Abstract
This article reports multiple metabolic pathways of amino acid production via phenol and naphthalene use by Corynebacterium glutamicum. Biodegradation of phenol and naphthalene by C. glutamicum occurred in a mineral salt medium containing 1% yeast extract without any additional carbon sources. Among the amino acids synthesized via the TCA-cycle, glutamate synthesis increased in C. glutamicum supplemented with 8.5 mM phenol or with 4.2 mM naphthalene. Aspartate synthesis significantly increased when cultured with 4.2 mM naphthalene, and increased synthesis of threonine and histidine was observed only with the addition of phenol. In addition, synthesis of valine and leucine decreased considerably under both conditions. Moreover, the bioconversion of glutamate from phenol and naphthalene is regulated by a transcriptional regulator, FarR, at the transcription level of the gltBD and gdh genes. In this study, we found that the utilization of phenol and naphthalene enhances biosynthesis of several amino acids and that this mechanism is controlled by a transcriptional regulator.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Various mono- and poly-cyclic aromatic compounds in the environment must be removed because they are toxic to cellular systems [7, 23]. Generally, the degradation of aromatic compounds in bacteria proceeds in complex stages, which involve preparation for the ring cleavage of aromatic compounds by a variety of ring modification reactions via mono- or dioxygenases [1]. The catabolism of aromatic compounds was studied in various gram-positive and soil bacteria (i.e., members of the genera Corynebacterium, Rhodococcus, and Streptomyces) that are able to use these components as a sole carbon source [5, 16, 22].
Among them C. glutamicum, which is widely used in the industrial production of many amino acids [13], was reported to assimilate various mono-cyclic aromatic compounds, such as phenol, gentisate, and benzene [5, 22]. For instance, phenol catabolism in C. glutamicum occurs through the β-ketoadipate pathway, and its ring fission is catalyzed by ring-cleavage dioxygenase (ortho-cleavage, catalyzed by intradiol dioxygenases) [7]. After processing the phenol catabolism, the degraded intermediates participate in the energy metabolism-related tricarboxylic acid (TCA) cycle that modulates bacterial physiology and metabolism [4, 17]. For instance, a previous study showed that phenol degradation continued to increase some useful metabolites, and amino acids were produced from an intermediate in the TCA-cycle [14]. In contrast, metabolic pathways for poly-cyclic aromatic compound use are rare in C. glutamicum.
Here, we report our observations on the biodegradation of phenol—as a mono-cyclic aromatic compound—and naphthalene—as a poly-cyclic aromatic compound—in C. glutamicum. We also have proposed a metabolic relationship between phenol and naphthalene use as a carbon source and the production of various amino acids.
Materials and Methods
Bacterial Strains and Growth Conditions
Corynebacterium glutamicum ATCC 13032 (America Type Culture Collection, Manassas, VA) was cultivated routinely at 30°C on Luria–Bertani medium [21]. In order to test its ability to grow with phenol or naphthalene, C. glutamicum was inoculated into a mineral salt medium containing 5.9 mM KH2PO4; 75.7 mM (NH4)2SO4; 4.1 mM MgSO4·7H2O; 8.5 mM Na2HPO4; 11.8 μM MnSO4·H2O; 7.2 μM FeSO4·7H2O; 3.5 μM ZnSO4·7H2O; and 1% yeast extract [12]. The cells were cultivated on a rotary incubator at 150 rpm at 30°C.
Determination of Phenol and Naphthalene Concentration
Triplicate culture media were mixed with an equal volume of ethyl acetate, and residual phenol and naphthalene were extracted. The organic extractions were combined and dried with anhydrous sodium sulfate. The amount of residual phenol and naphthalene in the culture medium was determined by gas chromatography (GC) using an Acme 6000 GC (Young Lin Instrument Co., LTD., Korea) equipped with an HP-5ms capillary column (Agilent Technologies Inc., USA) and a flame ionization detector. The injector and detector were maintained at 200°C, and the column temperature was set to 70°C. All the results represent data from at least three independent experiments and include a mean value.
Analysis of Amino Acid Synthesis
High-performance liquid chromatography (HPLC; Waters Alliance 2690 Analytical HPLC system, Waters Co., USA) was used to identify intracellular amino acids. The system was equipped with a Nova-PakTM C14 column and a Waters 747 scanning fluorescence detector (Waters Co., USA). All the results represent data from at least three independent experiments and include a mean value.
Purification of Histidine-Tagged FarR
The farR coding region (ncgl 2794) was amplified from the chromosomal DNA of C. glutamicum ATCC 13032 by PCR using a primer pair, farRf (5′-ATAGGATCCATG CCTGACCAACCGCTC-3′, BamHI) and farRr (5′-CACAAGCTTGAGATCTTCGGAGCGTGT-3′, HindIII), to generate a fusion protein of FarR and a hexahistidine tag. Purification of the fusion protein was carried out by Ni–NTA affinity chromatography according to the instructions from Qiagen (Hilden, Germany). The purified fusion protein was used directly for the production of polyclonal rabbit antibodies for ChIP assays (Ab Frontier Inc., Korea).
Chromatin Immunoprecipitation Assays
Cultures of C. glutamicum were grown at 30°C for several generations in salt medium containing 1% yeast extract without chemicals, with 8.5 mM phenol, or with 4.2 mM naphthalene. Chromatin immunoprecipitation was carried out using a ChIP Assay Kit (UPSTATE, USA) according to a previously reported protocol [13]. Subsequent PCR amplification of the immunoprecipitated DNA was carried out for 27 cycles using the primer pair gltBD-F (5′-GGATCCATCGGC ACGAA-3′) and gltBD-R (5′-GGCTCCGAGAAACTCTT-3′), which were designed to amplify the upstream of the gltBD gene, and gdh-F (5′-GCACAGATATGACCACA-3′) and gdh-R (5′-TGAGGGCGCTCAATTGT-3′), which were designed to amplify the upstream of the gdh gene [13].
Results
Growth and Degradative Capabilities of C. Glutamicum in the Presence of Phenol and Naphthalene
In bacteria, the degradation of phenol and naphthalene often are limited by toxic effects in the cell (i.e., membrane or protein damage) and by low chemical reactivity due to a lack of functional groups, respectively [23, 24]. Therefore, the appropriate concentrations of phenol and naphthalene were selected to produce amino acids in C. glutamicum. In this study, we found that cell growth in mineral salts media with 1% yeast extract increased greatly with supplementation of phenol or naphthalene (data not shown). Moreover, we found that cell growth was not inhibited until the media were heavily supplemented (8.5 mM phenol and 4.2 mM naphthalene) (data not shown). Therefore, we used 8.5 mM phenol or 4.2 mM naphthalene to evaluate the degradative capabilities of C. glutamicum in all experiments.
In order to investigate the phenol and naphthalene degradative capabilities of C. glutamicum, growth and GC analysis were performed in the presence of 8.5 mM phenol or 4.2 mM naphthalene, and all experimental media were compounded with mineral salt and 1% yeast extract over 48 h (Fig. 1). After 48 h, final degradation efficiencies of phenol and naphthalene by C. glutamicum were ~66.8 and 100%, respectively. In contrast, neither phenol nor naphthalene was degraded without C. glutamicum (data not shown). In addition, the degradation of both phenol and naphthalene occurred rapidly, before the growth status of C. glutamicum reached the stationary phase (within 24 h after inoculation). This suggests that C. glutamicum can use phenol or naphthalene as a carbon source [14, 17, 18].
Time profiles of cell growth, residual phenol, and residual naphthalene during cultivation of C. glutamicum. The cells were grown in a mineral salt medium containing 1% yeast extract and supplemented with 8.5 mM phenol or 4.2 mM naphthalene as a carbon source. The cell growth (closed) and phenol concentration (open) in the presence of 8.5 mM phenol are indicated by circles; the cell growth (closed) and phenol concentration (open) in the presence of 4.2 mM naphthalene are indicated by triangles. The results are reported as the means ± SD (n = 3)
Effect of Amino Acid Synthesis on Phenol and Naphthalene Use
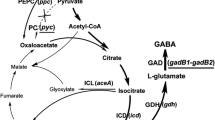
It has been reported that the degraded intermediates from phenol and naphthalene are used as a carbon sources in the TCA-cycle [17, 22]. In addition, most amino acids are derived from intermediates of the TCA cycle in C. glutamicum [4]. For this reason, amino acid production by C. glutamicum in medium containing 1% yeast extract and supplemented with 8.5 mM phenol or 4.2 mM naphthalene was analyzed 48-h post-inoculation (Table 1). Under the 8.5 mM phenol-supplemented condition, the production of threonine, glutamate, and histidine increased 1.4-, 1.2-, and 1.3-fold, respectively, compared to control conditions (no phenol). In addition, when cultured with 4.2 mM naphthalene, aspartate and glutamate production also increased 1.5- and 1.3-fold, respectively, compared to control (no naphthalene). Conversely, valine and leucine production were markedly reduced compared to the control condition when supplemented with both 8.5 mM phenol and 4.2 mM naphthalene. Interestingly, aspartate, threonine, and glutamate are derived from intermediates (e.g., oxaloacetate and 2-oxoglutarate) of the TCA-cycle [4, 8], while valine and leucine are derived from other starting metabolites (e.g., pyruvate) associated with gluconeogenesis [11, 18]. Therefore, it may be concluded that the levels of three metabolites (acetyl-CoA, succinyl-CoA, and fumarate) involved in the TCA-cycle are increased by phenol and naphthalene use, such that aspartate, threonine, and glutamate production are increased in C. glutamicum [17, 22].
Involvement of the Transcriptional Regulator Farr in the Biosynthesis of Glutamate from Phenol and Naphthalene
In contrast to the experiments investigating amino acid production, only glutamate production in C. glutamicum was increased in the presence of both 8.5 mM phenol and 4.2 mM naphthalene (Table 1). Glutamate is synthesized by two differential enzymes: glutamate dehydrogenase encoded by the gdh gene and glutamate synthase encoded by the gltBD gene from 2-oxglutarate of TCA-cycle in C. glutamicum [2, 3]. Recently FarR, which is an uncharacterized Hut/FarR-like transcription factor of the HTH GntR family, has been studied for its ability to repress glutamate biosynthesis in C. glutamicum [9, 20]. Therefore, this study examined the relationship between FarR and two genes encoding glutamate metabolism (gltBD and gdh) cultured with phenol or naphthalene. ChIP assays measured the level of FarR binding upstream of the two genes to determine the regulatory role of FarR in the use of phenol and naphthalene by C. glutamicum (Fig. 2). Putative recognition sites for FarR-DNA binding upstream of the gltBD and gdh genes were selected according to results from our previous study [14]. FarR binds upstream of the gltBD gene when C. glutamicum is grown under control conditions (without chemicals). In contrast, the DNA-binding of FarR completely disappeared in the presence of phenol or naphthalene, but FarR strongly bound upstream of the gdh gene when C. glutamicum was grown in the control conditions. Compared with the control conditions, the DNA-binding affinity of FarR was reduced slightly by supplementation with phenol. Moreover, this phenomenon disappeared under the naphthalene-supplemented condition. When comparing amino acid production (Table 1), the FarR-DNA binding affinity under naphthalene-supplemented conditions was reduced significantly upstream of gdh, compared to the phenol-supplemented conditions. Therefore, glutamate production increased more with naphthalene than with phenol in C. glutamicum. Overall, the combined results strongly support the hypothesis that the biotransformation from phenol and naphthalene to glutamate is regulated by FarR at the transcriptional levels of the gltBD and gdh genes.
ChIP assays of FarR binding upstream of the gltBD and gdh genes. The immunoprecipitate was obtained from culture media at 36 h. The DNA band signals are from DNA binding of FarR in C. glutamicum grown in mineral salt medium containing 1% yeast extract. C control conditions (no chemicals), P supplementation with 8.5 mM phenol, N supplementation with 4.2 mM naphthalene
Discussion
Herein, we reported the amino acid production related to the metabolic pathway for the utilization of phenol and naphthalene by C. glutamicum. Based on these results, the biosynthesis of amino acids derived from the TCA-cycle was enhanced under culture conditions incorporating phenol or naphthalene, and glutamate production from phenol or naphthalene was regulated by the transcriptional factor FarR.
In this study, the degradation of 8.5 mM phenol and 4.2 mM naphthalene was observed in C. glutamicum grown in mineral medium containing 1% yeast extract (Fig. 1). Phenol is degraded through the catechol branch of the β-ketoadipate pathway, and the genes involved in the catechol branch are organized in a single cluster in C. glutamicum [22]. Phenol degradation required the phenol hydroxylase gene (ncgl 2588) for converting phenol into catechol and the cat genes for degrading the resulting catechol [5]. Conversely, the naphthalene catabolic pathway is composed of two processes in naphthalene-degrading bacteria: the upper pathway that converts naphthalene to salicylate by a meta-cleavage catalyzed by extradiol dioxygenases [17] and the lower pathway that converts salicylate to pyruvate and fumarate through the gentisate pathway [1, 17]. The relevant ring-cleavage activity of this pathway is provided by the gentisate 1,2-dioxygenase gene (ncgl 2920) in C. glutamicum [5]. Therefore, it is suggested that naphthalene is used as the carbon source through a degradative process in the gentisate pathway, even though the upper pathway for conversion of naphthalene into salicylate has been not discovered in C. glutamicum. There are some clues regarding naphthalene use in C. gutamicum: (i) it has been reported that C. renale, of the genus Corynebacterium, has naphthalene oxygenase for forming cis-1,2-dihydroxy-1,2-dihydronaphthalene from naphthalene and (ii) a PSI-BLAST search (http://www.ncbi.nlm.nih.gov) of the C. glutamicum ATCC 13032 genome sequence [10] revealed several proteins that are not demonstrated by experiments but probably related to extradiol dioxygenase (i.e., Ncgl2007).
The production of amino acids derived from pyruvate was reduced compared to the control conditions when phenol or naphthalene were added (Table 1). It was demonstrated in an earlier study that the abundance of pyruvate dehydrogenase and citrate synthase increased greatly in C. glutamicum grown with various aromatic compounds (i.e., benzoate, 4-cresol, gentisate, resorcinol, phenol) [18]. This suggests that the pyruvate produced by utilizing aromatic compounds did not convert to amino acids but to citrate, which is an intermediate in the TCA-cycle; thus, the level of intracellular citrate is increased. It also was described that the abundance of enzymes involved in the pentose phosphate pathway (PPP) changes with phenol [18]. Marx et al. [15] reported that increased metabolic activities within PPP could involve metabolite (i.e., histidine and rivoflavin) overproduction from phospho-ribosyl-pyrophosphate. Therefore, we suggest that enhanced histidine production is caused by phenol supplementation in C. glutamicum (Table 1).
As indicated by this study, FarR function might regulate the connecting node of glutamate biosynthesis and central carbon metabolism in C. glutamicum [9]. ChIP experiments showed that the DNA-binding affinity of FarR was stronger on the upstream region of the gdh gene than the gltBD gene (Fig. 2). In E. coli, FarR has been identified as a fatty acid and fatty acyl-CoA-responsive DNA-binding protein, and it autoregulates farR expression [19]. Indeed, multiple sequence alignments of putative FarR-binding regions for three genes (E. coli farR, C. glutamicum farR, and gdh gene) contained a consensus sequence: 5′-CTGGT(T/N)AN(N/A)(T/C)(G/A)ACC(A/T)G-3′ (generated by WebLogo 2.8.2 software [6]). However, the conserved sequence matched less well upstream of the gltBD gene (data not shown). This implies that the activity of glutamate dehydrogenase enzyme is more tightly governed in C. glutamicum by FarR via the glutamate biosynthesis pathway than the activity of glutamate synthase.
Our results show new metabolic processes for the production of amino acids by bio-degradative mechanisms of harmful aromatic compounds (phenol and naphthalene) in C. glutamicum. The production of essential amino acids (i.e., aspartate, glutamate, threonine, and histidine) was enhanced by phenol and naphthalene utilizations. Further, glutamate biosynthesis was regulated by the transcriptional regulator FarR during phenol and naphthalene use by C. glutamicum. The results of this study might be applicable to bioremediation of environments contaminated with polycyclic aromatic hydrocarbons or aromatic compounds.
References
Annweiler E, Richnow HH, Antranikian G et al (2000) Naphthalene degradation and incorporation of naphthalene-derived carbon into biomass by thermophile Bacillus thermoleovarans. Appl Environ Microbiol 66:518–523
Asakura A, Kimura E, Usuda Y et al (2007) Altered metabolic flux due to deletion of odhA causes l-glutamate overproduction in Corynebacterium glutamicum. Appl Environ Microbiol 73:1308–1319
Beckers B, Nolden L, Burkovski A (2001) Glutamate synthase of Corynebacterium glutamicum is not essential for glutamate synthesis and is regulated by the nitrogen status. Microbiology 147:2961–2970
Bott M (2007) Offering surprises: TCA cycle regulation in Corynebacterium glutamicum. Trends Microbiol 15:417–425
Brinkrolf K, Brune I, Tauch A (2006) Transcriptional regulation of catabolic pathways for aromatic compounds in Corynebacterium glutamicum. Genet Mol Res 5:773–789
Crooks GE, Hon G, Chandonia JM et al (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Díaz E (2004) Bacteriol degradation of aromatic pollutants: a paradigm of metabolic versatility. Int Microbiol 7:173–180
Eggeling L, Bott M (2005) Handbook of Corynebacterium glutamicum. CRC Press, Taylor & Francis Group, Boca Raton, FL
Hänßler E, Müller T, Jeßberger N et al (2007) FarR, a putative regulator of amino acid metabolism in Corynebacterium glutamicum. Appl Environ Microbiol 76:625–632
Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62:99–109
Kalinowski J, Bathe B, Bartels D et al (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25
Karigar C, Mahesh A, Nagenahalli M et al (2006) Phenol degradation by immobilized cells of Arthrobacter citreus. Biodegradation 17:47–55
Lee SY, Kim Y-H, Min J (2010) ArgR binding upstream of argB in Corynebacterium glutamicum: DNA-binding affinity of ArgR under proline-supplemented conditions. Appl Microbiol Biotechnol 86:235–242
Lee SY, Shin HS, Park J-S et al (2010) Conversion of phenol to glutamate and proline in Corynebacterium glutamicum is regulated by transcriptional regulator ArgR. Appl Microbiol Biotechnol 85:713–720
Marx A, Striegel K, Graaf AA et al (1997) Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol Bioeng 56:168–180
Meer JR, de Vos WM, Harayama S et al (1992) Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev 56:677–694
Peng R-H, Xiong A-S, Xue Y et al (2008) Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol Rev 32:927–955
Qi S-W, Chaudhry MT, Zhang Y et al (2007) Comparative proteomes of Corynebacterium glutamicum grown on aromatic compounds revealed novel proteins involved in aromatic degradation and a clear link between aromatic catabolism and gluconeogenesis via fructose-1,6-bisphosphatase. Proteomics 7:3775–3787
Quail MA, Dempsey CE, Guest JR (1994) Identification of a fatty acyl responsive regulator (FarR) in Escherichia coli. FEMS Lett 356:183–187
Rigali S, Derouaux A, Giannotta F et al (2002) Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem 277:12507–12515
Sambrook J, Fritsch EF, Maniatis T (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Shen X-H, Huang Y, Liu S-J (2005) Genomic analysis and identification of catabolic pathways for aromatic compounds in Corynebacterium glutamicum. Microbes Environ 20:160–167
Tam LT, Eymann C, Albrecht D et al (2006) Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ Microbiol 8:1408–1427
Widdel F, Rabus R (2001) Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr Opin Biotechnol 12:259–276
Acknowledgments
This work was supported by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korea government (MEST) (No. R01-2008-000-20773-0). The authors are grateful for their support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yang-Hoon Kim is the co-corresponding author of this article.
Rights and permissions
About this article
Cite this article
Lee, S.Y., Le, TH., Chang, ST. et al. Utilization of Phenol and Naphthalene Affects Synthesis of Various Amino Acids in Corynebacterium glutamicum . Curr Microbiol 61, 596–600 (2010). https://doi.org/10.1007/s00284-010-9658-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9658-6