Abstract
Cytoplasmic male sterile (CMS) lines are valuable resources in plant breeding for heterosis utilization, guaranteeing quality of hybrid seeds, and genetic integrity. Nta(sua)S K326, which contains the nuclei of Nicotiana tabacum cv. K326 and cytoplasm of N. suaveolens, is currently the dominant CMS source used in tobacco leaf production in Yunnan, China. Therefore, the alternative CMS lines are required. Here, the Nta(gla)S K326 with cytoplasm of N. glauca has been generated from asymmetric somatic hybridization between protoplasts of K326 and N. glauca followed by backcrossing with K326. Flow cytometry analysis revealed that Nta(gla)S has almost identical ploidy level as that in K326. Moreover, Nta(gla)S, Nta(sua)S, and K326 plants showed similar relative growth rate, yield, and contents of several chemical components in the field. Further scaning electron microscopic observation of floral organs revealed that the Nta(gla)S has shortened filaments and shrivelled stamens, whereas the Nta(sua)S has severe degenerated and carpel-liked stamens, compared with fertile K326 plants. In addition, the expression patterns of several mitochondrial genes in Nta(gla)S, Nta(sua)S, and K326 plants were compared. Furthermore, the cytoplasm source of Nta(gla)S and Nta(sua)S could be distinguished by cleaved amplified polymorphic sequence (CAPS) analysis with chloroplast-specific universal primers. Taken together, these results showed the development of a Nicotiana CMS hybrid via asymmetric somatic hybridization, suggesting that flow cytometry analysis at early stage could facilitate and accelerate the selection process of generating CMS lines. Importantly, the Nta(gla)S provides a valuable germplasm as an alternative CMS line instead of Nta(sua)S in Nicotiana hybrid breeding.
Key message
The Nicotiana CMS hybrid Nta(gla)S has been generated by asymmetric protoplast hybridization. The Nta(gla)S provides a valuable germplasm as an alternative CMS line instead of Nta(sua)S in Nicotiana hybrid breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytoplasmic male sterility (CMS), a maternally inherited trait from mitochondrial-nuclear gene interactions, has been proved to be of great importance in plant breeding for heterosis utilization, quality of hybrid seeds, and genetic integrity, and widely applied for efficient production of hybrids in agricultural production (Chen and Liu 2014; Guo and Liu 2012; Kim and Zhang 2018; Wang et al. 2013). CMS in plant leads to stamen and anther abnormity, even pollen abortion. Thus, CMS is widely used for large-scale hybrid seed production to avoid labor-intensive emasculation or chemical treatment of self-pollinating plant species. In many plant species, CMS has been found from either artificial or spontaneous mutations (Guo and Liu 2012). Additionally, CMS can also be generated through experimental methods such as wide-crossing, sexual introgression, symmetric or asymmetric somatic hybridization of protoplasts, and genetic engineering (Hernould et al. 1993, 1998; Pelletier and Budar 2007).
Among these methods, the asymmetric somatic hybridization (donor-recipient fusion) could alleviate the genome instability resulted from genome incompatibility in symmetric somatic hybridization (Bates 1990; Liu and Deng 1999; Liu and Xia 2014; Matibiri and Mantell 1994). Asymmetric protoplast fusion hybridizes the cytoplasm donor protoplasts, which are normally treated by UV light or irradiation prior to fusion to eliminate the chromosomes in nucleus with the recipient protoplasts or protoplasts that are treated by chemicals such as iodoacetic acid (IOA) prior to fusion for inactivating the mitochondria metabolically in protoplast cytoplasm (Liu and Deng 1999; Trick et al. 1994; Xia et al. 2003; Yamagishi et al. 2002). In some of somatic hybrid plants regenerated from asymmetric somatic hybridization, most of the mitochondria and nucleus originate from cytoplasm donor and recipient protoplasts, respectively (Liu and Deng 1999). Therefore, the CMS trait could be obtained through the newly generated interactions between donor mitochondria and recipient nuclei (Chase 2007; Luo et al. 2013; Xia et al. 2003; Yan et al. 2004).
In most plants, mitochondria and chloroplast are maternally inherited. Due to complex mitochondrial genome (Gualberto and Newton 2017; Hanson 1991; Hu et al. 2014), the chloroplast DNA (cpDNA) molecular markers have been developed to obtaining clear results and overcome the limitations of mitochondrial DNA (mtDNA) markers in Raphanus sativus (Kim et al. 2009). Analysis of ninety accessions of R. sativus with both cpDNA and mtDNA markers revealed that PCR patterns from different CMS origins are perfect match, which suggests that strict co-transmission of mitochondria and chloroplast in radish (Kim et al. 2009). Similarly, the polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) markers, which locate within a chloroplast psbA gene region, could be used to distinguish male-fertile and -sterile cytoplasm in onions (Cho et al. 2006). Importantly, PCR amplification of mtDNA and cpDNA with their corresponding universal primers followed by electrophoresis of either entire or restriction enzyme-digested amplicons, namely cleaved amplified polymorphic sequence (CAPS) analysis, have been proved as an efficient and convenient method for distinguishing the organelle genome composition in cytoplasm (Bastia et al. 2001; Kim et al. 2009).
Flue-cured tobacco (Nicotiana tabacum L.) is one of the most important non-food crop in China, the Nicotiana CMS hybrids have been widely grown in Yunnan Province, which is the largest producing area that accounts for more than 40% of flue-cured tobacco leaf production in China. However, the Nta(sua)S that harbors the nuclei of N. tabacum with cytoplasm from a wild species N. suaveolens is almost the only commercial Nicotiana CMS source used for F1 hybrid seed and flue-cured tobacco leaf production in China (Zheng et al. 2018). The dominant application of Nta(sua)S results in narrowed genetic variation and potential vulnerability to diseases, as the case had been reported in maize carrying CMS-T cytoplasm (Dewey et al. 1988; Levings 1990). Thus, more attempts should be made to develop new Nicotiana CMS sources for reducing the potential risks mentioned above.
Here, we demonstrated generation of a new Nicotiana CMS source Nta(gla)S via flow cytometry analysis at the early stage (BC1R1) after interspecific asymmetric protoplast hybridization. The floral morphology and expression patterns of several mitochondrial genes in Nta(gla)S, Nta(sua)S, and K326 plants were compared by scanning electron microscopy (SEM) and PCR analyses, respectively. The cytoplasm source of two Nicotiana CMS lines was distinguished by CAPS analysis with chloroplast-specific universal primers. In addition, the morphological, agronomical, and chemical characteristics of Nta(gla)S, Nta(sua)S, and K326 plants were compared.
Materials and methods
Plant materials
Seeds of Nicotiana tabacum cv. K326 (fertile K326 as a cytoplasm recipient), N. glauca (as a cytoplasm donor), and male-sterile Nta(sua)S K326 (with cytoplasm from N. suaveolens), and N. suaveolens, which were kept by Yunnan Academy of Tobacco Agricultural Sciences (YATAS), were surface sterilized with 0.1% mercuric chloride for 10 min and then washed three times with sterilized water before germinating on MS basal medium. About 25 days later, the fully expanded leaves were used for mesophyll protoplast isolation.
Protoplast isolation, asymmetric fusion and plant regeneration
Protoplast isolation and treatments, PEG-mediated fusion of protoplasts, protoplast culture, and plant regeneration from micorocalli via tissue culture were conducted as described previously in Chen et al. (2004) with some modifications. The experiments related to protoplasts are described in the Supplementary Data (Methods for protoplast experiments).
UV irradiation and iodoacetic acid (IOA) treatments were used to fragment nuclear genome and inactivate cytoplasm metabolically in mesophyll protoplasts from N. glauca and K326, respectively (Li et al. 2014; Xia et al. 2003; Yamagishi et al. 2002). A treatment of 0.15 mM IOA for 10 min was used to metabolically inactivate cytoplasm of K326 protoplasts according to the previous study (Melchers et al. 1992; Trick et al. 1994; Yamagishi et al. 2002). Polyethylene glycol (PEG)-mediated protoplast fusion and plantlet regeneration were performed as described in Chen et al. (2004). Briefly, agarose beads were used to isolate PEG-mediated fusion of mesophyll protoplasts from the UV-treated N. glauca and IOA-treated K326.
Evaluation of several agronomic characteristics, chemical contents, and economic value
Field trial for several agronomic characteristics of Nta(gla)S K326, Nta(sua)S K326, and male-fertile K326 (MF K326) was conducted in the growth season of 2018–2019 at two field stations of YATAS. The experiment was set up as a randomized complete block design with three replicates/blocks. Each plot size is 40 m2 with 60 plants.
Thirty-seven days after transplanting of seedlings to the field, plant relative growth rate was measured by leaf number per plant and plant height every one week. After topping (72 days after transplanting), the plant height, stem girth, and internode distance were measured at leaf harvesting. After harvesting and flue-curing, the yield and value of tobacco leaves were calculated, contents of total soluble sugar and reducing sugar were measured by colorimetric method, and levels of nicotine were quantified by CORESTA recommended method N° 35: continuous flow analysis (https://www.coresta.org/determination-total-alkaloids-nicotine-tobacco-continuous-flow-analysis-29161.html).
Ploidy analysis
Ploidy analysis of leaves from BC1R1 tobacco plants was conducted via a flow cytometer (Partec PA, Munster, Germany) following the procedures described in Fu et al. (2003). Ploidy analysis of leaves from Nta(gla)S K326 (BC11R1), Nta(sua)S K326, and male-fertile K326 (MF K326) was carried out by a flow cytometer (BD C6, Becton, Dickinson and Company, USA) according to the manufacturer’s instructions.
Scanning electron microscopy
Floral organs collected from 10 plants of Nta(gla)S K326, Nta(sua)S K326, and male-fertile K326 (MF K326), respectively. Flower buds up to 10 mm in length were fixed in Sorensen's phosphate buffer (pH 7.2) containing 2.5% glutaraldehyde (v/v) over a week at 4 °C. The samples were gradually dehydrated in an ethanol series (10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90% (v/v), and absolute ethanol). Each change was for 10–15 min, and three changes of absolute ethanol were made after 1 h followed by three changes of absolute ethanol:amyl acetate [3:1, 1:1, 1:3 (v/v)] for 15 min each, then three times in pure amyl acetate for 15 min each.
Samples were dried by replacing the amyl acetate with CO2 in a critical point dryer. The dried samples were mounted on a brass stub using carbon adhesive tabs, and sputter-coated with gold. Observations were made with a KYKY-EM3200 (KYKY Technology Co., Ltd., China) scanning electron microscope at 25 kV, and the morphology of samples were photographed with a camera attached with the microscope.
Pollen germination analysis
Mature flowers were collected from MF K326 and Nta(gla)S K326 plants. Pollens were gently spread out by a hair on the pollen culture medium (10% sucrose + 0.02% H3BO3 + 2% agar) that was coated on the glass slides. The slides were placed on moist filter paper in a petri dish at 25 °C for 6 h. The germination of pollen tubes were observed under a light microscope (Axio Imager A2, Zeiss, Germany).
PCR assay
Total DNA was isolated from fresh leaves and young buds of six tobacco plants using the DNeasy Plant mini Kit (Qiagen, Germany) respectively. Mitochondria (mt)-specific primers for atp1, atp9, and orf274 were synthesized (Table S1). Fifty ng of DNA was amplified in a volume of 25 µl consisting of 10 mmol l−1 primers, dNTPs Mix (200 mmol l−1 each), 2.5 µl PCR reaction buffer, and 1 U Taq polymerase (Takara Biomedical Technology (Beijing) Co., Ltd., China).
PCR was performed in an Eppendorf 5332 thermocycler (Eppendorf, Germany). The PCR program is: preheating at 94 °C for 2 min; followed by 35 cycles of 30 s at 94 °C for denaturation, 30 s at 56 °C for annealing, and 45 s at 72 °C for extension; and finally ended by 1 cycle of 5 min at 72 °C for extension. The PCR products were separated on 1.2% agarose gel with 5 μg ml−1 ethidium bromide in 1 × TAE buffer, and photographed under UV light.
Transcriptional RT-PCR analysis
Total RNA was isolated using Invitrogen TRIzol reagents (Thermo Fisher Scientific, USA) from six biological replicates of young leaves and buds of Nta(sua)S K326, Nta(gla)S K326 and MF K326, respectively. RNA was digested with RNase-free DNase (Promaga, Madison USA), and 3 µg of total RNA was used for reverse transcription of cDNA synthesis by Invitrogen SuperScript II reverse transcriptase (Thermo Fisher Scientific, USA) following the manufacturer’s instructions.
For transcriptional Reverse Transcription-Polymerase Chain Reaction (RT-PCR) analysis, mitochondria-specific primers were used, and expression levels of Actin gene was analyzed as a loading control for RT-PCR (Table S2). Additionally, a control without addition of reverse transcriptase was also carried out. PCR reactions were performed with 30 cycles for all genes tested in an Eppendorf 5332 thermocycler (Eppendorf, Germany). The PCR-running parameters and visualization of amplified products were identical to those described in PCR assay.
Cleaved amplified polymorphic sequence (CAPS) analysis
The total DNA from six biological replicates of Nta(gla)S K326, Nta(sua)S K326, and male-fertile K326 was extracted with a DNeasy Plant Mini Kit (QIAGEN, Germany). PCR was performed in an Eppendorf 5332 thermocycler (Eppendorf, Germany) using a chloroplast universal primer pair (trnH-trnK, Table S3) as described previously (Chen et al. 2010). The PCR-running parameters and visualization of amplified products were identical to those described in PCR assay. The PCR products were subjected to Dalian for sequencing in Takara Biomedical Technology (Beijing) Co., Ltd. The sequence data was analyzed using the software DNASTAR Lasergene (DNASTAR, Inc., USA).
The PCR products (5 μl) were digested with restriction endonuclease Taq I (4 U) in a 20 μl volume at 65 °C for 4 h. The digested PCR products were electrophoresed in a 2.0% agarose gel with 5 μg ml−1 ethidium bromide in 1 × TAE buffer under 2.5 V cm−1 for 1.5 h, and photographed under UV light.
Data analysis
The data were analyzed using one-way ANOVA [Data Processing System (DPS), China], and the statistical significance of the results was determined using Duncan’s multiple range post hoc test (P < 0.05). The data are presented as mean value ± the standard deviation (SD).
Results
Protoplast isolation, fusion, and plant regeneration
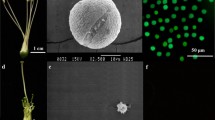
To develop Nicotiana CMS lines, N. glauca (2n = 24) (Fig. 1A) and N. tabacum cv. K326 (2n = 48, hereafter K326) (Fig. 1B) were chosen as cytoplasm donor and recipient, respectively. Increased dosage (treatment time) of UV irradiation resulted in more shrunk/broken N. glauca protoplasts and reduced division percentage of protoplasts (Fig. 1C). The N. glauca protoplasts irradiated with 75.6 J cm−2 UV divided at very low percentage (5%) in the early stage of culture but stopped development eventually. Thus, a UV dosage of 50.4 J cm−2 was chosen to inactivate the nuclear genome of N. glauca protoplasts. Similarly, extended treatment time of 0.15 mM IOA resulted in reduced differentiation percentage of K326 protoplasts (data not shown). Yellow white microcalli was visible after culture of the PEG-treated protoplasts for 25 days (Fig. 2A). The microcalli (2–3 mm in diameter) were transferred to the regeneration medium (Supplementary Data: Methods for protoplast experiments) (Fig. 2B). After another 10-day culture, enlarged callus could be observed (Fig. 2C). Followed by a 35-day culture, shoots and roots regenerated from the callus (Fig. 2D). The regenerated plantlets were transferred to soil for obtaining R1 (N. glauca + K326) plants.
The cytoplasm donor and recipient tobacco for asymmetric protoplast fusion, and effects of UV treatment on the N. glauca protoplasts. A The cytoplasm donor Nicotiana glauca. B The cytoplasm recipient N. tabacum cv. K326. Scale bars: A, 30 cm; B, 20 cm. C Dosage effect of UV treatment on the division percentage of N. glauca protoplasts. The “a, b, c, d” indicate significant difference among the 4 UV dosage treatments
Plantlet regeneration from microcalli after PEG-mediated asymmetric protoplast hybridization between K326 and N. glauca. A Microcalli generation after protoplast fusion of K326 and N. glauca. B Transferred micorocalli. C Enlarged callus from micorocalli. D Shoots and roots induced in regeneration media. Scale bars: A, 0.2 cm; B, 0.2 cm; C, 1.0 cm; D, 1.5 cm
Plant ploidy analysis
In total, 8 R1 (N. glauca + K326) plants were regenerated through tissue culture. Only 2 of the 8 R1 plants can be crossed with pollens from recurrent parent K326, and 64 BC1R1 (R1 × K326) plants were obtained finally. Therefore, we checked the ploidy levels of all the 64 progenies to select the best candidate plant from larger sample population for further efficiently backcrossing via flow cytometer.
To accurately select the candidate plants for further backcross (BC), flow cytometry (FCM) was used to analyze the ploidy levels of the 64 BC1R1 and K326 plants. In FCM assay, five of the 64 BC1R1 plants were found to have the similar fluorescence peaks compared with that in K326 (data not shown). Therefore, one BC1R1 plant D29, which showed the most similar ploidy level to K326, was selected for further backcross (Fig. 3B, E).
Ploidy analysis of BC1R1 and BC11R1 plants via flow cytometer. D29 and D58 are two progeny (BC1R1) obtained from R1 plants (regenerated from asymmetric protoplast hybridization of N. tabacum cv. K326 + N. glauca) backcrossed with K326. The BC1R1 plant (D29) was further backcrossed with recurrent parent K326 for 10 times to generate BC11R1 plants, named Nta(gla)S K326. Ploidy level (measured by a flow cytometer Partec PA) of a K326, the BC1R1 (D29), the BC1R1 (D58) plant were shown in (A–C), respectively. Ploidy level (measured using a flow cytometer BD C6) of a male fertile K326, Nta(gla)S K326 (BC11R1), and Nta(sua)S K326 plants are shown in (G–I), respectively. Growth of a K326, the BC1R1 (D29), and the BC1R1 (D58) plant were shown in (D–F), respectively. Growth of a male-fertile K326, Nta(gla)S K326 (BC11R1), and Nta(sua)S K326 plants were shown in (J–L), respectively. Scale bars in picture D, E, F, J, K, and L are 20 cm
Ploidy analysis revealed that, compared to the ploidy level (Figs. 3A; S1A) of K326 plants (Fig. 3D), the D29 plant (Fig. 3E) has the most similar ploidy level (Figs. 3B; S1B), whereas other BC1R1 plants (for example, the D58 plant, Fig. 3F) showed obviously different ploidy level (Figs. 3C; S1C). A source of CMS, named Nta(gla)S K326 (BC11R1), was achieved after further 10 times of backcrossing the BC1R1 plant D29 with K326. Following ploidy analysis confirmed that the K326, Nta(gla)S K326, and Nta(sua)S K326 plants have the same ploidy level (Fig. 3G–I, respectively). Additionally, the K326, Nta(gla)S K326, and Nta(sua)S K326 plants showed similar growth phenotypes (Fig. 3J–L, respectively).
Evaluating field performances of Nta(gla)S K326
To compare the field performances of Nta(gla)S K326, Nta(sua)S K326, and male-fertile (MF) K326, several agronomic and chemical traits, and economic value were investigated in the field. No significant difference could be found in relative growth rate of Nta(gla)S K326, Nta(sua)S K326, and K326 plants in a successive five-week observation. In the field, Nta(gla)S K326, Nta(sua)S K326, and K326 plants showed similar leaf number per plant, plant height before topping (Fig. 4A, B), and plant height, stem girth, internode distance after topping (Fig. 4C).
Comparison of several agronomic traits of K326, Nta(gla)S K326, and Nta(sua)S K326 in the field. Relative growth of male-fertile (MF) K326 (white column) and two male sterile lines Nta(gla)S K326 (column with slash) and Nta(sua)S K326 (black column) measured by A leaf number per plant, B plant height before topping, and C plant height, stem girth, and internode distance after topping. The “a” indicates there is no significant difference among the traits measured
Importantly, the yield and economic value of Nta(gla)S K326 are higher than those of K326 and Nta(sua)S K326. Nta(gla)S K326 showed higher contents of total soluble sugar, and similar reducing sugar and nicotine levels compared with K326. Moreover, Nta(gla)S K326 has higher contents of total soluble sugar and nicotine, and closer ratio of total soluble sugar/nicotine to K326 than Nta(gla)S does (Table 1). These results suggest that Nta(gla)S K326 performances better than Nta(sua)S K326, and at least as good as the K326, a commercial flue-cured tobacco cultivar grown worldwide regarding to the investigated field traits.
Scanning electron microscopy analysis
Comparison of floral morphology revealed that Nta(gla)S K326 has shortened filaments and shriveled anthers (Figs. 5A, middle; S2A, middle), and Nta(sua)S K326 contains shortened and shrinking filaments, and degenerated carpelloid anthers (Figs. 5A, left; S2A, right), whereas male-fertile (MF) K326 has filaments as long as the style, and anthers covered with mature pollens (Figs. 5A, right; S2A, left).
Morphology and SEM observation of floral buds of Nta(sua)S K326, Nta(gla)S K326, and K326. A Comparison of flower morphology (from left to right: Nta(sua)S K326, Nta(gla)S K326, and K326). B–J Scanning electron microscopic (SEM) observation of the reproductive organs of Nta(sua)S K326 (B, E, H), Nta(gla)S K326 (C, F, I), and the male-fertile maintainer line N. tabacum cv. K326 (K326) (D, G, J). B Carpel-like stamens surround pistil of Nta(sua)S K326. C Wizened stamens (shortened filaments and shriveled anthers) surround pistil of Nta(gla)S K326. D normal stamens surround pistil of K326; E–G ovaries of Nta(sua)S K326, Nta(gla)S K326, and K326, respectively; H splitting stigma of Nta(sua)S K326; I stigma of Nta(gla)S K326; J normal stigma of maintainer line K326. Scale bars: A, 1.0 cm; B–J, 1.0 mm
To investigate the floral morphology of two CMS lines Nta(gla)S K326 and Nta(sua)S K326 in detail, structure of floral organs was further observed by scanning electron microscopy (SEM). The fine architecture of floral organs at late stage of floral development revealed that the Nta(gla)S K326 has wizened stamens (shortened filaments and shriveled anthers) (Fig. 5C), whereas Nta(sua)S K326 has carpel-liked stamens, severe degenerated filaments and carpelliod anthers (Fig. 5B), and split stigma (Fig. 5H), compared with those in MF K326 (Fig. 5D, J). Interestingly, surface of K326 anthers are covered with shining tomentum (Fig. 5D), which is not observed on the anthers of Nta(gla)S K326 and Nta(sua)S K326 (Fig. 5B, C). In addition, the surface structure of ovaries is similar among Nta(gla)S K326, Nta(sua)S K326, and MF K326. However, Nta(sua)S K326 seems has relative shorter vertical length and smaller size of ovary (Fig. 5E), compared with those in Nta(gla)S K326 and MF K326 (Fig. 5F, G).
Furthermore, to confirm the male sterility trait of Nta(gla) S K326, pollen germination assay were performed and pollen germination were observed under light microscope (Zeiss). Pollen tubes could germinate from pollens of MF K326 (Fig. S3, left), but not from pollens of Nta(gla)S K326 (Fig. S3, right).
Analysis of mitochondria-specific genes
Expression of a wheat unedited mitochondria gene atp9 (subunit 9 of the ATP synthase complex) resulted in male sterility such as smaller stamens and anthers, and pollen abortion in Nicotiana (Hernould et al. 1998). In addition, co-transcripts of orf274-atp1 were detected by northen hybridization only in male-sterile Nta(rep)S plants (Bergman et al. 2000). These studies indicates orf274, atp1 (or atpA, F1-ATPase subunitαgene) and atp9 might involve in alloplasmic CMS.
Therefore, three mitochondria genes (atp1, atp9, and orf274) related to CMS were analyzed in leaf and bud tissues of Nicotiana CMS lines. Gene of atp1, atp9, and orf274 were amplified with mitochondria-specific primers (Table S1). Electrophoresis analysis revealed that the sizes of atp9 and orf274 PCR products amplified from total DNA of leaf and bud tissues of K326, Nta(gla)S K326, and Nta(sua)S K326 were similar. Notably, sizes of PCR products for atp1 from leaf and bud tissues of Nta(sua)S K326 seem smaller than those of K326 and Nta(gla)S K326. (Fig. 6A).
Analysis of three mitochondrial genes related to CMS. A The electrophoresis of PCR products of three mitochondrial genes related to CMS. Total DNA amplified with the mitochondrial-specific primers for atp1, atp9, and orf274. Line 1 and 2: leaf and bud samples of MF K326, respectively; Line 3 and 4: leaf and bud samples of Nta(gla)S K326, respectively; Line 5 and 6: leaf and bud samples of Nta(sua)S K326, respectively. M 100 bp DNA ladder. B RT-PCR analysis of the relative expression levels of three mitochondrial genes in leaf and bud samples from MF K326, Nta(gla)S K326, and Nta(sua)S K326, respectively. NtActin gene was served as the control of cDNA input
Transcript analysis of mitochondrial-specific atp1, atp9, and orf274 in K326 and the Nta(gla)S K326, Nta(sua)S K326) and K326 was conducted by Reverse Transcription-Polymerase Chain Reaction (RT-PCR) with mitochondrial-specific primers (Table S2). The expression levels of atp1 and orf274 in leaves and buds of Nta(sua)S K326 and Nta(gla)S K326 are relatively higher and lower than those of MF K326, respectively. The expression levels of atp1 and orf274 in leaves and buds of Nta(sua)S K326 are relatively higher than those of Nta(gla)S K326. The transcripts of atp1 or orf274 accumulate to a similar level in leaves and buds of Nta(gla)S K326. The transcripts of atp1 or orf274 accumulate to a similar level in leaves and buds of Nta(sua)S K326 as well. In K326, the expression levels of atp1 in leaves and buds seems similar, whereas the expression levels of atp9 and orf274 in buds are higher than those in leaves.
For atp9, the amounts of transcripts in leaves of Nta(gla)S K326 and Nta(sua)S K326 is similar and less than those of K326, respectively. Expression levels of atp9 in buds of Nta(gla)S K326 and Nta(sua)S K326 are lower than those of K326. Particularly, the amount of atp9 transcripts in K326 buds is higher than that in K326 leaves, while the opposite trend and no difference was observed in Nta(gla)S K326 and Nta(sua)S K326, respectively. Notably, the product size of atp1 transcripts in leaves and buds of Nta(sua)S K326 seems smaller than those from K326 and Nta(gla)S K326 (Fig. 6B).
Cleaved amplified polymorphic sequence (CAPS) analysis
Due to complexity of Nicotiana mitochondria (mt) genomes, mtDNA markers have limitations for obtaining clear results. In radish, molecular markers were developed based on variation of chloroplast (cp) genome sequences, and analysis of 90 accessions using both mtDNA and cpDNA markers suggested that strict co-transmission of mitochondria and chloroplast (Kim et al. 2009). Thus, universal cpDNA-specific primers were screened for cleaved amplified polymorphic sequence (CAPS) analysis.
Chloroplast-specific primers trnH and trnK were selected for CAPS analysis to distinguish cytoplasm origin of Nta(gla)S K326 from Nta(sua)S K326 (Table S3). Total DNA of Nta(gla)S K326, Nta(sua)S K326, N. glauca, N. suaveolens, and K326 were amplified by PCR with trnH and trnK, and subjected to Sanger sequencing. PCR amplification of total DNA from leaf and bud tissues of K326, Nta(gla)S K326 and Nta(sua)S K326, and leaf samples from N. suaveolens and N. glauca resulted in similar size of bands about 1.7 kb by electrophoresis analysis (Fig. 7A). However, CAPS analysis revealed that PCR products digested by Taq I resulted in three bands, in which the middle band showed different size by electrophoresis, indicating polymorphism of the PCR products (Fig. 7B).
Cleaved Amplified Polymorphic Sequence (CAPS) analysis. Electrophoresis of the PCR products from A total DNA amplified with the chloroplast-specific universal primer pair trnH-trnK, and B the Taq I-digested PCR products in (A). Lane 1 and 2: leaf and bud samples of MF K326 respectively; Lane 3 and 4: leaf and bud samples of Nta(gla)S K326, respectively; Lane 5 and 6: leaf and bud samples of Nta(sua)S K326, respectively; Lane 7: leaf samples of N. suaveolens; Lane 8: leaf samples of N. gluaca. M: 100 bp DNA ladder. C Part of the sequence alignment of PCR products in (A), the NCBI GenBank accession number of PCR products from Nicotiana tabacum cv. K326, N. glauca, and N. suaveolens are HQ602917.1, HQ602920.1, and HQ602931.1, respectively. The digestion sites of Taq I (5′-T/CGA-3′) are labeled by red boxes. (Color figure online)
Sanger sequencing analysis of the PCR products revealed that the size of PCR products with trnH and trnK is around 1.7 kb. Further analysis of sequence alignment showed that PCR products of Nta(sua)S K326 and N. suaveolens harbor a 32-bp deletion and 31-bp deletion (at the position of 161–192 in the consensus sequence) compared with those of Nta(gla)S K326/N. glauca and K326, respectively (Fig. 7C). Furthermore, the PCR products of Nta(sua)S K326, N. suaveolens, Nta(gla)S K326, and N. glauca harbor a 4-bp deletion (5′-GTTA-3′, at the position of 65–68 in the consensus sequence) compared with that of K326 (Fig. 7C).
In addition, some SNPs and three Taq I digestion sites were found within the PCR products of K326, Nta(sua)S K326, N. suaveolens, Nta(gla)S K326, and N. glauca (Fig. S4). The sizes of the fragments between cutting site 1 and 2 of Taq I (the middle band that showed polymorphism, in Fig. 7B) are 394, 396, and 363 bp for K326, Nta(gla)S K326/N. glauca, and Nta(sua)S K326/N. suaveolens, respectively. The sizes of the fragments before cutting site 1 of Taq I (the bottom band in Fig. 7B) are 93 and 89 bp for K326 and Nta(gla)S K326/N. glauca/Nta(sua)S K326/N. suaveolens, respectively. The size of the fragments between cutting site 2 and 3 of Taq I (the top band in Fig. 7B) is 1241 bp (Fig. S4). The Genebank ID deposited in NCBI for the sequences of PCR products from K326, N. glauca, and N. suaveolens are HQ602917.1, HQ602920.1, and HQ602931.1, respectively.
Discussion
In this study, the Nicotiana CMS line Nta(gla)S K326 was generated by asymmetric hybridizing the protoplasts of UV-treated N. glauca and IOA-treated N. tabacum cv. K326 followed by backcrossing with K326. Ploidy levels of the BC1R1 and BC11R1 plants were further analyzed via flow cytometer, and several agronomic, economic, chemical characteristics were evaluated in the field. Moreover, floral morphology of Nta(gla)S K326, Nta(sua)S K326, and K326 was compared by SEM. Additionally, expression pattern of three mitochondrial genes/orfs related to CMS has been investigated. Importantly, a pair of chloroplast-specific universal primers was selected for CAPS analysis to distinguish the cytoplasm origin of Nta(gla)S K326 and Nta(sua)S K326.
Difference in floral morphology between Nta(sua)S and Nta(gla)S
Sexually produced CMS cultivar Nta(sua)S resulted from the introgression of N. tabacum nucleus to the cytoplasm of N. suaveolens after backcrossing for 12 to 16 generations, and the morphological analysis of Nta(sua)S flowers showed normal (not split) corolla but no stamens (Kofer et al. 1990, 1991). However, Fitter et al. (2005) reported that the flowers of somatic hybrid, which derived from the symmetric hybridization of protoplasts from N. tabacum and N. suaveolens, are N. tabcum-like but with a split corolla, carpelloid stamens, and short style. Interestingly, the somatic hybrid plants, Nta(big)S + Nta(und)S and Nta(big)S + Nta(sua)S, derived from symmetric protoplast fusion of Nicotiana CMS cultivars have flowers with novel male-sterile morphology, including split corolla and petaloid stamens (Kofer et al. 1991).
In contrast to the sexual alloplasmic Nta(sua)S line that contain the nucleus of N. tabacum and cytoplasm of N. suaveolens, the N. tabacum + N. suaveolens symmetric somatic hybrid plants harbor mitochondria with recombined mtDNA from N. tabacum and N. suaveolens, chloroplasts from either of N. tabacum and N. suaveolens, and N. tabacum-type nuclear with variable amount of N. suaveolens nuclear DNA (Fitter et al. 2005). Given the similar nuclear background from N. tabacum in Nta(big)S, Nta(und)S, Nta(sua)S, and N. tabacum + N. suaveolens, and the various floral morphology of somatic hybrids is highly correlated with the types (parental or recombined bi-parental) of mitochondrial DNA, the modification of mitochondrial DNA seems to regulate development of petal and stamen in the context of the interactions between the same nucleus with different mitochondria (Fitter et al. 2005; Kofer et al. 1991). Furthermore, the reproductive abnormalities of CMS phenotypes resulted from homeotic changes such as conversion of stamens to petals or carpels have been associated with the transcriptional levels of nuclear genes encoding MADS-box class transcription factors that control floral organ identity, which suggests retrograde regulation of nuclear genes by mitochondrial CMS genes (Carlsson et al. 2008; Chase 2007; Zubko 2004).
In this report, the flower morphology analysis of Nta(sua)S K326, which consists of the cytoplasm of N. suaveolens and nucleus of N. tabacum cv. K326, revealed normal (not split) corolla but severe degenerated and feminized stamens in Nta(sua)S K326 flowers (Fig. 5A the left flower). Particularly, most of Nta(sua)S K326 flowers harbor five degenerated stamens (Fig. S2). The difference in existence/number of stamens and types (split or not) of corolla among the Nta(sua)S reported in Kofer et al. (1991), the Nta(sua)S developed in Fitter et al. (2005), and the Nta(sua)S K326 used in this study might due to the different nuclear-cytoplasmic interactions between the cytoplasm (particularly mitochondria) from N. suaveolens with nuclei from the N. tabacum variety used in report of Kofer et al. (1991), N. tabacum cv. Xanthi or N. tabacum cv. K326 used in this study. Compared with Nta(gla)S K326 and MF K326 plants, Nta(sua)S K326 plants suffered from split stigma and severe degeneration of the stamens, indicating more severe recession of reproductive organs in Nta(sua)S K326. To our knowledge, this is the first SEM observation to compare the floral morphology of the Nta(gla)S K326 and Nta(sua)S K326.
Moreover, the split stigma and more severe degenerated stamens (carpeliod filaments and stigma-like anthers) observed in Nta(sua)S K326 compared with shriveled anthers in Nta(gla)S K326 (Fig. 5A–C, H, I) should be resulted from the specific interactions of the nuclear genes of N. tabacum cv. K326 with N. suaveolens and N. glauca CMS genes, respectively. Thus, the Nta(sua)S K326 and Nta(gla)S K326 somatic hybrids provide valuable materials for deciphering the mitochondrial CMS genes in N. suaveolens and N. glauca with their corresponding nuclear restorers of male fertility (Rf) genes, plant mitochondrial-nuclear interactions, and the molecular mechanisms by which the CMS genes regulate the floral (especially stamen) identity as well (Carlsson et al. 2008; Chase 2007; Chen and Liu 2014; Fishman and Sweigart 2018; Gualberto and Newton 2017; Hanson 1991; Kim and Zhang 2018; Kofer et al. 1991; Meur et al. 2006; Touzet and Budar 2004; Zubko 2004).
CAPS analysis with chloroplast-specific universal primer to distinguish cytoplasmic origin of Nta(sua)S and Nta(gla)S
In most plants, mitochondria and chloroplast are co-inherited maternally. DNA markers for CMS have been developed base on the mitochondria (mt) and chloroplast (cp) genomic sequences (Bohra et al. 2016; Cho et al. 2006; Pelletier and Budar 2007). However, mtDNA markers have limitations for obtaining clear results, due to larger and recombined mitochondrial genome with diverse structure and various isoforms even in a single plant cell (Gualberto and Newton 2017; Hanson 1991; Hu et al. 2014). Therefore, polymorphic DNA sequences in the chloroplast genome could be used to develop CMS markers, based on the conserved sequences and distinctive structure of chloroplast genome (Cho et al. 2016; Kim et al. 2009).
In the study on asymmetric somatic hybridization of protoplasts between Lesquerella fendleri and Orychophragmus violaceus, the atpB-rbcL spacer and aadA16gfp gene fragments carried by L. fendleri chloroplast were amplified via PCR to confirm the chloroplast origin in the intergeneric hybrid plants (Ovcharenko et al. 2011). In onion (Allium cepa), male-fertile and male-sterile cytoplasm were distinguished by PCR–RFLP markers, which amplify a chloroplast psbA gene fragment that could be recognized by a specific restriction enzyme. Both the PCR–RFLP markers and CMS-specific Sequence Characterized Amplified Region (SCAR) markers, which amplify an intergenic spacer (IGS) region of the chloroplast genome, could identify the cytoplasmic male sterile type of 35 onion varieties, suggesting that the chloroplast-specific PCR–RFLP analysis is as reliable as the SCAR method (Cho et al. 2006). Consistently, Kim et al. (2009) reported that cpDNA-based molecular markers developed according to variation of chloroplast genome sequences could distinguish four cytoplasm types in radish (Raphanus sativus). The patterns of PCR products amplified with cpDNA or mtDNA markers showed perfect consistency in the analysis of 90 radish accessions, indicating strict co-transmission of mitochondria and chloroplast in radish.
Similarly, a pair of chloroplast-specific universal primers (trnH-trnK) were selected for CAPS analysis in this study (Fig. 7, Table S3). The PCR products amplified with trnH-trnK were subjected to electrophoresis analysis and Sanger sequencing. Separation by agarose gel could hardly discriminate the PCR products, but could distinguish Taq I-digested PCR products of K326/Nta(gla)S K326 with Nta(sua)S K326 (Fig. 7A, B). Analysis of sequencing results revealed that 31 bp- and 32 bp-deletion exist in amplicons of Nta(sua)S K326 compared with those in K326 and Nta(gla)S K326, respectively (Fig. 7C, Fig. S4). Thus, the size difference among Taq I-digested amplicons of Nta(sua)S K326 and Nta(gla)S K326/K326 could be used as the polymorphic marker for distinguishing the CMS cytoplasm types of Nta(sua)S K326 with Nta(gla)S K326 and K326, as shown in Fig. 7C. However, new molecular markers or CAPS analysis need to be developed for further discriminating the cytoplasm types between Nta(gla)S K326 and K326.
Polymorphic markers have been identified base on the analysis of the sequencing of the chloroplast genomes of male-fertile and CMS lines in soybean. Four from the 23 SNPs identified in gene coding region could be used for PCR–RFLP (CAPS) analysis to distinguish the cytoplasm of CMS with male-fertile lines (Lin et al. 2014). Since chloroplasts and mitochondria are strictly co-transmitted into next generation during reproduction, and the chloroplast genome is relatively stable and conserve compared to hypervariable mitochondrial genome (Cho et al. 2006; Kim et al. 2009), sequencing the chloroplast genomes of Nta(sua)S K326, Nta(gla)S K326 and K326 should be done in the future to discover SNPs for developing CMS molecular markers (Cho et al. 2016; Lin et al. 2014). The Nta(gla)S and Nta(sua)S plants were distinguished at molecular level by CAPS analysis of cpDNA similar to the case of a classical study in citrus (Xiao et al. 2014). However, further functional characterization of mitochondrial genes associated with CMS by analyzing transcriptome, RNA editing, proteome, and transgenic lines, and sequencing mitochondria genome need to be done in the future (Faddetta et al. 2018; Fang et al. 2016, 2020; Garcia et al. 2019; Gualberto and Newton 2017; Storchova 2017; Zheng et al. 2014, 2018).
Characterization of the alternative CMS source for Nicotiana breeding
Flow cytometry (FCM) analysis is a powerful technique frequently used for determining the ploidy level of plants (Xu et al. 2007; Yu et al. 2012). However, most of the reports used FCM to analyze ploidy level of somatic hybrids. Here, our results showed that FCM analysis of the early progenies from asymmetric somatic hybrids could facilitate the selection of regenerated plants for further backcross, and reduce labor intensity during the generation of CMS lines.
The field performance of Nta(sua)S K326, Nta(gla)S K326, and K326 revealed that no significant difference could be observed among these lines regarding to several agronomic, economic, chemical traits in the same K326 nuclear background (Fig. 4, Table 1). Particularly, Nta(gla)S K326 showed higher yield and value, similar contents of reducing sugar and nicotine, and closer ratio of total sugar/nicotine to those in K326 than Nta(sua)S K326 did. The comparison of these characteristics in the field suggests that Nta(gla)S K326 has the potential to be used as an alternative CMS line instead of Nta(sua)S K326 in Nicotiana hybrid producing. However, field experiments in multiple locations and production seasons should be done to further evaluate the performance of Nta(gla)S K326 in the field.
Furthermore, the CMS line Nta(gla)S K326 generated in this study, together with the CMS Nta(sua)S K326 plants, will be used for further molecular analyses of the mitochondria and chloroplast genome, and mitochondrial-nuclear interactions in Nta(gla)S and Nta(sua)S plants (Chase 2007; Cho et al. 2016; Faddetta et al. 2018; Fang et al. 2016, 2020; Gualberto and Newton 2017; Kim and Zhang 2018; Lin et al. 2014; Luo et al. 2013; Touzet and Budar 2004; Zheng et al. 2014, 2018).
Taken together, these results showed that early flow cytometry analysis after asymmetric somatic hybridization could facilitate accurate selecting the BC1R1 plant with the highest similarity of ploidy level to that of recurrent plant for further developing Nicotiana CMS lines efficiently. Importantly, the Nta(gla)S K326 generated in this study provides a valuable germplasm as an alternative CMS source for Nicotiana hybrid breeding.
Abbreviations
- 2, 4-D:
-
2,4-Dichlorophenoxyacetic acid
- BAP:
-
6-Benzylaminopurine
- CMS:
-
Cytoplasmic male sterility
- CAPS:
-
Cleaved amplified polymorphic sequence
- CPW:
-
Cell and protoplast washing solution
- IBA:
-
Indole-3-butyric acid
- IOA:
-
Iodoacetic acid
- MS:
-
Murashige and Skoog
- NAA:
-
Naphthalene acetic acid
- Kn:
-
Kinetin
References
Bastia T, Scotti N, Cardi T (2001) Organelle DNA analysis of Solanum and Brassica somatic hybrids by PCR with ’universal primers’. Theor Appl Genet 102:1265–1272. https://doi.org/10.1007/s001220000508
Bates GW (1990) Asymmetric hybridization between Nicotiana tabacum and N. repanda by donor recipient protoplast fusion: transfer of TMV resistance. Theor Appl Genet 80:481–487. https://doi.org/10.1007/BF00226749
Bergman P, Edqvist J, Farbos I, Glimelius K (2000) Male-sterile tobacco displays abnormal mitochondrial atp1 transcript accumulation and reduced floral ATP/ADP ratio. Plant Mol Biol 42:531–544. https://doi.org/10.1023/a:1006388814458
Bohra A, Jha UC, Adhimoolam P, Bisht D, Singh NP (2016) Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep 35:967–993. https://doi.org/10.1007/s00299-016-1949-3
Carlsson J, Leino M, Sohlberg J, Sundstrom JF, Glimelius K (2008) Mitochondrial regulation of flower development. Mitochondrion 8:74–86. https://doi.org/10.1016/j.mito.2007.09.006
Chase CD (2007) Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet 23:81–90. https://doi.org/10.1016/j.tig.2006.12.004
Chen L, Liu YG (2014) Male sterility and fertility restoration in crops. Annu Rev Plant Biol 65:579–606. https://doi.org/10.1146/annurev-arplant-050213-040119
Chen XJ, Chen MH, Ma WG, Xiao BG, Li YP (2004) A simple method for rapid calli regeneration from symmetric fusion of tobacco. J Yunnan Agric Univ (Nat. Sci.) 19:24–27 (in Chinese)
Chen XJ, Liu Y, Xiao BG, Li MY (2010) Evaluation of flue-cured tobacco varieties introduced from Brazil and Zimbabwe. J Plant Genet Resour 11:503–508 (in Chinese)
Cho KS, Yang TJ, Hong SY, Kwon YS, Woo JG, Park HG (2006) Determination of cytoplasmic male sterile factors in onion plants (Allium cepa L.) using PCR-RFLP and SNP markers. Mol Cells 21:411–417
Cho KS, Cheon KS, Hong SY et al (2016) Complete chloroplast genome sequences of Solanum commersonii and its application to chloroplast genotype in somatic hybrids with Solanum tuberosum. Plant Cell Rep 35:2113–2123. https://doi.org/10.1007/s00299-016-2022-y
Dewey RE, Siedow JN, Timothy DH, Levings CS III (1988) A 13-kilodalton maize mitochondrial protein in E. coli confers sensitivity to Bipolaris maydis toxin. Science 239:293–295. https://doi.org/10.1126/science.3276005
Faddetta T, Abbate L, Renzone G et al (2018) An integrated proteomic and metabolomic study to evaluate the effect of nucleus-cytoplasm interaction in a diploid citrus cybrid between sweet orange and lemon. Plant Mol Biol 98:407–425. https://doi.org/10.1007/s11103-018-0787-9
Fang YN, Zheng BB, Wang L et al (2016) High-throughput sequencing and degradome analysis reveal altered expression of miRNAs and their targets in a male-sterile cybrid pummelo (Citrus grandis). BMC Genom 17:591. https://doi.org/10.1186/s12864-016-2882-0
Fang YN, Yang XM, Jiang N et al (2020) Genome-wide identification and expression profiles of phased siRNAs in a male-sterile somatic cybrid of pummelo (Citrus grandis). Tree Genet Genomes. https://doi.org/10.1007/s11295-020-01437-z
Fishman L, Sweigart AL (2018) When two rights make a wrong: the evolutionary genetics of plant hybrid incompatibilities. Annu Rev Plant Biol 69:707–731. https://doi.org/10.1146/annurev-arplant-042817-040113
Fitter JT, Thomas MR, Niu C, Rose RJ (2005) Investigation of Nicotiana tabacum (+) N. suaveolens cybrids with carpelloid stamens. J Plant Physiol 162:225–235. https://doi.org/10.1016/j.jplph.2004.02.006
Fu CH, Guo WW, Liu JH, Deng XX (2003) Regeneration of Citrus sinensis (+) Clausena lansium intergenetic triploid and tetraploid somatic hybrids and their identification by molecular markets. In Vitro Cell Dev Biol-Plant 39:360–364. https://doi.org/10.1079/IVP2002407
Garcia LE, Zubko MK, Zubko EI, Sanchez-Puerta MV (2019) Elucidating genomic patterns and recombination events in plant cybrid mitochondria. Plant Mol Biol 100:433–450. https://doi.org/10.1007/s11103-019-00869-z
Gualberto JM, Newton KJ (2017) Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu Rev Plant Biol 68:225–252. https://doi.org/10.1146/annurev-arplant-043015-112232
Guo JX, Liu YG (2012) Molecular control of male reproductive development and pollen fertility in rice. J Integr Plant Biol 54:967–978. https://doi.org/10.1111/j.1744-7909.2012.01172.x
Hanson MR (1991) Plant mitochondrial mutations and male sterility. Annu Rev Genet 25:461–486. https://doi.org/10.1146/annurev.ge.25.120191.002333
Hernould M, Suharsono S, Litvak S, Araya A, Mouras A (1993) Male-sterility induction in transgenic tobacco plants with an unedited atp9 mitochondrial gene from wheat. Proc Natl Acad Sci USA 90:2370–2374. https://doi.org/10.1073/pnas.90.6.2370
Hernould M, Suharsono ZE, Carde JP, Litvak S, Araya A, Mouras A (1998) Impairment of tapetum and mitochondria in engineered male-sterile tobacco plants. Plant Mol Biol 36:499–508. https://doi.org/10.1023/a:1005946104983
Hu J, Huang W, Huang Q et al (2014) Mitochondria and cytoplasmic male sterility in plants. Mitochondrion 19(Pt B):282–288. https://doi.org/10.1016/j.mito.2014.02.008
Kim YJ, Zhang D (2018) Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci 23:53–65. https://doi.org/10.1016/j.tplants.2017.10.001
Kim S, Lee YP, Lim H, Ahn Y, Sung SK (2009) Identification of highly variable chloroplast sequences and development of cpDNA-based molecular markers that distinguish four cytoplasm types in radish (Raphanus sativus L.). Theor Appl Genet 119:189–198. https://doi.org/10.1007/s00122-009-1028-z
Kofer W, Glimelius K, Bonnett HT (1990) Modifications of floral development in tobacco induced by fusion of protoplasts of different male-sterile cultivars. Theor Appl Genet 79:97–102. https://doi.org/10.1007/BF00223793
Kofer W, Glimelius K, Bonnett HT (1991) Modifications of mitochondrial DNA cause changes in floral development in homeotic-like mutants of tobacco. Plant Cell 3:759–769. https://doi.org/10.1105/tpc.3.8.759
Levings CS III (1990) The Texas cytoplasm of maize: cytoplasmic male sterility and disease susceptibility. Science 250:942–947. https://doi.org/10.1126/science.250.4983.942
Li C, Cheng A, Wang M, Xia G (2014) Fertile introgression products generated via somatic hybridization between wheat and Thinopyrum intermedium. Plant Cell Rep 33:633–641. https://doi.org/10.1007/s00299-013-1553-8
Lin C, Zhang C, Zhao H et al (2014) Sequencing of the chloroplast genomes of cytoplasmic male-sterile and male-fertile lines of soybean and identification of polymorphic markers. Plant Sci 229:208–214. https://doi.org/10.1016/j.plantsci.2014.09.005
Liu J, Deng X (1999) Production of hybrid calluses via donor-recipient fusion between Microcitrus papuana and Citrus sinensis. Plant Cell Tissue Organ Cult 59:81–87
Liu S, Xia G (2014) The place of asymmetric somatic hybridization in wheat breeding. Plant Cell Rep 33:595–603. https://doi.org/10.1007/s00299-013-1552-9
Luo D, Xu H, Liu Z et al (2013) A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nature Genet 45:573–577. https://doi.org/10.1038/ng.2570
Matibiri EA, Mantell SH (1994) Cybridization in Nicotiana tabacum L. using double inactivation of parental protoplasts and post-fusion selection based on nuclear-encoded and chloroplast-encoded marker genes. Theor Appl Genet 88:1017–1022. https://doi.org/10.1007/BF00220810
Melchers G, Mohri Y, Watanabe K, Wakabayashi S, Harada K (1992) One-step generation of cytoplasmic male sterility by fusion of mitochondrial-inactivated tomato protoplasts with nuclear-inactivated Solanum protoplasts. Proc Natl Acad Sci USA 89:6832–6836. https://doi.org/10.1073/pnas.89.15.6832
Meur G, Gaikwad K, Bhat SR, Prakash S, Kirti PB (2006) Homeotic-like modification of stamens to petals is associated with aberrant mitochondrial gene expression in cytoplasmic male sterile Ogura Brassica juncea. J Genet 85:133–139. https://doi.org/10.1007/bf02729019
Ovcharenko O, Momot V, Cherep N, Sheludko Y, Komarnitsky I, Rudas V, Kuchuk N (2011) Transfer of transformed Lesquerella fendleri (Gray) Wats. chloroplasts into Orychophragmus violaceus (L.) O.E. Schulz by protoplast fusion. Plant Cell Tissue Organ Cult 105:21–27
Pelletier G, Budar F (2007) The molecular biology of cytoplasmically inherited male sterility and prospects for its engineering. Curr Opin Biotechnol 18:121–125. https://doi.org/10.1016/j.copbio.2006.12.002
Storchova H (2017) The role of non-coding RNAs in cytoplasmic male sterility in flowering plants. Int J Mol Sci. https://doi.org/10.3390/ijms18112429
Touzet P, Budar F (2004) Unveiling the molecular arms race between two conflicting genomes in cytoplasmic male sterility? Trends Plant Sci 9:568–570. https://doi.org/10.1016/j.tplants.2004.10.001
Trick H, Zelcer A, Bates GW (1994) Chromosome elimination in asymmetric somatic hybrids: effect of gamma dose and time in culture. Theor Appl Genet 88:965–972. https://doi.org/10.1007/BF00220803
Wang K, Peng X, Ji Y, Yang P, Zhu Y, Li S (2013) Gene, protein, and network of male sterility in rice. Front Plant Sci 4:92. https://doi.org/10.3389/fpls.2013.00092
Xia G, Xiang F, Zhou A, Wang H, Chen H (2003) Asymmetric somatic hybridization between wheat (Triticum aestivum L.) and Agropyron elongatum (Host) Nevishi. Theor Appl Genet 107:299–305. https://doi.org/10.1007/s00122-003-1247-7
Xiao SX, Biswas MK, Li MY, Deng XX, Guo WW (2014) Production and molecular characterization of diploid and tetraploid somatic cybrid plants between male sterile Satsuma mandarin and seedy sweet orange cultivars. Plant Cell Tissue Organ Cult 116:81–88
Xu XY, Hu ZY, Li JF, Liu JH, Deng XX (2007) Asymmetric somatic hybridization between UV-irradiated Citrus unshiu and C. sinensis: regeneration and characterization of hybrid shoots. Plant Cell Rep 26:1263–1273. https://doi.org/10.1007/s00299-007-0350-7
Yamagishi H, Landgren M, Forsberg J, Glimelius K (2002) Production of asymmetric hybrids between Arabidopsis thaliana and Brassica napus utilizing an efficient protoplast culture system. Theor Appl Genet 104:959–964. https://doi.org/10.1007/s00122-002-0881-9
Yan CQ, Qian KX, Yan QS et al (2004) Use of asymmetric somatic hybridization for transfer of the bacterial blight resistance trait from Oryza meyeriana L. to O. sativa L. ssp. japonica. Plant Cell Rep 22:569–575. https://doi.org/10.1007/s00299-003-0732-4
Yu XS, Chu BJ, Liu RE et al (2012) Characteristics of fertile somatic hybrids of G. hirsutum L. and G. trilobum generated via protoplast fusion. Theor Appl Genet 125:1503–1516. https://doi.org/10.1007/s00122-012-1929-0
Zheng BB, Fang YN, Pan ZY et al (2014) iTRAQ based quantitative proteomics analysis revealed alterations of carbohydrate metabolism pathways and mitochondrial proteins in a male sterile cybrid pummelo. J Proteome Res 13:2998–3015. https://doi.org/10.1021/pr500126g
Zheng Y, Liu Z, Sun Y, Liu G, Yang A, Li F (2018) Characterization of genes specific to sua-CMS in Nicotiana tabacum. Plant Cell Rep 37:1245–1255. https://doi.org/10.1007/s00299-018-2309-2
Zubko MK (2004) Mitochondrial tuning fork in nuclear homeotic functions. Trends Plant Sci 9:61–64. https://doi.org/10.1016/j.tplants.2003.12.001
Acknowledgements
This study was funded by the National Natural Science Foundation of China (Nos. 31760072 and 31860413) and the Yunnan Academy of Tobacco Agricultural Sciences (Nos. CNTC-110201302009 and YNTC-2017YN05).
Author information
Authors and Affiliations
Contributions
X-J C, D-H Y, and Y-P L designed the experiments. D-H Y and X-J C wrote the manuscript. X-J C, B-G X and F-C J carried out asymmetric fusion. D-H Y, Z-J T, D-H F, HX and GB carried out the analyses of nucleotide sequences, CAPS, and the expression patterns of several mitochondrial genes. X-F W and Y-H Z carried out the agronomic traits analysis for Nta(gla), Nta(sua), and MF K326 in the field. X-J C carried out flow cytometry analysis and the SEM observation. YL and TP carried out the quantification of chemical components. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Sergio J. Ochatt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, XJ., Tong, ZJ., Xiao, BG. et al. Identification and evaluation of tobacco cytoplasmic male sterile line Nta(gla)S K326 generated from asymmetric protoplast fusion of Nicotiana glauca and N. tabacum followed by backcrossing with N. tabacum K326. Plant Cell Tiss Organ Cult 142, 269–283 (2020). https://doi.org/10.1007/s11240-020-01855-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-020-01855-w