was also found between AOA abundance and composition and the measured sediment environmental variables in lakes on the northeastern Qinghai-Tibetan Plateau (Yang et al. 2013).
Auguet et al. (2012) found that water depth was a key factor regulating AOA community composition in water columns in a deep-glacial lake located in central Spanish Pyrenees. Vissers et al. (2013) suggested that the AOA/AOB ratio in water column varied with water depth in a perialpine lake located in Central Switzerland. In this study, the AOA/AOB ratio in lake sediment was found to be positively correlated with water depth.
The impact of geographic location on the distribution of aquatic microbial community remains controversial. Logares et al. (2013) showed a weak correlation between bacterioplankton community composition and geographical distance between lakes, while Xiong et al. (2012) revealed that geographic distance was a driving force for bacterial distribution in lake sediments across the Tibetan Plateau. In this study, the spatial dissimilarity in the abundance and structure of AOA and AOB communities was found among high-altitude freshwater lakes. Moreover, UniFrac clustering indicated that eastern and western plateau lakes generally had much different sediment AOB community structures. These results illustrated that geographic location might be a key factor influencing the inter-lake distribution of sediment AOB community on the Yunnan Plateau.
In conclusion, spatial dissimilarity in the abundance and structure of AOA and AOB communities occurred in 13 freshwater lakes on the Yunnan Plateau. AOB usually outnumber AOA, while AOA usually had higher community diversity than AOB. The AOA/AOB ratio was positively correlated with lake water depth. Geographic location affected lake sediment AOB community structure.
References
Abell GCJ, Ross DJ, Keane J, Holmes BH, Robert SS, Keough MJ, Eyre BD, Volkman JK (2014) Niche differentiation of ammonia-oxidising archaea (AOA) and bacteria (AOB) in response to paper and pulp mill effluent. Microb Ecol 67:758–768
Auguet JC, Triado-Margarit X, Nomokonova N, Camarero L, Casamayor EO (2012) Vertical segregation and phylogenetic characterization of ammonia-oxidizing archaea in a deep oligotrophic lake. ISME J 6:1786–1797
Glaser K, Hackl E, Inselsbacher E, Strauss J, Wanek W, Zechmeister-Boltenstern S, Sessitsch A (2010) Dynamics of ammonia-oxidizing communities in barley-planted bulk soil and rhizosphere following nitrate and ammonium fertilizer amendment. FEMS Microbiol Ecol 74:575–591
Han MQ, Li ZY, Zhang FL (2013) The ammonia oxidizing and denitrifying prokaryotes associated with sponges from different sea areas. Microb Ecol 66:427–436
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139
He J, Shen J, Zhang L, Zhu Y, Zheng Y, Xu M, Di HJ (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374
Herrmann M, Saunders AM, Schramm A (2009) Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl Environ Microbiol 75:3127–3136
Hou J, Song CL, Cao XY, Zhou YY (2013) Shifts between ammonia oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu). Water Res 47:2285–2296
Jiang HC, Dong HL, Yu BS, Lv G, Deng SC, Berzins N, Dai MH (2009) Diversity and abundance of ammonia-oxidizing archaea and bacteria in Qinghai Lake, Northwestern China. Geomicrobiol J 26:199–211
Kim JG, Jung MY, Park SJ, Rijpstra WIC, Damste JSS, Madsen EL, Min D, Kim JS, Kim GJ, Rhee SK (2012) Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I1b from an agricultural soil. Environ Microbiol 14:1528–1543
Liu S, Shen L, Lou L, Tian G, Zheng P, Hu B (2013) Spatial distribution and factors shaping the niche segregation of ammonia-oxidizing microorganisms in the Qiantang River, China. Appl Environ Microbiol 79:4065–4071
Liu Y, Zhang JX, Zhang XL, Xie SG (2014a) Depth-related changes of sediment ammonia-oxidizing microorganisms in a high-altitude freshwater wetland. Appl Microbiol Biotechnol 98:5697–5707
Liu Y, Zhang JX, Zhao L, Li YZ, Yang YY, Xie SG (2014b) Aerobic and nitrite-dependent methane-oxidizing microorganisms in sediments of freshwater lakes on the Yunnan Plateau. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-6141-5
Logares R, Lindstrom ES, Langenheder S, Logue JB, Paterson H, Laybourn-Parry J, Rengefors K, Tranvik L, Bertilsson S (2013) Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J 7:937–948
Lozupone C, Hamady M, Knight R (2006) UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinforma 7:371
Meyer A, Focks A, Radl V, Welzl G, Schoning I, Schloter M (2014) Influence of land use intensity on the diversity of ammonia oxidizing bacteria and archaea in soils from grassland ecosystems. Microb Ecol 67:161–166
Miazga-Rodriguez M, Han S, Yakiwchuk B, Wei K, English C, Bourn S, Bohnert S, Stein LY (2012) Enhancing nitrification at low temperature with zeolite in a mining operations retention pond. Front Microbiol 3:271
Mintie AT, Heichen RS, Cromack K, Myrold DD, Bottomley PJ (2003) Ammonia-oxidizing bacteria along meadow-to-forest transects in the Oregon Cascade Mountains. Appl Environ Microbiol 69:3129–3136
Park HD, Noguera DR (2007) Characterization of two ammonia-oxidizing bacteria isolated from reactors operated with low dissolved oxygen concentrations. J Appl Microbiol 102:1401–1417
Pereira E Silva E, Silva MC, Poly F, Guillaumaud N, van Elsas JD, Salles JF (2012) Fluctuations in ammonia oxidizing communities across agricultural soils are driven by soil structure and pH. Front Microbiol 3:77
Purkhold U, Wagner M, Timmermann G, Pommerening-Roser A, Koops HP (2003) 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the Nitrosomonads. Int J Syst Evol Microbiol 53:1485–1494
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425
Vissers EW, Anselmetti FS, Bodelier PLE, Muyzer G, Schleper C, Tourna M, Laanbroek HJ (2013) Temporal and spatial coexistence of archaeal and bacterial amoA genes and gene transcripts in Lake Lucerne. Archaea 2013:289478
Wang SY, Wang Y, Feng XJ, Zhai LM, Zhu GB (2011) Quantitative analyses of ammonia-oxidizing Archaea and bacteria in the sediments of four nitrogen-rich wetlands in China. Appl Microbiol Biotechnol 90:779–787
Wang Y, Zhu G, Ye L, Feng X, Op den Camp HJ, Yin C (2012) Spatial distribution of archaeal and bacterial ammonia oxidizers in the littoral buffer zone of a nitrogen-rich lake. J Environ Sci 24:790–799
Wang YF, Gu JD (2013) Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl Microbiol Biotechnol 97:7015–7033
Wang YF, Gu JD (2014) Effects of allylthiourea, salinity, and pH on ammonia/ammonium-oxidizing prokaryotes in mangrove sediment incubated in laboratory microcosm. Appl Microbiol Biotechnol 98:3257–3274
Wang XY, Wang C, Bao LL, Xie SG (2014a) Abundance and community structure of ammonia-oxidizing microorganisms in reservoir sediment and adjacent soils. Appl Microbiol Biotechnol 98:1883–1892
Wang J, Dong H, Wang W, Gu JD (2014b) Reverse-transcriptional gene expression of anammox and ammonia-oxidizing archaea and bacteria in soybean and rice paddy soils of northeast China. Appl Microbiol Biotechnol 98:2675–2686
Wu YC, Xiang Y, Wang JJ, Zhong JC, He JZ, Wu QLL (2010) Heterogeneity of archaeal and bacterial ammonia oxidizing communities in Lake Taihu, China. Environ Microbiol Rep 2:569–576
Xiong JB, Liu YQ, Lin XG, Zhang HY, Zeng J, Hou JZ, Yang YP, Yao TD, Knight R, Chu HY (2012) Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbiol 14:SI 2457–2466
Yang J, Jiang HC, Dong HL, Wang HY, Wu G, Hou WG, Liu WG, Zhang CL, Sun YJ, Lai ZP (2013) amoA-encoding archaea and thaumarchaeol in the lakes on the northeastern Qinghai-Tibetan Plateau, China. Front Microbiol 4:329
Zhao DY, Zeng J, Wan WH, Liang HD, Huang R, Wu QLL (2013) Vertical distribution of ammonia-oxidizing archaea and bacteria in sediments of a eutrophic lake. Curr Microbiol 67:327–332
Zhao DY, Luo J, Zeng J, Wang M, Yan WM, Huang R, Wu QLL (2014) Effects of submerged macrophytes on the abundance and community composition of ammonia-oxidizing prokaryotes in a eutrophic lake. Environ Sci Pollut Res 21:389–398
Zheng YL, Hou LJ, Liu M, Lu M, Zhao H, Yin GY, Zhou JL (2013) Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments. Appl Microbiol Biotechnol 97:8351–8363
Zheng Y, Hou L, Newell S, Liu M, Zhou J, Zhao H, You L, Cheng X (2014) Community dynamics and activity of ammonia-oxidizing prokaryotes in intertidal sediments of the Yangtze Estuary. Appl Environ Microbiol 80:408–419
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 41222002 and No. 51279001) and special fund of State Key Joint Laboratory of Environment Simulation and Pollution Control (14Y02ESPCP).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 182 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Zhang, J., Zhao, L. et al. Distribution of sediment ammonia-oxidizing microorganisms in plateau freshwater lakes. Appl Microbiol Biotechnol 99, 4435–4444 (2015). https://doi.org/10.1007/s00253-014-6341-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6341-z