Abstract
l-Amino acids find various applications in biotechnology. l-Glutamic acid and its salts are used as flavor enhancers. Other l-amino acids are used as food or feed additives, in parenteral nutrition or as building blocks for the chemical and pharmaceutical industries. l-amino acids are synthesized from precursors of central carbon metabolism. Based on the knowledge of the biochemical pathways microbial fermentation processes of food, feed and pharma amino acids have been developed. Production strains of Corynebacterium glutamicum, which has been used safely for more than 50 years in food biotechnology, and Escherichia coli are constantly improved using metabolic engineering approaches. Research towards new processes is ongoing. Fermentative production of l-amino acids in the million-ton-scale has shaped modern biotechnology and its markets continue to grow steadily. This review focusses on recent achievements in strain development for amino acid production including the use of CRISPRi/dCas9, genome-reduced strains, biosensors and synthetic pathways to enable utilization of alternative carbon sources.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
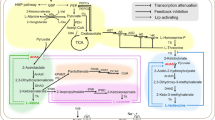
This review summarizes recent technological achievements in C. glutamicum strain development for amino acid production (Fig. 1) and applications to broadening the substrate spectrum of this bacterium to improve production of glutamate-family amino acids. C. glutamicum strain development has benefited from target gene identification by CRISPR interference, from riboswitch-based control of biosynthetic genes, from transcription factor-based screening and from synthetic biology approaches such as genome reduction to generate chassis strains. Moreover, this review highlights recent developments of metabolic engineering efforts for production of l-ornithine, l-citrulline, l-arginine, l-proline and gamma-aminobutyrate (GABA) as well as for access to alternative carbon sources.
Technologies recently applied in C. glutamicum strain development. Selected examples comprise genome reduction (Unthan et al. 2015b), riboswitch-based biosensors (Zhou and Zeng 2015a), transcriptional regulator-based biosensors (Binder et al. 2012) and CRISPRi/dCas9 gene repression (Cleto et al. 2016)
CRISPRi/dCas9-based engineering of l-lysine producing strains
C. glutamicum is used for the industrial production of l-glutamate, l-lysine and other amino acids. Strain development and improvement make use of classical mutagenesis and selection or screening (sometimes combined with omics analysis) and rational approaches such as metabolic engineering. Target identification often involves various parallel omics techniques, while target verification relies on more tedious methods such as deletion mutagenesis, which depends on rare homologous double-crossover events. Recently, CRISPR interference (CRISPRi) technology with the deactivated Cas9 protein (dCas9) was used to determine the effects of target gene repression (Fig. 1) on l-lysine and l-glutamate production (Cleto et al. 2016). Effects on amino acid production due to CRISPRi/dCas9-mediated target gene repression (pgi, pck, and pyk encoding phosphoglucoisomerase, PEP carboxykinase, and pyruvate kinase, respectively) were comparable to levels achieved by target gene deletion (Cleto et al. 2016). This fast approach may potentially be operated in parallel for many target genes.
On-demand flux engineering of l-lysine producing strains
An intracellular riboswitch-based l-lysine biosensor has been developed to control gene expression according to the metabolic demand (Fig. 1). Since C. glutamicum lacks a l-lysine riboswitch, the l-lysine riboswitches of E. coli and B. subtilis were used as l-lysine biosensors to control the citrate synthase gene gltA (Eikmanns et al. 1994) in l-lysine producing C. glutamicum strains (Zhou and Zeng 2015b). Previously, a low citrate synthase flux has been shown to improve l-lysine production (van Ooyen et al. 2012). At low intracellular l-lysine concentrations the riboswitch maintains a confirmation that allows free access of the ribosome binding site of gltA and citrate synthase is synthesized. At l-lysine concentrations exceeding 25–100 µM, the l-lysine riboswitch changes its conformation and in consequence the gltA ribosome binding site is masked and gltA translation ceases. Thus, a l-lysine-dependent translational OFF-switch was used for negative on-demand control of citrate synthase (Zhou and Zeng 2015b).
Positive on-demand control of l-lysine export was achieved by constructing a synthetic l-lysine ON-switch controlling translation of l-lysine export gene lysE (Zhou and Zeng 2015a). When the l-lysine concentration exceeded 0.1 mM, the lysE ribosome binding site was de-masked and lysE translation initiated. This increased the l-lysine yield on glucose by 89 % (Zhou and Zeng 2015a). Combining the OFF-switch to reduce gltA translation and the l-lysine ON-switch to increase l-lysine efflux further improved the l-lysine yield on glucose (Zhou and Zeng 2015a).
Repurposing natural genetic control circuits as shown for the lysine riboswitches may be extended to other genetic control mechanisms and in particular may include on-demand flux control by amino acid-responsive transcriptional regulators since these were also used as biosensors in strain screening.
Intracellular amino acid sensors in C. glutamicum
In C. glutamicum, two amino acid-responsive transcriptional regulators, Lrp (Mustafi et al. 2012) and the l-lysine regulator LysG (Binder et al. 2012), have been used to develop genetically-encoded biosensors (Fig. 1). Transcriptional fusions of one of their target promoters with a promoterless fluorescence reporter gene (Mustafi et al. 2014) is suitable to monitor intracellular amino acid concentrations of millions of single cells in fluorescence activated cell screening/sorting (FACS).
The transcriptional activator Lrp was used in a biosensor for development of strains overproducing branched-chain amino acids or l-methionine (Mustafi et al. 2012). At elevated intracellular concentrations of l-valine, l-leucine, l-isoleucine or l-methionine, Lrp activates the expression of the brnFE operon, which encodes an export system for these amino acids (Lange et al. 2012; Trotschel et al. 2005). The transcriptional fusion of the brnF promoter with promoterless gfp allowed intracellular detection and quantification of these amino acids (Mustafi et al. 2012) and led to improved l-valine producing strains (Mustafi et al. 2014).
The LysR-type transcriptional regulator LysG activates transcription of lysE, the gene for the l-lysine exporter (Vrljic et al. 1996), at elevated intracellular concentrations of l-lysine and basic amino acids in general (Bellmann et al. 2001). The l-lysine biosensor approach was used to screen populations of cells either expressing variants of murE-encoded UDP-N-acetylmuramoy-L-alanyl-d-glutamate:meso-diaminopimelate ligase (Binder et al. 2013) or lysC-encoded aspartate kinase (Schendzielorz et al. 2014). As the almost linear relationship between intracellular l-lysine concentration and biosensor response plateaus at about 25 mM (Binder et al. 2012), the dynamic range of the biosensor system needs to be expanded.
Genome-reduction of C. glutamicum wild type and a l-lysine producing strain
Bacteria have evolved in Nature to cope with changing conditions in their habitats (e.g. temperature, pH, etc.). By contrast, bacteria used as production hosts in biotechnology face more stable environments under production conditions and, thus, may benefit if freed from the metabolic burden of enzymes and genes not relevant under these conditions (e.g. for maintaining intracellular pH). Moreover, reduced metabolic and regulatory complexity may be beneficial for engineering bacterial hosts (Feher et al. 2007; Mampel et al. 2013).
For biotechnological applications, achieving a minimal genome, i.e. the smallest genome required for survival under optimal conditions, is not helpful since multiple auxotrophies (e.g. for amino acids and vitamins) preclude cost-efficient fermentation processes. Biotechnologically relevant genome-reduced chassis organisms have to retain robust growth and production characteristics under defined cultivation conditions. These conditions and characteristics have to be defined prior to initiating a genome reduction project.
Genome reduction by a bottom-up strategy, i.e. creating the entire genome from scratch (de Lorenzo 2015), has not been applied for biotechnologically relevant microorganisms. By contrast, genome reduction by a top-down strategy (Fig. 1), i.e. by gradually reducing genome size in either a targeted or untargeted manner, has been used and the genomes of biotechnologically relevant bacteria have been reduced by 7–22 % (Hashimoto et al. 2005; Leprince et al. 2012; Manabe et al. 2011). The relatively small genome of C. glutamicum strain R has been reduced by 5.7 % by deletion of 188 open reading frames in eight regions not shared with the wild-type C. glutamicum strain ATCC 13032 (Suzuki et al. 2005). The genome of the C. glutamicum wild-type strain ATCC 13032 was reduced by 6 % by deletion of prophage sequences (Baumgart et al. 2013). The resulting strain, MB001, did not show unfavorable properties under the previously defined growth conditions. Moreover, MB001 showed no defects under stress conditions and exhibited increased transformation efficiency and plasmid copy number. Subsequently, the genome-reduced C. glutamicum strain MB001 was engineered for the production of the carotenoids lycopene, decaprenoxanthin and astaxanthin (Heider et al. 2014), and the amino acids l-citrulline with a yield on glucose of 0.38 ± 0.01 g g−1 (Eberhardt et al. 2014). Similarly, MB001-based strains for the production of the amino acids l-ornithine, l-arginine, and l-proline and the diamine putrescine were developed (Jensen et al. 2015). High-level and uniform protein overproduction was achieved by introducing the DE3 region of E. coli BL21(DE3) harboring the T7 RNA polymerase gene under control of the lacUV5 promoter into the chromosome of C. glutamicum MB001 (Kortmann et al. 2015).
Corynebacterium glutamicum MB001 was also the starting point for further genome reduction. Several omics data sets, evolutionary considerations, and prior knowledge on the function of genes were used to classify all genes of C. glutamicum with respect to their essentiality and importance for maintaining fast growth in a previously defined medium. 36 of 41 gene clusters with a high content of non-essential or unclassifiable genes could be deleted (Unthan et al. 2015b). Genome reduction was not only reported for C. glutamicum wild-type strain ATCC 13032, but also for a l-lysine-producing model strain (Unthan et al. 2015a, b). The genome-reduced strains were phenotyped in multi-well plates in a mini pilot plant allowing for automatic separation of supernatants from biomass in a robotic workflow (Rohe et al. 2012; Unthan et al. 2015b). After at-line clarification, amino acids were quantified using the ninhydrin assay and the genome-reduced l-lysine producer strains were categorized with respect to growth rate and amino acid titer (Unthan et al. 2015b).
Production of the l-glutamate-derived amino acids l-ornithine, l-citrulline, l-arginine, l-proline and gamma-aminobutyrate (GABA)
Corynebacterium glutamicum, a bacterium discovered in 1956 for its natural capacity to accumulate l-glutamate extracellularly (Kinoshita et al. 2004), is currently used for the industrial production of l-glutamate (about 3 million tons in 2014) and, thus, is considered an excellent host for the production of glutamate-derived amino acids (Wendisch 2014) such as l-ornithine, l-citrulline, l-arginine, l-proline and GABA (Table 1).
l-Arginine biosynthesis (Fig. 2) initiates with l-glutamate and proceeds via the intermediates l-ornithine and l-citrulline. l-Ornithine is a non-essential amino acid that can be used in the food industry as a dietary supplement (Kim et al. 2015c) or in the pharmaceutical industry e.g. in the treatment of liver disease (Salvatore et al. 1964) or wound healing (Shi et al. 2002). A number of groups engineered C. glutamicum for l-ornithine production (Hwang et al. 2008; Jensen et al. 2015; Kim et al. 2015a, c; Matsui and Oikawa 2010; Schneider et al. 2011). l-ornithine is secreted as result of interrupting l-arginine biosynthesis by deletion of the ornithine carbamoyltransferase gene argF. Deletion of the arginine repressor gene argR leads to de-repression of the l-arginine biosynthesis genes. The pacemaker enzyme N-acetylglutamate kinase is alleviated from feedback inhibition by l-arginine by introducing point mutations into its gene, argB. Supply of l-glutamate and of NADPH for l-ornithine production was improved by increased promoter activity of l-glutamate dehydrogenase gene gdh and by decreased expression of pgi, which encodes the key enzyme phosphoglucoisomerase from the oxidative pentose phosphate pathway. After combining these changes with introduction of a second copy of the arginine biosynthesis operon in the genome-streamlined strain MB001 l-ornithine yields of about 0.52 g ornithine g−1 glucose were achieved (Jensen et al. 2015). Kim et al. 2015c reported an l-ornithine titer of 51.5 g L−1 and a volumetric productivity of 1.29 g L−1 h−1 (Kim et al. 2015c).
Biosynthetic pathways of the l-glutamate derived amino acids l-ornithine, l-citrulline, l-arginine, l-proline and gamma-aminobutyrate. Heterologous reactions are highlighted in boxes. Abbreviations: Gdh, glutamate dehydrogenase; ArgB, N-acetylglutamate kinase; ArgC, N-acetyl-gamma-glutamyl-phosphate reductase; ArgD, N-acetylornithine aminotransferase; ArgJ, ornithine acetyltransferase; ArgF, ornithine carbamoyltransferase; ArgG, argininosuccinate synthetase; ArgH, argininosuccinate lyase; GAD, glutamate decarboxylase (Escherichia coli); OCD, ornithine cyclodeaminase (Pseudomonas putida); 2-OXO, 2-oxoglutaric acid; l-glut, l-glutamic acid; CarP, carbamoyl phosphate; l-Asp, l-aspartic acid; Fum, fumaric acid; GABA, gamma-aminobutyrate
l-Citrulline is an intermediate of arginine biosynthesis (Fig. 2) derived from l-ornithine by ornithine carbamoyltransferase. l-Citrulline plays an important role in human health and nutrition (Eberhardt et al. 2014; Hao et al. 2015; Ikeda et al. 2009). l-citrulline is excreted as consequence of deletion of argG, which codes for argininosuccinate synthetase, the enzyme converting l-citrulline to N-argininosuccinate (Hao et al. 2015). Engineering an l-ornithine producing strain to an efficient l-citrulline producing strain required the overexpression of argF as well as deletion of argG. An l-citrulline titer of about 44 mM l-citrulline and a yield of 0.38 g l-citrulline g−1 glucose were achieved (Eberhardt et al. 2014).
l-Arginine is a semi-essential amino acid, i.e. there is no dietary requirement for healthy adults, but for infants and growing children or adults under catabolic stress or with dysfunction of the small intestine or kidney (Flynn et al. 2002), and it has applications in food, cosmetics and pharmaceutical industries (Ikeda et al. 2009; Park et al. 2014). Rationally engineered l-arginine producing strains were constructed in a similar manner as l-citrulline and l-ornithine producing strains, however, l-citrulline was a side-product of l-arginine production (Ikeda et al. 2009; Schneider et al. 2011; Jensen et al. 2015). This limitation was overcome by random mutagenesis and screening (Park et al. 2014). l-Arginine titers of about 90 g L−1 were achieved.
l-Proline is a secondary proteinogenic amino acid which finds applications in the feed, pharmaceutical or cosmetic industries (Jensen and Wendisch 2013). In a widespread biosynthetic pathway leading to l-proline, l-glutamate is converted to l-proline in three enzyme-catalysed and one spontaneous reaction (Eggeling and Bott 2005; Jensen and Wendisch 2013). Heterologous expression of the ornithine cyclodeaminase gene ocd from Pseudomonas putida in an l-ornithine production strain (Fig. 2) enabled production of about 13 g L−1 of proline with a yield of 0.36 g proline g−1 glucose (Jensen and Wendisch 2013).
GABA is a non-protein amino acid that can be used in the pharmaceutical and food industries. Moreover, the lactamization of GABA yields 2-pyrrolidone, which can be converted chemically to the bio-plastic polyamide 4 (Kawasaki et al. 2005; Park et al. 2013). l-Glutamate can be decarboxylated to GABA by glutamate decarboxylase (Fig. 2) and heterologous expression of glutamate decarboxylase genes usually from Escherichia coli or Lactobacillus brevis resulted in GABA production under conditions triggering l-glutamate production (Choi et al. 2015; Okai et al. 2014; Shi et al. 2013; Takahashi et al. 2012). GABA titers of about 39 g L−1 were achieved in fed-batch cultures (Choi et al. 2015).
Utilization of alternative carbon source derived from complex polymers
Corynebacterium glutamicum utilizes a relative broad spectrum of carbon sources for its growth and energy supply, including e.g. the monosaccharides glucose, fructose and ribose as well as the disaccharides sucrose and maltose (Blombach and Seibold 2010). However, C. glutamicum wild type is neither able to catabolise a number of polymeric carbon sources (e.g. starch, cellulose, hemicellulose, lignocellulose or chitin) nor their monomeric constituents such as xylose, arabinose, or N-acetylglucosamine. To realize a flexible feedstock concept (Table 2), C. glutamicum was engineered to catabolize e.g. starch, xylose, arabinose, glycerol or levoglucosan by heterologous gene expression (Kim et al. 2015b; Zahoor et al. 2012). Notably, when present in blends C. glutamicum utilizes many carbon sources simultaneously (Blombach and Seibold 2010; Gerstmeir et al. 2003), an ideal characteristic when considering hydrolysates of complex carbon sources such as lignocellulose. Substrate co-utilization is typically also observed for C. glutamicum strains engineered to catabolize non-native carbon sources (Gopinath et al. 2011; Meiswinkel et al. 2013; Rittmann et al. 2008; Seibold et al. 2006).
Starch is a major feedstock for industrial amino acid production, however, fermentations are based on glucose derived from enzymatic starch hydrolysis. Heterologous production and secretion of α-amylase from Streptomyces griseus in C. glutamicum enabled growth with starch as sole or combined carbon source (Seibold et al. 2006). Surface display of α-amylase from Streptococcus bovis anchored via pgsA from Bacillus subtilis enabled starch catabolism as well (Tateno et al. 2007). Nevertheless, complete utilization of starch was not observed since some starch degradation products still remained in the medium (Seibold et al. 2006). Pullulanase (EC 3.2.1.41), a debranching enzyme, enables a complete and efficient conversion of the branched polysaccharides into small fermentable sugars during saccharification process (Hii et al. 2012). Heterologous expression pullulanases has been reported in E. coli with expression of pul3YH5 from Exiguobacterium acetylicum YH5 (Qiao et al. 2015) and in Bacillus subtilis with expression of apuA from Delsulforococcus muscosus, whereas heterologous expression of pullulanases in C. glutamicum has not yet been reported.
Lignocellulose hydrolysates, which can be obtained from residual plant biomass, support growth of C. glutamicum since they contain natural carbon substrates of this bacterium such as cellulose-derived glucose or acetate (Gopinath et al. 2011; Meiswinkel et al. 2013). Efficient conversion of residual plant biomass to value-added products including biofuels or chemicals requires their complete utilization. Lignocellulose contains cellulose (40–50 %), hemicellulose (25–30 %) and lignin (10–20 %) (Anwar et al. 2014; Wyman 1999), and cannot be utilized directly by most microorganisms including C. glutamicum. Pre-treatment of lignocellulosic biomass e.g. by alkali/acidic or enzymatic hydrolysis into its monomeric components (Anwar et al. 2014; Rumbold et al. 2010) yields glucose, xylose, arabinose, acetate and galactose as major products of hydrolysis (Aristidou and Penttila 2000). Endoglucanases and exoglucanases depolymerize cellulose incompletely to the disaccharide cellobiose which is subsequently cleaved by β-glucosidase (Schwarz 2001). For an industrially scalable process, heterologous cellulases need to be produced and secreted by the host itself (Adham et al. 2001; Paradis et al. 1987) and synthetic microcellulosomes based on genes from Clostridium cellulovorans may increase efficiency (Hyeon et al. 2011; Lambertz et al. 2014). C. glutamicum R is able to use cellobiose as a result of a mutation in phosphotransferase permease subunit BglF which imports methyl β-glucoside, arbutin and salicin (Kotrba et al. 2003).
Recently, a combined approach of metabolic engineering and adaptive evolution of C. glutamicum ATCC 13032 has been reported with respect to cellobiose utilization as C-source. Genes for a cellodextrin transporter and β-glucosidase were expressed and the resulting strain was evolved. Secreted and cell-displayed forms of β-glucosidase led to complete, but slow consumption of 20 g L–1 cellobiose in 4 days (Lee et al. 2016). In a similar approach, 1 g L–1 of l-lysine was produced from 20 g L–1 of cellobiose after 4 days based on a strain harboring β-glucosidase Sde1394 from Saccharophagus degradans (Adachi et al. 2013).
The hemicellulose-derived pentose sugars xylose and arabinose are not natural C-sources for C. glutamicum. Since C. glutamicum possesses xylulokinase XylB, heterologous expression of xylose isomerase gene xylA from E. coli was sufficient to enable growth with xylose as sole carbon source (Kawaguchi et al. 2006). Faster growth was observed when endogenous xylB was overexpressed in combination with heterologous expression of xylA from Xanthomonas campestris (Meiswinkel et al. 2013). Xylose-based production has been described e.g. for l-glutamate and l-lysine (Gopinath et al. 2011; Meiswinkel et al. 2013), cadaverine (Buschke et al. 2011), and putrescine (Meiswinkel et al. 2013). Arabinose utilization was achieved by heterologous expression of the araBAD operon of E. coli, which encodes arabinose isomerase AraA, ribulokinase AraB, and ribulose 5-phosphate 4-epimerase AraD (Kawaguchi et al. 2008; Schneider et al. 2011). The arabinose uptake system from C. glutamicum strain ATCC 31831 was not only useful to accelerate import and catabolism of arabinose, but also utilization of xylose (Sasaki et al. 2009).
Growth with galactose, a component of lignocellulosic hydrolysates, became possible for C. glutamicum only via heterologous expression of genes from Lactococcus lactis subsp. cremoris MG1363 encoding for galactokinase (galK), UDP-glucose-1-P-uridylyltransferase (galT), UDP-galactose-4-epimerase (galE) and aldose-1-epimerase (galM) (Barrett et al. 2004).
Lignin is synthesized via radical reactions and is recalcitrant towards degradation. Its constituents are the aromatic coniferyl-, sinapyl- and p-coumaryl-alcohols. C. glutamicum is able to catabolize a wide spectrum of aromatic compounds, e.g. benzoate, phenol (Shen et al. 2005a), 3-hydroxybenzoate, gentisate (Shen et al. 2005b), and naphthalene (Lee et al. 2010; Shen et al. 2005a). Catabolism of various aromatic compounds via peripheral pathways involving ring modification reactions converges in the formation of the dihydroxylated benzene derivatives gentisate (from naphthalene and 3-hydrobenzoate; (Lee et al. 2010; Shen et al. 2005b), catechol (from benzyl alcohol, benzoate and phenol; (Shen et al. 2004), and protocatechuate (from ferulic acid, vanillin, vanillate, 4-hydroxybenzoate, caffeate, 4-cresol, p-coumarate, quinate, shikimate; (Merkens et al. 2005; Qi et al. 2007; Shen et al. 2005b). These key aromatic intermediates are then converted to pyruvate, fumarate or succinyl-CoA after aromatic ring cleavage by the dioxygenases catechol 1,2-dioxygenase, protocatechuate 3,4-dioxygenase, gentisate 1,2-dioxygenase, and hydroxyquinol 1,2-dioxygenase. This metabolic potential of C. glutamicum may be used in bioremediation, for bioconversion of aromatic pollutants or for production of amino acids from aromatic feedstocks (Shen et al. 2012).
The amino sugars glucosamine and N-acetyl glucosamine, which have the potential to serve as combined carbon and nitrogen sources, are the monomeric components of chitin and chitosan. Chitin, which can be found in exoskeletons of arthropods, in the cell walls of fungi, in the radula of gastropods, is an underutilized feedstock for fermentation. Consequently, C. glutamicum strains have been constructed to enable access to glucosamine and to N-acetylglucosamine (Matano et al. 2014; Uhde et al. 2013). Glucosamine uptake is mediated by the glucose specific permease PtsG, but fast growth on glucosamine as sole source of carbon and nitrogen required that the endogenous glucosamine-6-phosphate deaminase NagB was overproduced (Uhde et al. 2013). To enable growth of C. glutamicum with N-acetyl-glucosamine, the N-acetyl-glucosamine-specific PTS gene nagE from the related C. glycinophilum had to be expressed in addition to overexpression of endogenous nagB and endogenous nagA which encodes the cytoplasmic N-acetylglucosamine-6-phosphate deacetylase (Matano et al. 2014). Glucosamine-based as well as N-acetyl-glucosamine-based l-lysine production was reported (Matano et al. 2014; Uhde et al. 2013).
Outlook
Since its discovery more than 50 years ago, C. glutamicum has been optimized for amino acid production. New technologies and concepts ranging from metabolic engineering, genomics, transcriptomics, metabolomics, fluxomics to systems and synthetic biology have been applied to C. glutamicum strain development and developed further. This has also boosted engineering C. glutamicum for the production of other value-added products such as diamines, organic acids, carotenoids, proteins and biopolymers. New technologies like the CRISPR/Cas9 system are expected to contribute to improving production of amino acids (Cleto et al. 2016) and other value-added chemicals by the workhorse of industrial biotechnology C. glutamicum.
References
Adachi N, Takahashi C, Ono-Murota N, Yamaguchi R, Tanaka T, Kondo A (2013) Direct l-lysine production from cellobiose by Corynebacterium glutamicum displaying beta-glucosidase on its cell surface. Appl Microbiol Biotechnol 97:7165–7172. doi:10.1007/s00253-013-5009-4
Adham SA, Honrubia P, Diaz M, Fernandez-Abalos JM, Santamaria RI, Gil JA (2001) Expression of the genes coding for the xylanase Xys1 and the cellulase Cel1 from the straw-decomposing Streptomyces halstedii JM8 cloned into the amino-acid producer Brevibacterium lactofermentum ATCC13869. Arch Microbiol 177:91–97. doi:10.1007/s00203-001-0365-3
Anwar Z, Gulfraz M, Irshad M (2014) Agro-industrial lignocellulosic biomass a key to unlock the future bio-energy: a brief review. J Radiat Res Appl Sci 7:163–173
Aristidou A, Penttila M (2000) Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol 11:187–198
Barrett E, Stanton C, Zelder O, Fitzgerald G, Ross RP (2004) Heterologous expression of lactose- and galactose-utilizing pathways from lactic acid bacteria in Corynebacterium glutamicum for production of lysine in whey. Appl Environ Microbiol 70:2861–2866
Baumgart M, Unthan S, Ruckert C, Sivalingam J, Grunberger A, Kalinowski J, Bott M, Noack S, Frunzke J (2013) Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl Environ Microbiol 79:6006–6015. doi:10.1128/AEM.01634-13
Bellmann A, Vrljic M, Patek M, Sahm H, Kramer R, Eggeling L (2001) Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765–1774. doi:10.1099/00221287-147-7-1765
Binder S, Schendzielorz G, Stabler N, Krumbach K, Hoffmann K, Bott M, Eggeling L (2012) A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol 13:R40. doi:10.1186/gb-2012-13-5-r40
Binder S, Siedler S, Marienhagen J, Bott M, Eggeling L (2013) Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res 41:6360–6369. doi:10.1093/nar/gkt312
Blombach B, Seibold GM (2010) Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Appl Microbiol Biotechnol 86:1313–1322. doi:10.1007/s00253-010-2537-z
Buschke N, Schroder H, Wittmann C (2011) Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol J 6:306–317. doi:10.1002/biot.201000304
Choi JW, Yim SS, Lee SH, Kang TJ, Park SJ, Jeong KJ (2015) Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb Cell Fact 14:21. doi:10.1186/s12934-015-0205-9
Cleto S, Jensen JV, Wendisch VF, Lu TK (2016) Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth Biol. doi:10.1021/acssynbio.5b00216
de Lorenzo V (2015) Chassis organism from Corynebacterium glutamicum: the way towards biotechnological domestication of Corynebacteria. Biotechnol J 10:244–245. doi:10.1002/biot.201400493
Eberhardt D, Jensen JV, Wendisch VF (2014) l-citrulline production by metabolically engineered Corynebacterium glutamicum from glucose and alternative carbon sources. AMB Express 4:85. doi:10.1186/s13568-014-0085-0
Eggeling L, Bott M (2005) Handbook of Corynebacterium glutamicum. CRC, Boca Raton
Eikmanns BJ, Thum-Schmitz N, Eggeling L, Ludtke KU, Sahm H (1994) Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140(Pt 8):1817–1828. doi:10.1099/13500872-140-8-1817
Feher T, Papp B, Pal C, Posfai G (2007) Systematic genome reductions: theoretical and experimental approaches. Chem Rev 107:3498–3513. doi:10.1021/cr0683111
Flynn NE, Meininger CJ, Haynes TE, Wu G (2002) The metabolic basis of arginine nutrition and pharmacotherapy. Biomed Pharmacother 56:427–438
Gerstmeir R, Wendisch VF, Schnicke S, Ruan H, Farwick M, Reinscheid D, Eikmanns BJ (2003) Acetate metabolism and its regulation in Corynebacterium glutamicum. J Biotechnol 104:99–122
Gopinath V, Meiswinkel TM, Wendisch VF, Nampoothiri KM (2011) Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl Microbiol Biotechnol 92:985–996. doi:10.1007/s00253-011-3478-x
Hao N, Mu J, Hu N, Xu S, Yan M, Li Y, Guo K, Xu L (2015) Improvement of l-citrulline production in Corynebacterium glutamicum by ornithine acetyltransferase. J Ind Microbiol Biotechnol 42:307–313. doi:10.1007/s10295-014-1561-x
Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, Katayama T, Kato J (2005) Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol Microbiol 55:137–149. doi:10.1111/j.1365-2958.2004.04386.x
Heider SA, Wolf N, Hofemeier A, Peters-Wendisch P, Wendisch VF (2014) Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum. Front Bioeng Biotechnol 2:28. doi:10.3389/fbioe.2014.00028
Hii SL, Tan JS, Ling TC, Ariff AB (2012) Pullulanase: role in starch hydrolysis and potential industrial applications. Enzyme Res 2012:921362. doi:10.1155/2012/921362
Hwang JH, Hwang GH, Cho JY (2008) Effect of increased glutamate availability on l-ornithine production in Corynebacterium glutamicum. J Microbiol Biotechnol 18:704–710
Hyeon JE, Jeon WJ, Whang SY, Han SO (2011) Production of minicellulosomes for the enhanced hydrolysis of cellulosic substrates by recombinant Corynebacterium glutamicum. Enzyme Microb Technol 48:371–377. doi:10.1016/j.enzmictec.2010.12.014
Ikeda M, Mitsuhashi S, Tanaka K, Hayashi M (2009) Reengineering of a Corynebacterium glutamicum l-arginine and l-citrulline producer. Appl Environ Microbiol 75:1635–1641. doi:10.1128/AEM.02027-08
Jensen JV, Wendisch VF (2013) Ornithine cyclodeaminase-based proline production by Corynebacterium glutamicum. Microb Cell Fact 12:63. doi:10.1186/1475-2859-12-63
Jensen JV, Eberhardt D, Wendisch VF (2015) Modular pathway engineering of Corynebacterium glutamicum for production of the glutamate-derived compounds ornithine, proline, putrescine, citrulline, and arginine. J Biotechnol 214:85–94. doi:10.1016/j.jbiotec.2015.09.017
Kawaguchi H, Vertes AA, Okino S, Inui M, Yukawa H (2006) Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol 72:3418–3428. doi:10.1128/AEM.72.5.3418-3428.2006
Kawaguchi H, Sasaki M, Vertes AA, Inui M, Yukawa H (2008) Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Appl Microbiol Biotechnol 77:1053–1062. doi:10.1007/s00253-007-1244-x
Kawasaki N, Nakayama A, Yamano N, Takeda S, Kawata Y, Yamamoto N, Aiba S (2005) Synthesis, thermal and mechanical properties and biodegradation of branched polyamide 4. Polymer 46:9987–9993. doi:10.1016/j.polymer.2005.06.092
Kim DJ, Hwang GH, Um JN, Cho JY (2015a) Increased l-ornithine production in Corynebacterium glutamicum by overexpression of a gene encoding a putative aminotransferase. J Mol Microbiol Biotechnol 25:45–50. doi:10.1159/000375124
Kim EM, Um Y, Bott M, Woo HM (2015b) Engineering of Corynebacterium glutamicum for growth and succinate production from levoglucosan, a pyrolytic sugar substrate. FEMS Microbiol Lett. doi:10.1093/femsle/fnv161
Kim SY, Lee J, Lee SY (2015c) Metabolic engineering of Corynebacterium glutamicum for the production of l-ornithine. Biotechnol Bioeng 112:416–421. doi:10.1002/bit.25440
Kinoshita S, Udaka S, Shimono M (2004) Studies on the amino acid fermentation. Part 1. Production of l-glutamic acid by various microorganisms. J Gen Appl Microbiol 50:331–343
Kortmann M, Kuhl V, Klaffl S, Bott M (2015) A chromosomally encoded T7 RNA polymerase-dependent gene expression system for Corynebacterium glutamicum: construction and comparative evaluation at the single-cell level. Microb Biotechnol 8:253–265. doi:10.1111/1751-7915.12236
Kotrba P, Inui M, Yukawa H (2003) A single V317A or V317M substitution in Enzyme II of a newly identified beta-glucoside phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiology 149:1569–1580. doi:10.1099/mic.0.26053-0
Lambertz C, Garvey M, Klinger J, Heesel D, Klose H, Fischer R, Commandeur U (2014) Challenges and advances in the heterologous expression of cellulolytic enzymes: a review. Biotechnol Biofuels 7:135. doi:10.1186/s13068-014-0135-5
Lange C, Mustafi N, Frunzke J, Kennerknecht N, Wessel M, Bott M, Wendisch VF (2012) Lrp of Corynebacterium glutamicum controls expression of the brnFE operon encoding the export system for l-methionine and branched-chain amino acids. J Biotechnol 158:231–241. doi:10.1016/j.jbiotec.2011.06.003
Lee SY, Le TH, Chang ST, Park JS, Kim YH, Min J (2010) Utilization of phenol and naphthalene affects synthesis of various amino acids in Corynebacterium glutamicum. Curr Microbiol 61:596–600. doi:10.1007/s00284-010-9658-6
Lee J, Saddler JN, Um Y, Woo HM (2016) Adaptive evolution and metabolic engineering of a cellobiose- and xylose- negative Corynebacterium glutamicum that co-utilizes cellobiose and xylose. Microb Cell Fact 15:20. doi:10.1186/s12934-016-0420-z
Leprince A, de Lorenzo V, Voller P, van Passel MW, Martins dos Santos VA (2012) Random and cyclical deletion of large DNA segments in the genome of Pseudomonas putida. Environ Microbiol 14:1444–1453. doi:10.1111/j.1462-2920.2012.02730.x
Mampel J, Buescher JM, Meurer G, Eck J (2013) Coping with complexity in metabolic engineering. Trends Biotechnol 31:52–60. doi:10.1016/j.tibtech.2012.10.010
Manabe K, Kageyama Y, Morimoto T, Ozawa T, Sawada K, Endo K, Tohata M, Ara K, Ozaki K, Ogasawara N (2011) Combined effect of improved cell yield and increased specific productivity enhances recombinant enzyme production in genome-reduced Bacillus subtilis strain MGB874. Appl Environ Microbiol 77:8370–8381. doi:10.1128/AEM.06136-11
Matano C, Uhde A, Youn JW, Maeda T, Clermont L, Marin K, Kramer R, Wendisch VF, Seibold GM (2014) Engineering of Corynebacterium glutamicum for growth and l-lysine and lycopene production from N-acetyl-glucosamine. Appl Microbiol Biotechnol 98:5633–5643. doi:10.1007/s00253-014-5676-9
Matsui D, Oikawa T (2010) Detection of d-ornithine extracellularly produced by Corynebacterium glutamicum ATCC 13032:argF. Biosci Biotechnol Biochem 74:2507–2510
Meiswinkel TM, Gopinath V, Lindner SN, Nampoothiri KM, Wendisch VF (2013) Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb Biotechnol 6:131–140. doi:10.1111/1751-7915.12001
Merkens H, Beckers G, Wirtz A, Burkovski A (2005) Vanillate metabolism in Corynebacterium glutamicum. Curr Microbiol 51:59–65. doi:10.1007/s00284-005-4531-8
Mustafi N, Grunberger A, Kohlheyer D, Bott M, Frunzke J (2012) The development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metab Eng 14:449–457. doi:10.1016/j.ymben.2012.02.002
Mustafi N, Grunberger A, Mahr R, Helfrich S, Noh K, Blombach B, Kohlheyer D, Frunzke J (2014) Application of a genetically encoded biosensor for live cell imaging of l-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains. PLoS One 9:e85731. doi:10.1371/journal.pone.0085731
Okai N, Takahashi C, Hatada K, Ogino C, Kondo A (2014) Disruption of pknG enhances production of gamma-aminobutyric acid by Corynebacterium glutamicum expressing glutamate decarboxylase. AMB Express 4:20. doi:10.1186/s13568-014-0020-4
Paradis FW, Warren RA, Kilburn DG, Miller RC Jr (1987) The expression of Cellulomonas fimi cellulase genes in Brevibacterium lactofermentum. Gene 61:199–206
Park SJ, Kim EY, Noh W, Oh YH, Kim HY, Song BK, Cho KM, Hong SH, Lee SH, Jegal J (2013) Synthesis of nylon 4 from gamma-aminobutyrate (GABA) produced by recombinant Escherichia coli. Bioprocess Biosyst Eng 36:885–892. doi:10.1007/s00449-012-0821-2
Park SH, Kim HU, Kim TY, Park JS, Kim SS, Lee SY (2014) Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun 5:4618. doi:10.1038/ncomms5618
Qi SW, Chaudhry MT, Zhang Y, Meng B, Huang Y, Zhao KX, Poetsch A, Jiang CY, Liu S, Liu SJ (2007) Comparative proteomes of Corynebacterium glutamicum grown on aromatic compounds revealed novel proteins involved in aromatic degradation and a clear link between aromatic catabolism and gluconeogenesis via fructose-1,6-bisphosphatase. Proteomics 7:3775–3787. doi:10.1002/pmic.200700481
Qiao Y, Peng Q, Yan J, Wang H, Ding H, Shi B (2015) Gene cloning and enzymatic characterization of alkali-tolerant type I pullulanase from Exiguobacterium acetylicum. Lett Appl Microbiol 60:52–59. doi:10.1111/lam.12333
Rittmann D, Lindner SN, Wendisch VF (2008) Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl Environ Microbiol 74:6216–6222. doi:10.1128/AEM.00963-08
Rohe P, Venkanna D, Kleine B, Freudl R, Oldiges M (2012) An automated workflow for enhancing microbial bioprocess optimization on a novel microbioreactor platform. Microb Cell Fact 11:144. doi:10.1186/1475-2859-11-144
Rumbold K, van Buijsen HJ, Gray VM, van Groenestijn JW, Overkamp KM, Slomp RS, van der Werf MJ, Punt PJ (2010) Microbial renewable feedstock utilization: a substrate-oriented approach. Bioeng Bugs 1:359–366. doi:10.4161/bbug.1.5.12389
Salvatore F, Cimino F, Dayello Caracciolo M, Cittadini D (1964) Mechanism of the protection by l-Ornithine-l-Aspartate mixture and by l-Arginine in ammonia intoxication. Arch Biochem Biophys 107:499–503
Sasaki M, Jojima T, Kawaguchi H, Inui M, Yukawa H (2009) Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl Microbiol Biotechnol 85:105–115. doi:10.1007/s00253-009-2065-x
Schendzielorz G, Dippong M, Grunberger A, Kohlheyer D, Yoshida A, Binder S, Nishiyama C, Nishiyama M, Bott M, Eggeling L (2014) Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth Biol 3:21–29. doi:10.1021/sb400059y
Schneider J, Niermann K, Wendisch VF (2011) Production of the amino acids l-glutamate, l-lysine, l-ornithine and l-arginine from arabinose by recombinant Corynebacterium glutamicum. J Biotechnol 154:191–198. doi:10.1016/j.jbiotec.2010.07.009
Schwarz WH (2001) The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol 56:634–649
Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ (2006) Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J Biotechnol 124:381–391. doi:10.1016/j.jbiotec.2005.12.027
Shen XH, Liu ZP, Liu SJ (2004) Functional identification of the gene locus (ncg12319 and characterization of catechol 1,2-dioxygenase in Corynebacterium glutamicum. Biotechnol Lett 26:575–580
Shen X-H, Huang Y, Liu S-J (2005a) Genomic analysis and identification of catabolic pathways for aromatic compounds in Corynebacterium glutamicum. Microb Environ 20(3):160–167. doi:10.1264/jsme2.20.160
Shen XH, Jiang CY, Huang Y, Liu ZP, Liu SJ (2005b) Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. Appl Environ Microbiol 71:3442–3452. doi:10.1128/AEM.71.7.3442-3452.2005
Shen XH, Zhou NY, Liu SJ (2012) Degradation and assimilation of aromatic compounds by Corynebacterium glutamicum: another potential for applications for this bacterium? Appl Microbiol Biotechnol 95:77–89. doi:10.1007/s00253-012-4139-4
Shi HP, Fishel RS, Efron DT, Williams JZ, Fishel MH, Barbul A (2002) Effect of supplemental ornithine on wound healing. J Surg Res 106:299–302
Shi F, Jiang J, Li Y, Li Y, Xie Y (2013) Enhancement of gamma-aminobutyric acid production in recombinant Corynebacterium glutamicum by co-expressing two glutamate decarboxylase genes from Lactobacillus brevis. J Ind Microbiol Biotechnol 40:1285–1296. doi:10.1007/s10295-013-1316-0
Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa H (2005) New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microbiol 71:8472–8480. doi:10.1128/AEM.71.12.8472-8480.2005
Takahashi C, Shirakawa J, Tsuchidate T, Okai N, Hatada K, Nakayama H, Tateno T, Ogino C, Kondo A (2012) Robust production of gamma-amino butyric acid using recombinant Corynebacterium glutamicum expressing glutamate decarboxylase from Escherichia coli. Enzyme Microb Technol 51:171–176. doi:10.1016/j.enzmictec.2012.05.010
Tateno T, Fukuda H, Kondo A (2007) Direct production of l-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl Microbiol Biotechnol 77:533–541. doi:10.1007/s00253-007-1191-6
Trotschel C, Deutenberg D, Bathe B, Burkovski A, Kramer R (2005) Characterization of methionine export in Corynebacterium glutamicum. J Bacteriol 187:3786–3794. doi:10.1128/JB.187.11.3786-3794.2005
Uhde A, Youn JW, Maeda T, Clermont L, Matano C, Kramer R, Wendisch VF, Seibold GM, Marin K (2013) Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl Microbiol Biotechnol 97:1679–1687. doi:10.1007/s00253-012-4313-8
Unthan S, Baumgart M, Radek A, Herbst M, Siebert D, Bruhl N, Bartsch A, Bott M, Wiechert W, Marin K, Hans S, Kramer R, Seibold G, Frunzke J, Kalinowski J, Ruckert C, Wendisch VF, Noack S (2015a) Chassis organism from Corynebacterium glutamicum—a top-down approach to identify and delete irrelevant gene clusters. Biotechnol J 10:290–301. doi:10.1002/biot.201400041
Unthan S, Radek A, Wiechert W, Oldiges M, Noack S (2015b) Bioprocess automation on a mini pilot plant enables fast quantitative microbial phenotyping. Microb Cell Fact 14:32. doi:10.1186/s12934-015-0216-6
van Ooyen J, Noack S, Bott M, Reth A, Eggeling L (2012) Improved l-lysine production with Corynebacterium glutamicum and systemic insight into citrate synthase flux and activity. Biotechnol Bioeng 109:2070–2081. doi:10.1002/bit.24486
Vrljic M, Sahm H, Eggeling L (1996) A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol Microbiol 22:815–826
Wendisch VF (2014) Microbial production of amino acids and derived chemicals: synthetic biology approaches to strain development. Curr Opin Biotechnol 30:51–58. doi:10.1016/j.copbio.2014.05.004
Wyman CE (1999) Biomass ethanol: technical progress, opportunities, and commercial challenges. Ann Rev Energy Environ 24(1):189–226
Zahoor A, Lindner SN, Wendisch VF (2012) Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products. Comput Struct Biotechnol J 3:e201210004. doi:10.5936/csbj.201210004
Zhou LB, Zeng AP (2015a) Engineering a lysine-ON riboswitch for metabolic control of lysine production in Corynebacterium glutamicum. ACS Synth Biol 4:1335–1340. doi:10.1021/acssynbio.5b00075
Zhou LB, Zeng AP (2015b) Exploring lysine riboswitch for metabolic flux control and improvement of l-lysine synthesis in Corynebacterium glutamicum. ACS Synth Biol 4:729–734. doi:10.1021/sb500332c
Funding information
João M.P. Jorge and Fernando Pérez-García are fellows of the CLIB2021 graduate cluster at Bielefeld University. Support by Grant KF2969004SB4 from ZIM, BMWi, is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Wendisch, V.F., Jorge, J.M.P., Pérez-García, F. et al. Updates on industrial production of amino acids using Corynebacterium glutamicum . World J Microbiol Biotechnol 32, 105 (2016). https://doi.org/10.1007/s11274-016-2060-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2060-1