Abstract
An annual investigation into the abundance of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in fresh water aquaculture ponds was performed by quantitative PCR of the amoA gene. The results showed that AOB were the main ammonia-oxidizing microorganisms in water, and significantly higher copy numbers of the AOB amoA gene were observed in the summer (Aug 2012), while no significant differences were detected among the other three seasons. AOA showed low abundances throughout the year. The predominance of AOB in aquaculture water was suggested to be related to photoinhibition. Both the AOB and AOA amoA genes in aquaculture pond sediments showed typical seasonal patterns. The maximum density of AOB was observed in the autumn (Nov 2012) and winter (Jan 2013), while the maximum density of AOA was observed in winter. The minimum densities of both AOA and AOB occurred in the summer. The concentration of the AOA amoA gene was higher than that of the AOB amoA gene in sediments by almost one order of magnitude, which indicates that AOA are the dominant ammonia-oxidizing microorganisms in the aquaculture pond sediments. Dissolved oxygen is suggested to be the key factor determining the predominance of AOA in pond sediments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Aquaculture production has been increasing at an average growth rate of 8.3 % per year, which makes aquaculture one of the fastest growing segments of the food economy in modern times (FAO 2012). According to the Fishery Bureau of the Chinese Ministry of Agriculture (MOA), the total aquaculture production of China in 2012 amounted to 59.1 million metric tons, and the production of freshwater aquaculture was 26.4 million metric tons (Fishery National Bureau of Statistics of China 2013, http://www.stats.gov.cn/english/statisticaldata/Quarterlydata/). Undoubtedly, China’s freshwater aquaculture plays an important role in the global supply of aquatic products.
Currently, there are 2,566,900 ha freshwater aquaculture ponds, and pond culture is the major freshwater culture method in China (National Bureau of Statistics of China 2013, http://www.stats.gov.cn/english/statisticaldata/Quarterlydata/). However, along with increasing development, concerns are evoked about the possible effects of ever-increasing aquaculture waste, both on the productivity of aquaculture systems and on the ambient aquatic ecosystem. It was estimated that 9.5 kg phosphorous (P) and 78 kg of nitrogen (N) are released into the aquaculture environment per ton of fish produced. Approximately 72 % of N and 70 % of P in the feed are released into the aquaculture environment (Ackefors and Enell 1994). Ammonia is one of the most toxic substances produced by intensive fish farming (Handy and Poxton 1993). Ammonia is toxic to all vertebrates and can cause convulsions, coma and death, probably because elevated ammonium (NH4 +) ions displace potassium (K+) and depolarizes neurons, causing activation of the N-methyl-d-aspartate (NMDA) type glutamate receptor, which leads to an influx of excessive calcium (Ca2+) ions and subsequent cell death in the central nervous system (Randall and Tsui 2002).
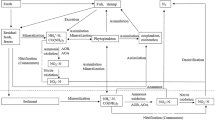
Nitrification is a key process in the cycling of nitrogen in aquatic ecosystems (Merbt et al. 2012). The oxidation of ammonia (NH3) to nitrite (NO2 −)—the first and rate-limiting step of nitrification—has traditionally been considered to be carried out solely by ammonia-oxidizing bacteria (AOB) (Koops and Pommerening-Röser 2001), which fall into two phylogenetic lineages within the β- and γ-Proteobacteria (Kowalchuk and Stephen 2001). However, this notion has changed owing to the recent discovery of ammonia-oxidizing archaea (AOA). Based on genomic level comparisons, AOA were classified into the newly proposed phylum Thaumarchaeota (Pester et al. 2011). AOA play important roles in the nitrogen cycle and have been found in various habitats, including hot springs (Hatzenpichler et al. 2008), oceans (Wuchter et al. 2006; Horak et al. 2013), freshwater (Auguet et al. 2011), and soil (Leininger et al. 2006). Although AOB and AOA are affiliated with different phylogenetic domains, they both contain homologous ammonia monooxygenase (AMO), which is responsible for catalyzing the first step in ammonia oxidation. The amoA gene, encoding the alpha subunit of AMO, has been used widely as a functional gene marker for tracking ammonia oxidizers in environmental samples. Nonetheless, there is a great lack of studies on AOA and AOB in aquaculture ponds. In this paper, we report on the seasonal dynamics of the abundance of crenarchaeotal and betaproteobacterial amoA genes, and investigated changes in temperature and concentration of ammonium (NH4 +), nitrite (NO2 −) and nitrate (NO3 −) in aquaculture water and sediment, aiming to achieve a better understanding of the nitrogen cycle in the aquaculture environment.
Materials and methods
Sample collection and hydration index determination
Experimental samples were collected from an aquaculture farm [29°55′ N, 112°(17–18)′ E], Gong’an, Hubei, China. Ten aquaculture ponds were selected for investigation. The average depth of the sampling ponds (350 m × 20 m) was about 1.5 m, in which grass carp, silver carp, bighead carp and soft-shelled turtle were raised for commercial use since the 1980s. The ponds have never been dredged and are typical of a eutrophic environment. The samples were collected on 14 April, 22 August, 2 November 2012, and 15 January 2013. The water samples were collected with a sterile glass bottle about 30 cm below the water surface. The top 0–5 cm of black color sediment samples were collected using a Peterson Grab Sampler in the center of the aquaculture ponds, and the homogenized sediment was transferred immediately to a sterile plastic bag on the fishing boat. The water and sediment samples were stored at 4 °C and treated within 48 h. The sediment was centrifuged at 7,000 g for 10 min to extract bulk pore water. The pore water was then filtered through a 0.22-μm membrane. Field measurements of NH4 +-N, NO3 −-N and NO2 −-N concentrations were performed by the Nessler reagent photometric method, photometry with phenol-2-sulfonic acid method, and the N (1-naphty1)-ethylenediamine dihydrochloride spectrophotometric method, respectively, according to the protocols described in Chinese Standard Methods (Editorial Board of Monitoring and Analytical Method of Water and Wastewater 2002).
DNA extraction from fresh water and sediments
DNA from water- and sediment-dwelling microorganisms was extracted as described by Lu et al. (2012) with some modifications. Water (50 mL) from each pond was filtered directly through a 0.22-μm membrane using negative pressure. After filtration, the membranes were cut into pieces and placed in a 2-mL sterile centrifuge tube and stored at −20 °C until DNA extraction. The pieces of filtered membranes or 0.3 g sediment were lysed using 0.7 μL lysis buffer (Zhou et al. 1996), 100 μL 10 % SDS, and two glass beads. The lysate was homogenized for 2 min on a vortexer, and then incubated at 70 °C for 1 h. After incubation, the tube was centrifuged at 13,000 g for 10 min and 0.7 mL supernatant was transferred into a new tube. Then, 0.6 mL phenol:chloroform:isoamyl alcohol (25:24:1) was added and shaken gently for 1 min, and the tube was centrifuged at 13,000 g for 5 min. After centrifugation, the supernatant was transferred to a new tube, followed by the addition of a 0.5 volume of PEG 6000 solution (50 % in m/v) and incubated for 60 min at ambient temperature. The tube was then centrifuged at 13,000 g for 15 min, the supernatant was discarded, and the pellet was re-dissolved in 0.6 mL distilled water before being extracted with 0.4 mL chloroform by several gentle end-over-end inversions. After extraction, the tube was centrifuged at 12,000 g for 5 min and 0.45 mL of the supernatant was transferred to a new tube and mixed with 0.9 mL ethanol and 45 μL NaCl solution (5 M) and precipitated for 15 min on ice. After precipitation, the tube was centrifuged at 13,000 g for 15 min, and the pellet was washed twice with 70 % ethanol and air-dried. The final pellet was dissolved in 50 μL TE buffer. DNA concentrations were quantified using a ND-2000 UV-vis spectrophotometer (http://www.nanodrop.com).
qPCR analysis of bacterial and archaeal amoA
Quantitative PCR (qPCR) was used to estimate the abundance of bacterial and archaeal amoA genes. qPCR was performed using a Qiagen Q thermocycler (Qiagen, Hilden, Germany) and primers amoA-1F/amoA-2R (specific for the AOB amoA gene) (Rotthauwe et al. 1997) and CrenamoA23f/CrenamoA616r (specific for the AOA amoA gene) (Tourna et al. 2008). The qPCR was conducted in a total volume of 20 μL containing 10 μL SYBR Premix Ex Taq II for AOB or SYBR Premix Ex Taq for AOA (Takara, Dalian, China), 1 μL DNA template, 0.2 μM of each primer and 0.2 mg mL−1 bovine serum albumin (BSA). The qPCR thermocycling steps were as follows: an initial denaturation of 95 °C for 30 s, followed by 35 cycles of 95 °C for 5 s, 54 °C for 35 s or 53 °C for 60 s, and 72 °C for 60 s. Plates were read at 81 °C for AOB or 80 °C for AOA, respectively. The negative control without DNA template was subjected to the same procedures to exclude or detect any possible contamination. After qPCR, the specificity of amplification was verified by melting curve analysis and agarose gel electrophoresis. All measurements were performed in triplicate.
Standard curves for qPCR were developed as described previously with some modifications (He et al. 2007). The bacterial or archaeal amoA gene was PCR-amplified from the extracted DNA using primers amoA-1F/amoA-2R or CrenamoA23f/CrenamoA616r, and the PCR amplification products were purified and cloned into the pMD18-T Easy Vector (Takara, Dalian, China). Plasmid extracted from the correct clones was used as a standard for quantitative analysis. The plasmid DNA concentration was determined using a ND-2000 UV-vis Spectrophotometer and the copy numbers of the target genes were calculated directly from the concentration of the extracted plasmid DNA. Ten-fold serial dilutions of plasmid DNA ranging from 2.0 × 102 to 2.0 × 107 copies of the AOA amoA gene or 2.0 × 101 to 2.0 × 106 copies of the AOB amoA gene were subjected to qPCR in triplicate to generate an external standard curve.
Two controls were performed to estimate the possible inhibition of qPCR performance by the co-extracted polyphenolic compounds or humic acids in sediment or water extracts: (1) different amounts of amoA standards were mixed with equal amounts of sediment or water DNA extract, and (2) two different volumes of DNA extract from each sediment or water sample used in this study were applied for the quantification of amoA copy number. In most cases, inhibition was not detected or was negligible.
Results and discussion
Seasonal changes and abundance of amoA gene in ammonia-oxidizing microorganisms in aquaculture water
Figure 1a–e shows the seasonal changes in temperature, NH4 +, NO2 − and NO3 − concentrations and AOB amoA gene copy number in aquaculture water in April, August, November 2012, and January 2013, respectively. The AOB content in water showed a typical seasonal pattern: the highest copy number of the AOB amoA gene, 8.56 ± 11.3 × 104 copy mL−1, was observed in summer (August 2012), and was significantly higher than in other seasons (1.55 ± 8.41 × 102 in spring, 1.04 ± 2.28 × 103 copy mL−1 in autumn and 3.83 ± 2.24 × 102 copy mL−1 in winter) (Fig. 1e; P < 0.01, one-way ANOVA). No significant differences were detected between the other three seasons. AOB amoA gene copy numbers showed significant positive correlation with temperature (r = 0.597, P < 0.01; nonparametric correlations). These results indicate that temperature plays a more important role in the seasonal dynamics of AOB than aquaculture activity, including the application of probiotics and fish medicine and aeration, etc. A similar phenomenon was observed in a constructed wetland, where the amoA gene copy numbers of AOB ranged from 5.3 ± 0.6 × 104 to 8.1 ± 0.5 × 106 copy mL−1 in water samples in summer, while AOB was not detected at the outlet of the first wetland treatment unit in the winter season (Sims et al. 2012a).
a Water temperatures in April, August, November 2012 and January 2013. b–e Results are means ± standard deviations of the NH4 +, NO2 −, NO3 − and AOB amoA gene concentrations in the overlying water of ten aquaculture ponds in April, August, November 2012 and Jan 2013, respectively. Identical letters above the bars indicate means with no statistically significant differences. Different letters indicate statistically significant differences at the 95 % confidence level
In the aquaculture water, the concentration of NH4 + correlated roughly with the copy numbers of the AOB amoA gene throughout the year (r = 0.251, P < 0.01), which indicates that other routes of NH4 + removal in aquaculture water, such as absorption by phytoplankton, also play a very important role in addition to the oxidation by AOB. No significant correlation was observed between the copy numbers of the AOB amoA gene and the concentrations of NO2 − and NO3 − in water, which was probably due to the fact that NO2 − and NO3 − levels are not only influenced by the amount and activity of nitrifying bacteria, but are also determined by denitrification intensity.
The AOA amoA gene was detected by PCR throughout the year in the aquaculture water based on our research over 2 years. However, because the copy number of the AOA amoA gene was below the minimum level of detection (20 copy mL−1 water) in the freshwater column, the exact copy number of the AOA amoA gene could not be determined. The availability of NH4 + does not appear to be the limiting ecological factor for this phenomenon. It was shown that enriched AOA cultures could grow well when the NH4 + concentration ranged from 0.22 to 70 mg mL−1 (French et al. 2012). In this study, the NH4 + concentrations of aquaculture water ranged from 1.02 ± 0.86 mg L−1 to 2.80 ± 0.89 mg L−1 during the year, which falls within the aforementioned range of NH4 + concentrations.
The fact that AOB were the predominant ammonia-oxidizer in the surface water could be explained by photoinhibition because the sampling point was at a depth of only 30 cm. A similar study, in which water samples were collected from 50 cm below the surface, found that AOB were the only ammonia-oxidizer in surface water in freshwater of Lake Taihu, which is hypertrophic and has a mean depth of 1.89 m (Ye et al. 2009). Another study found a greater abundance of amoA genes at lower light intensities in the ocean (Church et al. 2010). Merbt et al. (2012) showed that both AOA and AOB could be completely inhibited by continuous illumination at high intensity (500 μmol photons m−2 s−1) in the laboratory. However, at lower light intensities, archaeal growth was much more photosensitive than bacterial growth, with greater inhibition occurring at 60 μmol photons m−2 s−1 than at 15 μmol photons m−2 s−1, whereas bacteria were unaffected by these intensities. Archaeal ammonia oxidizers also were more sensitive to cycles of 8-h light/16-h darkness at two light intensities (60 and 15 μmol photons m−2 s−1) and, unlike bacterial strains, showed no evidence of recovery during dark phases (Merbt et al. 2012). In addition, French et al. (2012) showed that white light (30 μmol photons m−2 s−1) strongly inhibited the growth of AOA, but had no effect on the growth of AOB. Furthermore, it was shown that variations in light intensity ranged from 50 to 2,000 μmol photons m−2 s−1 between 07:00 and 19:00 hours on sunny days during the summer at Lake Taihu (30°56′–31°34′ N, 119°54′–120°36′ E) (Wang et al. 2011), which is located at the same latitude as the aquaculture farm investigated in our study. Therefore, AOA in the surface water of aquaculture ponds was probably inhibited by light.
Seasonal changes and abundance of the amoA gene of ammonia-oxidizing microorganisms in aquaculture pond sediments
Figure 2a–c shows the seasonal changes of NH4 +, NO2 − and NO3 − concentrations in sediment pore water in April, August and November 2012, and January 2013, respectively. The concentrations of the AOB amoA gene in the aquaculture pond sediments ranged from 4.05 ± 3.83 × 104 to 3.11 ± 1.65 × 105 copy g−1 (Fig. 2d). In sediments, the maximum densities of the AOB amoA gene were observed in autumn (November 2012) and winter (January 2013). There was no significant difference in the density of the AOB amoA gene between autumn and winter (P = 0.432, one-way ANOVA). The lowest density was observed in summer, which was in contrast with the high densities observed in aquaculture water during this season. Similar seasonal changes have been observed in estuarine and eelgrass zone studies (Bernhard et al. 2007; Ando et al. 2009). The low copy number of the AOB amoA gene in summer could be attributed to the decrease in the availability of NH4 + in the eelgrass zone studies (Ando et al. 2009). Based on AOB growth experiments, increasing NH4 + concentrations could increase the growth rates and shorten the lag phases of AOB. However, high concentrations of NH4 + pore water were observed during the summer in both the estuarine study (Bernhard et al. 2007) and this study (Fig. 2a). Obviously, NH4 + concentration is not a key factor for the growth of AOB in pond sediment. It is possible that AOB experience a greater oxygen limitation in summer when sediment oxygen demand peaks. Oxygen dynamics in aquaculture ponds differ substantially from those in natural aquatic systems, since pond environments are smaller in area and shallower in depth, have limited water circulation, and are subject to large depositions of feeding debris. The hypolimnion dissolved oxygen concentration was rarely greater than 2 mg L−1 in summer, although the super-saturation of oxygen usually occurs during the daylight period (Chang and Ouyang 1988). Therefore it can be speculated that the pond sediment is an extremely oxygen-deficient environment.
a–e Results are means ± standard deviations of the NH4 +, NO2 −, NO3 −, AOB and AOA amoA gene concentrations in the sediment of ten aquaculture ponds in April, August, November 2012 and January 2013, respectively. Identical letters above the bars indicate means with no statistically significant differences. Different letters indicate statistically significant differences at the 95 % confidence level
There were also distinct seasonal changes in the abundance of the AOA amoA gene in pond sediment. AOA amoA gene copy numbers showed significant negative correlation with temperature (r = − 0.637, P < 0.01; nonparametric correlations). The highest AOA content (1.71 ± 0.76 × 106) was detected in winter, and the lowest AOA content (4.21 ± 2.00 × 105) in summer. Thus, it can be stated that temperature is an important factor that regulates AOA populations in aquaculture pond sediments. The AOA amoA gene was more abundant in winter and spring than in summer and autumn. Similar results were was also observed in the sand of an eelgrass zone (Ando et al. 2009). The fact that the maximum abundance of AOA was observed in winter is in accordance with the findings in estuarine sediments and subtropical coastal mangrove sediments (Caffrey et al. 2007; Wang et al. 2013). In addition, it has been found that AOA is the major driver of nitrification in four cold-water sponges (Radax et al. 2012).
As shown in Fig. 2d, e, the concentration of the AOA amoA gene was greater than that of the AOB amoA gene in sediments by almost one order of magnitude. This result contradicts some previous observations, which suggested that AOB outnumbered AOA in the sediments of Eastern Taihu Bay, which was likely due to the presence of rich organic substances resulting from intensive pen aquaculture (Wu et al. 2010). Moreover, AOA tend to grow in oligotrophic environments with low NH4 + concentrations (Stahl and de la Torre 2012). Significant negative correlations were observed between the copy numbers of the AOA amoA gene and NH4 + concentrations in sediments in Lake Taihu, China (Wu et al. 2010). AOA were more abundant than AOB in freshwater ecosystems and nutrient-depleted oligotrophic wetlands, where NH4 + was quite limited (Sims et al. 2012b; Hugoni et al. 2013). However, under conditions with rich organic substances and high NH4 + concentrations in the aquaculture pond sediments, AOA, as opposed to AOB, were the predominant ammonia-oxidizing organisms. This also can be attributed to the fact that AOA are more resistant to low levels of dissolved oxygen (Coolen et al. 2007; Molina et al. 2010; Bouskill et al. 2012).
Conclusion
The annual investigation of freshwater aquaculture ponds showed that AOB were the main ammonia-oxidizing microorganisms in water, while the AOA were predominant in sediments. Both AOB and AOA densities showed typical seasonal changes in both water and sediments, and photoinhibition and dissolved oxygen levels were suggested to be the main regulating factors.
References
Ackefors H, Enell M (1994) The release of nutrients and organic matter from aquaculture systems in Nordic countries. J Appl Ichthyol 10:225–241
Ando Y, Nakagawa T, Takahashi R, Yoshihara K, Tokuyama T (2009) Seasonal changes in abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria and their nitrification in sand of an eelgrass zone. Microbes Environ 24:21–27
Auguet J-C, Nomokonova N, Camarero L, Casamayor EO (2011) Seasonal changes of freshwater ammonia-oxidizing archaeal assemblages and nitrogen species in oligotrophic alpine lakes. Appl Environ Microbiol 77:1937–1945
Bernhard AE, Tucker J, Giblin AE, Stahl DA (2007) Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ Microbiol 9:1439–1447
Bouskill NJ, Eveillard D, Chien D, Jayakumar A, Ward BB (2012) Environmental factors determining ammonia-oxidizing organism distribution and diversity in marine environments. Environ Microbiol 14:714–729
Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT (2007) Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J 1:660–662
Chang WYB, Ouyang H (1988) Dynamics of dissolved oxygen and vertical circulation in fish ponds. Aquaculture 74:263–276
Church MJ, Wai B, Karl DM, DeLong EF (2010) Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol 12:679–688
Coolen MJL, Abbas B, Bleijswijk JV, Hopmans EC, Kuypers MMM, Wakeham SG, Damsté JSS (2007) Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ Microbiol 9:1001–1016
Editorial Board of Monitoring and Analytical Method of Water and Wastewater, State Environmental Protection Administration of China (2002) Monitoring and analytical method of water and wastewater, 4th edn (in Chinese). China Environmental Science Press, Beijing
FAO (2012) The state of world fisheries and aquaculture 2012. Food and Agricultural Organization of the United Nations, Rome, pp 3–4
French E, Kozlowski JA, Mukherjee M, Bullerjahn G, Bollmann A (2012) Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater. Appl Environ Microbiol 78:5773–5780
Handy RD, Poxton MG (1993) Nitrogen pollution in mariculture: toxicity and excretion of nitrogenous compounds by marine fish. Rev Fish Biol Fish 3:205–241
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA 105:2134–2139
He J-Z, Shen J-P, Zhang L-M, Zhu Y-G, Zheng Y-M, Xu M-G, Di HJ (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374
Horak REA, Qin W, Schauer AJ, Armbrust EV, Ingalls AE, Moffett JW, Devol AH (2013) Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J 7:2023–2033
Hugoni M, Etien S, Bourges A, Lepere C, Domaizon I, Mallet C, Bronner G, Debroas D, Mary I (2013) Dynamics of ammonia-oxidizing Archaea and Bacteria in contrasted freshwater ecosystems. Res Microbiol 164:360–370
Koops H-P, Pommerening-Röser A (2001) Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol Ecol 37:1–9
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Lu SM, Liao MJ, Zhang M, Qi PZ, Xie CC, He XG (2012) A rapid DNA extraction method for quantitative real-time PCR amplification from fresh water sediment. J Food Agric Environ 10:1252–1255
Merbt SN, Stahl DA, Casamayor EO, Marti E, Nicol GW, Prosser JI (2012) Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett 327:41–46
Molina V, Belmar L, Ulloa O (2010) High diversity of ammonia-oxidizing archaea in permanent and seasonal oxygen-deficient waters of the eastern South Pacific. Environ Microbiol 12:2450–2465
Pester M, Schleper C, Wagner M (2011) The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306
Radax R, Hoffmann F, Rapp HT, Leininger S, Schleper C (2012) Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ Microbiol 14:909–923
Randall DJ, Tsui TKN (2002) Ammonia toxicity in fish. Mar Pollut Bull 45:17–23
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Sims A, Gajaraj S, Hu ZQ (2012a) Seasonal population changes of ammonia-oxidizing organisms and their relationship to water quality in a constructed wetland. Ecol Eng 40:100–107
Sims A, Horton J, Gajaraj S, McIntosh S, Miles RJ, Mueller R, Reed R, Hu ZQ (2012b) Temporal and spatial distributions of ammonia-oxidizing archaea and bacteria and their ratio as an indicator of oligotrophic conditions in natural wetlands. Water Res 46:4121–4129
Stahl DA, de la Torre JR (2012) Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol 66:83–101
Tourna M, Freitag TE, Nicol GW, Prosser JI (2008) Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol 10:1357–1364
Wang ZC, Zuo M, Wang YC, Liu YD, Li DH (2011) Dynamics of chlorophyll fluorescence and eco-morphological properties of Microcystis bloom in Meiliang Bay of Lake Taihu, China. Fresenius Environ Bull 20:2295–2305
Wang Y-F, Feng Y-Y, Ma XJ, Gu J-D (2013) Seasonal dynamics of ammonia/ammonium-oxidizing prokaryotes in oxic and anoxic wetland sediments of subtropical coastal mangrove. Appl Microbiol Biotechnol 97:7919–7934
Wu YC, Xiang Y, Wang JJ, Zhong JC, He JZ, Wu QL (2010) Heterogeneity of archaeal and bacterial ammonia-oxidizing communities in Lake Taihu, China. Environ Microbiol Rep 2:569–576
Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Damste JSS (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103:12317–12322
Ye WJ, Liu XL, Lin SQ, Tan J, Pan JL, Li DT, Yang H (2009) The vertical distribution of bacterial and archaeal communities in the water and sediment of Lake Taihu. FEMS Microbiol Ecol 70:263–276
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Acknowledgments
This study was supported financially by the Special Fund for National Technology System for Conventional Freshwater Industries (No. nycytx-49-09), Agro-scientific Research in the Public Interest Project (No. 201203083), and National Key Technology R&D Program (2012BAD25B01). We also thank Prof. Liu Zuo-xiong for reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shimin Lu and Mingjun Liao contributed equally to this work.
Rights and permissions
About this article
Cite this article
Lu, S., Liao, M., Xie, C. et al. Seasonal dynamics of ammonia-oxidizing microorganisms in freshwater aquaculture ponds. Ann Microbiol 65, 651–657 (2015). https://doi.org/10.1007/s13213-014-0903-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-014-0903-2