Abstract
Carthamus tinctorius L. (safflower) is an important oilseed crop that is cultivated in several countries. The present study investigates the genetic diversity and population structure of 531 safflower accessions from 43 countries representing all safflower growing regions of the world. Diversity analysis was performed using ten informative EcoRI/MseI amplified fragment length polymorphism primer pairs that were identified by screening 150 primer combinations. The selected primer pairs generated 381 fragments of which 157 were polymorphic among the analyzed accessions. The genetic diversity indices obtained for the entire collection (I = 0.4536, H = 0.2955) indicated high levels of molecular variability. The distance-based, neighbor-joining method classified the accessions into six clusters with internal subgroupings that were in consonance with 19 clusters obtained using Bayesian model-based BAPS analysis. Clusters obtained through STRUCTURE analysis (at K = 4) could not be correlated with their geographically diverse origins, while BAPS analysis (at K = 19) revealed geographical delineation with low admixture levels among most of the studied accessions. Accessions from Far East and Egypt clustered in distinct groups, indicating conserved nature of their gene pools. The Near East and Iran–Afghanistan regions were collectively found to harbor maximum diversity in accordance with earlier reports. Accessions from the Indian subcontinent showed substantial diversity that was previously undetected. The American accessions showed low molecular variability in contrast to earlier studies. Genetic sub-structuring within gene pools and inter-relationships between accessions belonging to different regional pools was also observed. To the best of our knowledge, this is the first comprehensive study of existing genetic variability in a large collection of safflower germplasm with a global distribution, which provides a more accurate representation of genetic structuring in the crop. This information will facilitate selection of elite genotypes for broadening the genetic base of various breeding programs in safflower.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vegetable oil is an essential component of human diet fulfilling the basic requirement of dietary fats. Soybean, rapeseed, cotton, groundnut, sunflower, palm kernels, and copra are the major oilseed crops constituting 97.7 % of global edible oil production (FAO 2013: http://www.fao.org/). A rapidly increasing population, rising health concerns, and geographical and environmental factors have led to a significant increase in edible oil demand. The gap between demand and supply of edible oil is acute in many countries including India, which currently imports more than 50 % of its edible oil requirement (GAIN Report, USDA Foreign Agriculture Service 2013: http://gain.fas.usda.gov/). Exploring alternative sources of edible oil and improving productivity of oilseed crops is therefore, essential to meet the requirements of an increasing global population.
Safflower (Carthamus tinctorius L.), belonging to family Asteraceae, produces high quality edible oil rich in unsaturated fatty acids. The crop is also used as a source of dye for coloring fabrics, flavoring food, and for prevention and treatment of cardiovascular and thrombotic diseases (Weiss 1983; Bankey et al. 1989; Van Zeist and Waterbolk-Van Rooijen 1992; Dajue and Mündel 1996). Its use as an oilseed crop was considered to be relatively recent beginning from the first half of the twentieth century (Dajue and Mündel 1996). However, archaeo-botanical evidence suggests its cultivation as a major oil crop in the Near East dating back to the Bronze Age (Marinova and Riehl 2009). It has also been used as a platform for production of plant-derived pharmaceuticals (McPherson et al. 2009; Flider 2013; Carlsson et al. 2014), biofuels, and as a source of industrial oil (Velasco et al. 2005; Ilkılıç et al. 2011). Safflower is cultivated in arid and semi-arid conditions in various agricultural zones across the world (Weiss 1983). India was the leading producer of safflower for the past two decades with maximum average production and largest area under cultivation. In 2012, Mexico surpassed India in safflower production with an improved average yield of 2503 kg/ha by the use of high yielding cultivars developed through breeding programs, while the average yield in India remained substantially lower at 660 kg/ha (FAO 2012). The high prospects of yield enhancement of safflower favor increasing its use as an alternative to other oilseed crops. However, low oil content, spiny nature of many cultivars, susceptibility to various biotic and abiotic stresses, and high input and purchase cost discourage large-scale cultivation of safflower (Nimbkar 2008), leading to drastic fluctuations in annual acreage and productivity of the crop (Dajue and Mündel 1996; Sujatha 2008). The above limitations can be resolved by concerted breeding programs using diverse genetic stocks and elite genotypes, which will be facilitated by the analysis of genetic diversity and population structure in safflower.
Pioneering studies in safflower focused on use of morphological traits and geographical origin for analysis of germplasm diversity and structure. Vavilov (1951) proposed three regions—India, Afghanistan, and Ethiopia—as ‘centers of origin’ of safflower on the basis of observed variability, distribution of wild species, and cultural representation of the crop. Safflower is considered to have a Near Eastern origin from where it spread to different geographical regions across the world (Ashri and Knowles 1960; Smith 1996). Knowles (1969) classified the available genotypes of safflower into seven “centers of similarity” based on morphological descriptors. Differences among genotypes for yield components, oil composition, and response towards diseases were also analyzed in subsequent studies (Ashri 1971a,b, 1975; Ashri et al. 1974). Application of morphological descriptors and geographical origin to assess the variability of around 2000 lines of safflower revealed a wide range of phenotypic variation for several traits, based on which safflower accessions were classified into 10 “regional gene pools” (Ashri 1975). Several recent studies used molecular markers, viz., random amplified polymorphic DNA (RAPD), inter-simple sequence repeat (ISSR), amplified fragment length polymorphism (AFLP), sequence-related amplified polymorphism (SRAP), and simple sequence repeat (SSR), for variation analysis either exclusively or in conjunction with morphological data for the crop (Sehgal and Raina 2005; Johnson et al. 2007; Yang et al. 2007; Amini et al. 2008; Peng et al. 2008; Chapman et al. 2009, 2010; Khan et al. 2009; Mahasi et al. 2009; Hamdan et al. 2011; Barati and Arzani 2012; Mokhtari et al. 2013; Lee et al. 2014). However, low sample size from selected regional gene pools and lack of a global representation of available germplasm were limitations in most of these studies, which may therefore not provide an accurate representation of the available genetic diversity and population structure in safflower. The sample size in these studies ranged from a minimum of 14 to a maximum of 100 accessions/cultivars.
Sehgal and Raina (2005) used RAPD, ISSR, and AFLP markers to characterize 14 Indian cultivars and found AFLP to be the most effective system among the three types of markers for assessing safflower variability. Although the dominant nature of AFLP markers under-represents the allelic frequencies, its advantages, viz., high repeatability, generation of large amount of data, multi-locus analysis, and ease of automation in the absence of prior sequence information, makes it a reliable tool for less explored crops like safflower. Johnson et al. (2007) utilized AFLP data to evaluate genetic diversity among 96 accessions and were able to validate three centers of similarity out of the seven centers proposed by Knowles (1969). No distinct correlation could be derived between the other described groups and their genetic components. Chapman et al. (2010) used 24 EST-SSR markers to survey a representative germplasm collection of 76 accessions belonging to ten centers proposed by Ashri (1975) and re-designated them to five distinct clusters. Recently, Lee et al. (2014) proposed three clusters along with few admixture groupings by analyzing 100 safflower accessions from Far East (mainly Korea), India–Pakistan, and Middle East regional gene pools using 30 microsatellite markers. These admixtures indicated that gene flow occurred between gene pools as a consequence of natural selection and human intervention.
In the present study, we analyzed the genetic structure of a global collection of safflower germplasm and studied the extent of diversity in different regional accessions. A novel set of AFLP markers were generated and utilized for diversity assessment on a large panel of 531 safflower accessions representing all the regional pools described by Ashri (1975). We also included accessions from regions of secondary introduction of safflower. Grouping of individuals into well-defined clusters was done using both distance-based and Bayesian model-based methods. Association between genetic differentiation and geographical distinction of safflower accessions was analyzed. To the best of our knowledge, this is the first comprehensive study involving the largest number of accessions representing all the safflower growing regions of the world. Results obtained from this study will broaden the available genetic base for selection of elite parental lines for crop improvement programs.
Materials and Methods
Plant Material
A collection of C. tinctorius L. seed samples comprising 2300 accessions representing the ten regional gene pools (Ashri 1975) across the globe was obtained from USDA-ARS, WRPIS, Pullman, WA, USA. Detailed information on these accessions is available at the Germplasm Resources Information Network (GRIN; http://www.ars-grin.gov/npgs/). In this study, we selected 531 accessions representing 43 countries including regions of secondary introduction (Fig. 1). It also includes five Indian cultivars (Sharda, Manjira, Annigeri, PBNS 12, and TSF-1), which were obtained from the National Bureau of Plant Genetic Resources, New Delhi, and University of Agricultural Sciences, Dharwad, India. Two accessions were obtained from IPK Gene Bank, Germany (Table 1). The accessions were selected based on USDA passport data for geographical distribution and diversity of phenotypic traits. A list of accessions with their PI number and country of origin are given in Supplementary Table 1.
Geographical representation of safflower growing regions evaluated in the current study. (http://wiki.alternatehistory.com/doku.php/blank_map_directory/world_gallery_4)
Genomic DNA Extraction
Young leaves were harvested from 10-week-old plants of each accession and freeze-dried. Genomic DNA was isolated using the cetyltrimethylammonium bromide (CTAB) method (Doyle 1991). The quality and quantity of extracted DNA was assessed by gel electrophoresis and spectrophotometry. A final working concentration of 500 ng/μL of DNA was prepared and used for further studies.
AFLP Fingerprinting
AFLP fingerprinting was performed following Vos et al. (1995). Briefly, 500 ng/μL of genomic DNA was digested for 16 h at 37 °C with 10 U EcoRI and 5 U MseI (New England Bio Labs, USA) in a final reaction volume of 12.5 μL. Reactions were terminated by heating the samples at 72 °C for 15 min. EcoRI (5 pmol/μL) and MseI (50 pmol/μL) adaptors were ligated to 5 μL of digested DNA with 5 U T4 DNA ligase (Promega, Switzerland) in 1× ligase buffer at 20 °C for 16 h. For pre-amplification, primers complementary to the adapters and restriction site with one additional selective base (EcoRI, 5′-GACTGCGTACCAATTC-A-3′; MseI, 5′- GATGAGTCCTGAGTAA-C-3′; Integrated DNA Technology, USA) were used. The thermo-cycler was programmed at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min for 30 cycles. The reaction mixture consisted of 10× PCR buffer (2 μL), 25 mM MgCl2 (1.6 μL), 10 mM dNTP mix (1 μL), 10 μM EcoRI primer (1 μL), 10 μM MseI primer (1 μL), and Taq polymerase (3 U; Intron, Korea) in a final reaction volume of 20 μL. Pre-amplified products were analyzed on 1.5 % agarose gel and diluted 40-fold. The diluted template (2.5 μL) was used for selective amplification with different combinations of IR-700 labeled EcoRI primer and unlabeled MseI primer (Integrated DNA Technology, USA) with three to four bases selection. The reaction mixture comprised of 10× PCR buffer (1 μL), 25 mM MgCl2 (0.8 μL), 10 mM dNTP mix (0.25 μL), 1 μM labeled EcoRIprimer (0.5 μL), 10 μM MseI primer (0.5 μL), and Taq polymerase (2.5 U; Intron, Korea) in a final reaction volume of 10 μL. A touchdown PCR program was used for selective amplification at 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 1 min followed by decrement of 0.8 °C per cycle in the annealing temperature for the next 12 cycles and final amplification for 25 cycles at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min. The amplified products were resolved on a 6 % denaturing polyacrylamide gel at 1000 W, 40 °C in 1× TBE for 3.5 h using the LICOR-4300 DNA analyzer system. A preliminary study with 150 different primer combinations of EcoRI/MseI was done on a panel of 10 selected accessions representing all the regional gene pools described by Ashri (1975) to check for amplification efficiency and frequency of polymorphism. Ten most informative primer combinations generating highest number of polymorphic bands among ten accessions were selected from the above set and used for diversity analysis of 531 accessions.
Genetic Analysis and Construction of Distance-Based Dendrogram
The AFLP genotyping data was assembled into a binary matrix by scoring unambiguous polymorphic bands manually. Presence of a band was denoted as “1,” and absence was marked as “0”. The extent of overall genetic diversity and within-region genetic diversity was assessed through computation of percentage of polymorphic loci (P), Nei’s expected gene diversity (I), and Shannon’s information index (H) using POPGENE version 1.32 (Yeh et al. 1999). Genetic distances between regions were evaluated by Nei’s unbiased genetic distance implemented in POPGENE. The binary data were also used to determine pairwise distance between accessions according to Dice’s similarity coefficient. The distance matrix was used to construct a dendrogram by neighbor-joining (NJ) method with 1000 bootstrap replicates, and for principal coordinate analysis (PCoA) using DARwin Version 5.0.155 (http://darwin.cirad.fr/darwin; Perrier and Jacquemoud-Collet 2006). Analysis of molecular variance (AMOVA) within and between gene pools on the basis of (a) geographical origin and (b) groups derived from population structure analysis (see below) was performed using GENALEX version 6.5 (Peakall and Smouse 2012).
Population Structure Analysis Using Bayesian Clustering Methods
Population stratification for estimation of the number of hypothetical sub-populations (K) within the studied germplasm was performed using Bayesian methods implemented in two software packages, STRUCTURE version 2.3.4 (Pritchard et al. 2000) and BAPS version 6.0 (Corander et al. 2004). Regional classification of the accessions was not taken into consideration during analysis in order to develop independent groups based solely on molecular marker data. In STRUCTURE, hypothetical sub-populations ranging from K = 1 to 25 were tested, with five independent runs at each K value. An admixture ancestry model with correlated allele frequencies between populations was followed (Falush et al. 2003). The burn-in period for each run was set at 100,000 generations and 200,000 MCMC repeats, and the optimum K value for the dataset was calculated as per Evanno et al. (2005). A threshold value of 80 % or greater shared ancestry was considered optimal for classifying an accession as pure, while accessions with lower values were considered as admixtures (Carrasco et al. 2009; Lee et al. 2014). The program BAPS v 6.0 was used for the identification of hidden population structures (Corander and Marttinen 2006). A non-spatial model at the individual level was used for mixture analysis to estimate the sub populations (K = 1–25) with five repeats for each run following the user manual. The mixture analysis was used as input for admixture analysis followed by estimation of gene flow among the derived clusters. In gene flow graph, the threshold value for the significant P values was set to 0.01 for pruning of insignificant edges. In both the programs, identified clusters with more than 50 % of representative population from a particular geographical region were assigned to that region (Johnson et al. 2007).
Results
AFLP Fingerprinting
Ten most informative primer combinations identified from preliminary studies (see “Materials and Methods”) produced 381 discrete amplified fragments of which 157 (41 %) were polymorphic (Table 2). The amplified bands were in a size range of 60–700 base pairs with an average of 38 fragments per primer combination. The total number of bands per primer pair varied between 30 to 48, and the number of polymorphic bands per primer combination ranged from a minimum of 10 (E-ACG/M-CCCA) to a maximum of 21 (E-AAC/M-CGAC). The primer pair E-AAC/M-CGAC was most successful in discriminating the genotyped individuals with a polymorphism rate of 53.9 %. The least discriminatory among the primers was E-AAC/M-CCCA with a polymorphism rate of 32.4 % (Table 2). To check the reproducibility of the AFLP profile, 62 accessions ( ̴ 12 % of the total panel size) were tested in independent replicates with the ten primer combinations starting from the initial step of restriction digestion of genomic DNA. The resulting average reproducibility of the bands was found to be 98 % among the replicated accessions (data not shown).
Genetic Analysis and Construction of Distance-Based Dendrogram
Genetic analysis of 531 accessions using 157 polymorphic markers resulted in Nei’s expected heterozygosity (I) values ranging from 0.1549 to 0.2850, with an average of 0.2955. The Shannon information index (H) ranged from 0.2359 to 0.4364 and averaged at a value of 0.4536 (Table 3). The percentage of polymorphic loci was found to be highest among accessions from the Indian subcontinent (IS) (93.63 %) followed by Iran–Afghanistan (IA) and American (US) regions (92.99 %). The markers were least informative for evaluation of genetic diversity of genotypes from Ethiopian (ET) region with the percentage polymorphism restricted to 39.49 %. The values for both Shannon information index and Nei’s gene diversity were highest in Iran–Afghanistan (0.4364 and 0.2850, respectively) and lowest in Sudan (SU) (0.2359 and 0.1549, respectively). The pairwise genetic distance value calculated through AFLP analysis ranged from 0.0405 to 0.2372 (Table 4). Near East (NE) and Europe (EU) were most similar with a minimum genetic distance of 0.0405, while Kenya (KE) and Australia (AUS) were most distant with a genetic distance value of 0.2372.
Analysis of the AFLP data using neighbor-joining method grouped the sampled accessions into six major clusters (designated as NJcl I–VI) with further internal subgroupings (Fig. 2). A low bootstrap support for majority of the branches was obtained. The NJcl I cluster (58 accessions) has a major proportion of individuals belonging to Far East (FE) region (51) with minor representation from Indian subcontinent (5), Near East (1), and European region (1). The NJcl II cluster consisting of 100 accessions had representation from regional pools of Indian subcontinent (52), Iran–Afghanistan (27), Near East (10), Europe (10), and America (1). NJcl III (30 accessions) consisted of a majority of germplasm from Egypt (EG) (19) and a few accessions from Indian subcontinent (4), Europe (4), Ethiopia (1), America (1), and Unknown (1). Iran–Afghanistan, Near East, and Far East accessions were completely absent from this cluster. NJcl IV (126 accessions) grouped individuals from most of the regional gene pools. The Indian subcontinent dominated this cluster (86) along with representation from Far East (14), Europe (9), Iran–Afghanistan (7), America (4), Near East and Australia (2 each), Turkey (TU), and Ethiopia (1 each). Similar to NJcl IV, NJcl V (75 accessions) also showed a mixed representation from Turkey (20), Indian subcontinent (18), Europe (11), Sudan (10), Near East (6), Iran–Afghanistan (4), 2 accessions each from Far East and America, and 1 each from Egypt and Kenya. Four of the five Indian cultivars (PBNS-12, Sharda, Manjira, Annigeri) used in this study also clustered in NJcl V. The largest group, NJcl VI (142 accessions) contained 104 accessions of America along with accessions from Iran–Afghanistan (15), Near East (7), Turkey (8), Europe (2), Unknown (2), and Australia (1). The Indian subcontinent was poorly represented in this cluster with only two accessions (including the Indian cultivar TSF-1), while Far East was completely absent.
In PCoA, axes 1 and 2 explained 30 and 26 % of the total existing variance, respectively, in the studied germplasm. The accessions were distributed in all the four quadrants, indicating that the global collection of safflower germplasm harbors substantial genetic diversity. Geographical clustering was also evident in the distributed accessions (Fig. 3a). Majority of Indian accessions were distributed in quadrants I and IV. Most accessions from Far East were detected in quadrant IV with minor representation in the other three quadrants. Accessions from Iran–Afghanistan and Turkey were represented in quadrants I and II, while those from Near East segregated in quadrants II and III. Sudanese accessions were present mainly in quadrant II, and accessions from Europe showed a wide distribution among all the quadrants. Most of the American and Egyptian accessions clustered together in quadrant III. However, axes 1 and 4, representing 30 and 14 % of variance, separated the Egyptian and American accessions into quadrants III and II, respectively (Fig. 3b). Six major clusters obtained by NJ analysis (NJcl I–VI) are indicated in PCoA to understand the congruence between the two distance-based methods.
Principal coordinate analysis on the basis of 157 polymorphic AFLP markers: a axes 1 and 2 and b axes 1 and 4. Percent variance explained by each axis is provided. NJ clusters are represented as indicative boundaries in PCoA. Cluster NJcl V is not marked with a boundary due to its wide distribution among different quadrants. Region wise color codes are provided in the figure
Population Structure Analysis Using Bayesian Clustering Methods
Cluster Formation Using STRUCTURE
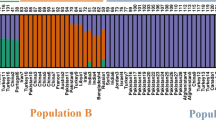
The relatedness among genotypes as well as their rooting with the geographical designation was studied by population structure analysis. Determination of log mean probability and change in log probability (∆K) following Evanno et al. (2005) provided two estimates of K at K = 2 and K = 4 (Fig. 4). Population structure inferred using STRUCTURE v2.3.4 at K = 2 distributed most accessions from Far East and Indian subcontinent into group I while accessions from all the other regions clustered in group II (data not shown). Analysis at K = 4 (Fig. 5a) delineated the studied germplasm into four groups (STRcl I, STRcl II, STRcl III, and STRcl IV). STRcl I, STRcl III, and STRcl IV were dominated by genotypes belonging to Far East (93 %), America (61 %), and the Indian sub-continent (92 %), respectively. STRcl II exhibited mixed allelic frequency clustering individuals primarily from Iran–Afghanistan (23 %), Europe (20 %), Turkey (16 %), and Indian subcontinent (13 %). The detailed contribution of each regional pool with number of accessions in each cluster is provided in Table 5 and Fig. 5a. The mean alpha value (an estimate of the degree of admixtures) for the analyzed samples was 0.1946.
Population structure of 531 safflower accessions at K = 4, derived using the model based Bayesian algorithm implemented in a STRUCTURE and b BAPS. Each sub-population is represented by a different color code. Each vertical bar represents a single safflower accession. Admixed individuals have bars composed of multiple colors
Cluster Formation Using BAPS
BAPS analysis was initially performed at K = 4 using the ∆K value obtained by STRUCTURE as described in earlier studies (Fang et al. 2013). BAPS analysis at K = 4 was done with five replications, and the results were compared with those obtained using STRUCTURE analysis (Fig. 5b). The four sub-populations were titled as BAPS I–IV. A major shifting of accessions (mainly from the Indian subcontinent) from STRcl IV into BAPS I was observed. BAPS I brought together accessions mainly from the Indian subcontinent (76 including 3 Indian cultivars) and Far East (41). Of the total number of accessions present in this cluster, 42 accessions belonging to Far East were seen in STRcl I. It also consisted of 82 accessions of STRcl IV, 2 accessions of STRcl III, and 1 accession from STRcl II. BAPS II provided a heterogeneous clustering of germplasm lines from America (97), Egypt (19), Far East (14), Near East (8), Indian subcontinent (6 including 1 Indian cultivar), Europe (4), Sudan (3), Iran–Afghanistan (3), Ethiopia (2), and Australia (1). This cluster corresponded to STRcl III except for 11 accessions that grouped in other clusters of STRUCTURE analysis. The third group, BAPS III, coincided with cluster STRcl II and comprised of accessions from different regions, viz., Iran–Afghanistan (51), Indian subcontinent (29 including 1 Indian cultivar), Turkey (28), Europe (28), Near East (17), America (14), Far East (12), Sudan (7), and 1 accession each from Egypt, Kenya, Australia, and unknown region. BAPS IV consisted mainly of accessions from the Indian subcontinent (56) and was similar to STRcl IV of STRUCTURE analysis except of one accession from STRcl II of STRUCTURE analysis. A comparative analysis of the distribution of individuals belonging to respective regional pools in different clusters of BAPS and STRUCTURE is given in Table 5. Thus, population differentiation at K = 4 using both STRUCTURE and BAPS was not able to discern correlation between the studied accessions and their geographical origins.
BAPS analysis was therefore repeated without pre-defining a fixed value of K in order to develop a better understanding of the internal structuring. K values ranging from 1 to 25 were tested for the entire panel, which resulted in formation of 19 optimal clusters designated as BAcl 1–19 (Fig. 6a; Table 6). Genotypes from Far East assembled in BAcl 1, 2, and 15 with few accessions grouping out in BAcl 5, 7, 12, and 17. BAcl 8 had a homogenous constitution grouping Turkish genotypes together. A few accessions from Turkey were also present in BAcl 11 and 12. Another regional pool for safflower, Egypt clustered primarily in BAcl 5 with only one individual segregating in BAcl 12. Similar to the results derived from NJ analysis, most of the genotypes belonging to the American region isolated into distinct clusters (BAcl 6, 10, 13, and 14). Interestingly, accessions from the Indian subcontinent were most widespread and dominated four clusters (BAcl 4, 7, 16, and 18). Some accessions from the Indian subcontinent shared the gene pool with accessions from other regions in BAcl 5, 9, 10, 11, 12, 17, and 19. The clusters BAcl 9, 11, 12, 17, and 19 were highly heterogeneous and showed the presence of accessions from different regions. Sudanese accessions were restricted to BAcl 12. Accessions from Iran–Afghanistan region were the major constituent of BAcl 3 and also had a diffused presence in most of the clusters. The Near East and European regions too followed a similar pattern of distribution as Iran–Afghanistan and could not be assigned to a specific cluster (Table 6).
BAPS analysis of 531 safflower accessions at K = 19. a Admixture analysis of 19 sub-populations. In the graph, each color represents a population group. The derived 19 groups are designated as Bacl1-19. Admixtures depending on the proportion of genetic blending have bars composed of multiple colors. b Gene flow network identified between the derived sub-populations. In the graph, bold arrows show fraction of variation arising within clusters, while dashed arrows indicate gene flow between clusters
The BAPS program was also used to identify mosaic groups, which contained alleles contributed by more than one sub-population and to estimate the pattern of gene flow between and within the clusters. In order to derive useful information from the gene flow analysis, the network was pruned by removing gene inflow or outflow representing <1 % allelic movement (Fig. 6b). BAcl 6, 10, 13, and 19 showed no input gene flow from any other cluster. BAcl 8 consisted of accessions exclusively from Turkey and contributed alleles to BAcl 2, 11, and 12. The groups BAcl 1, 3, 4, 11, 12, 14, 16, 17, and 18 showed gene flow ≥95 % within the respective cluster. The heterogeneous BAPS clusters BAcl 9, 15, 2, 5 and 7 show 94, 93, 89, 89, and 80 % gene flow, respectively, within the clusters. The most heterogeneous BAPS cluster was BAcl 7 with 80 % gene flow within the cluster and inflow from BAcl 4 (6.7 %), 1 (4.5 %), 17 (1.6 %), 5 (1.3 %), 18 (1.2 %), and 16 (1 %). This cluster is composed of accessions mainly from the Indian subcontinent and European region. BAcl 2 (accessions from Far East), with 89 % gene flow within the cluster, had inflow from BAcl 1 (7.6 %), 8 (1.2 %), and 17 (1.1 %). BAcl 5, dominated by accessions from Egypt along with Indian subcontinent and Europe, showed 89 % gene flow within the cluster and had inflow from BAcl 16 (5.6 %) and 1 (1.9 %).
AMOVA Analysis
The molecular variance within and between the regional pools as well as the sub-populations derived from STRUCTURE and BAPS analysis was evaluated (Table 7). For regional gene pools, percent variance within and among populations was found to be 80 and 20 % of the total variation, respectively. The four sub-populations derived from STRUCTURE showed 79 and 21 % of total variance within and between the groups. BAPS at K = 4 provided a variance of 18 and 82 % within and between sub-populations, respectively. The 19 sub-populations deduced from BAPS have a minimum within-population variance of 70 % and maximum variance of 30 % among populations indicating that the derived nineteen sub-populations are fairly structured groups for the panel under consideration.
Discussion
The present study was initiated with the objective of assessing the genetic diversity and population structure of a global collection of safflower germplasm. Effective assessment of population structure requires a good representative sample population along with a significant number of molecular markers (Ramu et al. 2013). In the current study, we used an exhaustive reference set of 531 accessions belonging to all the ten regional pools reported for safflower (Ashri 1975) along with accessions from regions of secondary introduction. Although India is one of the major producers of the crop, germplasm from the Indian subcontinent has not been comprehensively evaluated for its genetic diversity. Therefore, we included 167 accessions belonging to the Indian subcontinent (IS) in our study. The development of 157 novel polymorphic markers through AFLP fingerprinting in this study strengthened the available repertoire of molecular markers in the crop (Johnson et al. 2007; Chapman et al. 2009; Hamdan et al. 2011; Lee et al. 2014; Pearl et al. 2014) and successfully established high levels of genetic distinctness (I = 0.4536, H = 0.2955) among the studied safflower genotypes. Accessions from Iran–Afghanistan and Near East were genetically most diverse, while Sudan was found to have the least molecular diversity (Table 3). Safflower, owing to its widespread geographical distribution (Fig. 1), is expected to harbor significant genetic diversity. This was also demonstrated in PCoA analysis in which accessions analyzed were widely distributed in all the quadrants (Fig. 3).
A key objective of the present study was to identify correspondence, if any, between genetic diversity of the safflower germplasm and geographical zones. Analysis of population structure was performed using distance-based (NJ and PCoA) and Bayesian model-based clustering methods. PCoA supported groupings obtained through NJ method. However, structuring of the germplasm at K = 4 (based on estimated ∆K value; Evanno et al. 2005) using STRUCTURE and BAPS were unable to segregate the accessions based on geographical distinction. BAPS analysis was therefore performed using a non-spatial approach with K values ranging from 1 to 25, which identified the most appropriate clustering of accessions at K = 19. This was also in accordance with results obtained through NJ analysis. Bayesian clustering also discerned the gene flow network among the derived subpopulations (Fig. 6b) and complemented the analysis of distance-based method and population structure. Similar results were obtained in earlier studies where BAPS was able to discern meaningful genetic structure in comparison to STRUCTURE (Hanage et al. 2009; Tollefsrud et al. 2009; Smýkal et al. 2011). The possible explanation for the observed discrepancy between the outputs of the two programs may be due to the different algorithms used in the programs for analysis. STRUCTURE utilizes Gibbs sampler algorithm, which is known to face convergence problems during analysis of large data set with complex population structure (Rosenberg et al. 2002; Hanage et al. 2009). On the other hand, the BAPS algorithm is known to be stable during such analysis (Corander and Marttinen 2006).
Most accessions belonging to the Far East region assembled into isolated clusters in both NJ and BAPS analyses, indicating a relatively homogeneous population (Table 6). The Far East accessions were also identified in earlier studies to be one of the most conserved centers for safflower (Knowles 1969; Lee et al. 2014). Chapman et al. (2010) identified genetic similarity between accessions of the Indian subcontinent and Far East and grouped them in a single regional pool. We observed some association between the Far East and Indian subcontinent accessions using STRUCTURE and BAPS analysis at K = 4 (Table 5). However, NJ and BAPS (K = 19) analyses showed considerable genetic dissimilarity between these two regions. Therefore, we believe that Far East should be considered as a conserved, separate gene pool from the Indian subcontinent.
The American germplasm is considered as a region of secondary introduction consisting of safflower accessions collected from different regions of the world. However, in our derivations using both distance-based and Bayesian-based methods, we obtained distinct cluster(s) representing most of the American accessions (Fig. 2 and Table 6). A limited number of accessions from other geographical regions grouped within these clusters indicating a narrow genetic base of the American germplasm. American accessions used in our study included 23 breeding lines (Supplementary Table 1) all of which grouped in a single NJ cluster (NJcl VI) and in BAPS clusters, which are dominated by American accessions (BAcl 6, 10, and 13). Sehgal et al. (2009) also reported grouping of American accessions in a single cluster. In contrast, Johnson et al. (2007) reported rich diversity in safflower genotypes of the American region based on AFLP data, which could be due to limited sample size and presence of the highly variable Arizona Wild Composite in their study. An earlier report indicated blending of Egyptian accessions with Sudanese germplasm due to human migration following construction of the Aswan dam in Nubia (Knowles 1969). Other reports indicated clustering of Egypt and American accessions due to mixing during field trials in Egypt (Knowles 1969; Chapman et al. 2010). However, in our study, accessions from the Egyptian region assembled in a separate cluster in both NJ and BAPS analysis (Fig. 2 and Table 6) and did not show any clustering with American or Sudanese accessions.
Accessions from Near East and Iran–Afghanistan were distributed among all the NJ clusters (Fig. 2). This observation is in congruence with results obtained from population structure analysis through BAPS (Table 6). The Near East region, proposed as a “center of origin” by Ashri and Knowles (1960) and Chapman et al. (2010), is expected to share common genetic framework with accessions of other regions and therefore showed diffuse distribution across multiple clusters. The Iran–Afghanistan region was also proposed by Vavilov (1951) as a “center of origin” owing to the rich phenotypic diversity of safflower germplasm in this region. Knowles (1969) also acknowledged the diversity prevalent in this region and attributed it to the region being an active trading route that favored mixing of germplasm belonging to different regions. Increased genetic diversity of these accessions could be due to increased phenotypic variability as outlined in earlier studies and/or due to the presence of wild relatives in the region that might have facilitated exchange of genetic material between wild relatives and cultivated germplasm (Ashri 1975; Chapman and Burke 2007; Chapman et al. 2010). In our study, BAPS and NJ analyses consistently grouped accessions from these two regions across multiple clusters. The Near East and Iran–Afghanistan regions also showed higher values of genetic diversity indices (Table 3). Therefore, we propose that Near East and Iran–Afghanistan regions be collectively considered as a single region with significant genetic variability for safflower.
In our study, accessions from Turkey showed fragmentation into two main groups in both BAPS and NJ analyses. In BAPS analysis, one set of accessions clustered with Near East and Iran–Afghanistan in BAcl 11 and BAcl 12, respectively, while another set formed a different cluster in BAcl 8 (Table 6). In NJ analysis, most accessions from Turkey clustered in NJcl V along with a few accessions from Iran–Afghanistan (Fig. 2), but accessions corresponding to BAcl 8 formed a distinct sub-cluster within NJcl V. While our data was partly in consonance with Chapman et al. (2010) who considered accessions of Turkey to be genetically similar to those from Iran–Afghanistan, we also detected few accessions with unique genetic identity. Another regional gene pool, Europe, showed diffuse distribution in several clusters of NJ and BAPS analysis contrary to the findings of Chapman et al. (2010). Our study included 37 accessions representing 17 European countries, while Chapman et al. (2010) studied eight accessions from seven countries. Variations in our results could be due to differences in representation of European nations between the two studies. Due to poor representation of accessions from Kenya, Ethiopia, and Australia in our study, no significant conclusions could be made on the genetic diversity of these regions.
The current study also analyzed molecular diversity among safflower accessions from the Indian subcontinent and a few Indian cultivars, which were not assessed in earlier studies. Accessions from this regional pool were distributed in all NJ clusters (Fig. 2) and 12 out of 19 clusters of BAPS analysis (Table 6) indicating a high level of genetic diversity in the Indian subcontinent. The clustering of Indian cultivars analyzed in this study also gave interesting leads. Cultivar TSF-1 was found to be genetically similar to American accessions in both NJ and BAPS analysis, while the other four cultivars (Sharda, Manjira, Annigeri, and PBNS-12) grouped together with few Indian accessions in NJ analysis. In BAPS analysis, PBNS-12 shared a cluster with accessions from Iran–Afghanistan, Turkey, and a few Indian accessions in BAcl 11 (Table 6). The other Indian cultivars, Sharda, Manjira, and Annigeri clustered with Indian accessions in BAcl 4 (Table 6). The relatively isolated grouping of Indian cultivars used in this study from majority of Indian germplasm, which showed extensive diversity, indicates a significantly different genetic constitution and highlights the enormous potential for crop improvement by introgression of desirable alleles from the unexploited genotypes of Indian origin.
Safflower is characterized by substantial phenotypic variations and the present study demonstrates a high level of genetic variability in the germplasm. Geographical patterning was evident among the studied accessions. Evaluation of genetic diversity revealed untapped gene pools, which could be explored for use in plant breeding programs. For example, Indian accessions showed a high level of genetic variability, but the Indian cultivars analyzed exhibited a narrow genetic constitution. Therefore, there is significant potential for improvement of Indian cultivars using the untapped Indian germplasm. On the other hand, American breeding lines clustered with American accessions, indicating that further improvement of American germplasm would be facilitated by selection of promising genotypes from other geographical regions. Similar analysis of other regional cultivars, in conjunction with their phenotypic data, would help in the identification of accessions that could be used to broaden the genetic base of safflower. The markers generated in this study, along with markers developed by other groups, will be useful for genetic stratification, construction of linkage maps, mapping of agronomic traits and assigning parentage, which would be useful in marker-assisted breeding programs for crop improvement.
References
Amini F, Saeidi G, Arzani A (2008) Study of genetic diversity in safflower genotypes using agro-morphological traits and RAPD markers. Euphytica 163:21–30
Ashri A (1971a) Evaluation of the world collection of safflower, Carthamus tinctorius L. I. Reaction to several diseases and associations with morphological characters in Israel. Crop Sci 11:253–257
Ashri A (1971b) Evaluation of the world collection of safflower, Carthamus tinctorius L. II. Resistance to the safflower fly, Acanthophilus helianthi R. Euphytica 20:410–415
Ashri A (1975) Evaluation of the germplasm collection of safflower, Carthamus tinctorius L. V. Distribution and regional divergence for morphological characters. Euphytica 24:651–659
Ashri A, Knowles P (1960) Cytogenetics of safflower (Carthamus L.) species and their hybrids. Agron J 52:11–17
Ashri A, Zimmer D, Urie A, Cahaner A, Marani A (1974) Evaluation of the world collection of safflower, Carthamus tinctorius L. IV. Yield and yield components and their relationships. Crop Sci 14:799–802
Bankey P, Billiar T, Wang W, Carlson A, Holman R, Cerra F (1989) Modulation of Kupffer cell membrane phospholipid function by n-3 polyunsaturated fatty acids. J Surg Res 46:439–444
Barati M, Arzani A (2012) Genetic diversity revealed by EST-SSR markers in cultivated and wild safflower. Biochem Syst Ecol 44:117–123
Carlsson AS, Zhu L-H, Andersson M, Hofvander P (2014) Platform crops amenable to genetic engineering—a requirement for successful production of bio-industrial oils through genetic engineering. Biocatal Agric Biotechnol 3:58–64
Carrasco B, Avila P, Perez-Diaz J, Munoz P, García R, Lavandero B, Zurita-Silva A, Retamales JB, Caligari PD (2009) Genetic structure of highland papayas (Vasconcellea pubescens (Lenné et C. Koch) Badillo) cultivated along a geographic gradient in Chile as revealed by inter simple sequence repeats (ISSR). Genet Resour Crop Evol 56:331–337
Chapman MA, Burke JM (2007) DNA sequence diversity and the origin of cultivated safflower (Carthamus tinctorius L.; Asteraceae). BMC Plant Biol 7(1):60
Chapman MA, Hvala J, Strever J, Matvienko M, Kozik A, Michelmore RW, Tang S, Knapp SJ, Burke JM (2009) Development, polymorphism, and cross-taxon utility of EST–SSR markers from safflower (Carthamus tinctorius L.). Theor Appl Genet 120:85–91
Chapman MA, Hvala J, Strever J, Burke JM (2010) Population genetic analysis of safflower (Carthamus tinctorius L.; Asteraceae) reveals a Near Eastern origin and five centers of diversity. Am J Bot 97:831–840
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843
Corander J, Waldmann P, Marttinen P, Sillanpää MJ (2004) BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics 20:2363–2369
Dajue L, Mündel H (1996) Safflower (Carthamus tinctorius L.) promoting the conservation and use of underutilized and neglected crops. 7. Inst. Plant Genetic Resources Institute (IPGRI), Rome
Doyle J (1991) DNA protocols for plants—CTAB total DNA isolation. In: Hewitt GM, Johnston A (eds) Molecular techniques in taxonomy. Springer, Berlin, pp 283–293
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
FAO (2012). http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor. Accessed Apr 2014
FAO (2013) Oilseeds market summary. Food outlook June 2013. http://www.fao.org/fileadmin/templates/est/COMM_MARKETS_MONITORING/Oilcrops/Documents/Food_outlook_oilseeds/Food_Outlook_June_13.pdf. Accessed Apr 2014
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fang J-Y, Chung J-D, Chiang Y-C, Chang C-T, Chen C-Y, Hwang S-Y (2013) Divergent selection and local adaptation in disjunct populations of an endangered conifer, Keteleeria davidiana var. formosana (Pinaceae). PLoS One 8:e70162
Flider FJ (2013) Development and commercialization of GLA safflower oil. Lipid Technol 25:227–229
Hamdan Y, García‐Moreno MJ, Redondo‐Nevado J, Velasco L, Pérez‐Vich B (2011) Development and characterization of genomic microsatellite markers in safflower (Carthamus tinctorius L.). Plant Breed 130:237–241
Hanage WP, Fraser C, Tang J, Connor TR, Corander J (2009) Hyper-recombination, diversity, and antibiotic resistance in Pneumococcus. Science 324:1454–1457
Ilkılıç C, Aydın S, Behcet R, Aydin H (2011) Biodiesel from safflower oil and its application in a diesel engine. Fuel Process Technol 92:356–362
Johnson RC, Kisha T, Evans M (2007) Characterizing safflower germplasm with AFLP molecular markers. Crop Sci 47:1728–1736
Khan MA, von Witzke-Ehbrecht S, Maass BL, Becker HC (2009) Relationships among different geographical groups, agro-morphology, fatty acid composition and RAPD marker diversity in safflower (Carthamus tinctorius L.). Genet Resour Crop Evol 56:19–30
Knowles P (1969) Centers of plant diversity and conservation of crop germ plasm: safflower. Econ Bot 23:324–329
Lee GA, Sung JS, Lee SY, Chung JW, Yi JY, Kim YG, Lee MC (2014) Genetic assessment of safflower (Carthamus tinctorius L.) collection with microsatellite markers acquired via pyrosequencing method. Mol Ecol Resour 14:69–78
Mahasi M, Wachira F, Pathak R, Riungu T (2009) Genetic polymorphism in exotic safflower (Carthamus tinctorious L.) using RAPD markers. J Plant Breed Crop Sci 1:008–012
Marinova E, Riehl S (2009) Carthamus species in the ancient Near East and south-eastern Europe: archaeobotanical evidence for their distribution and use as a source of oil. Veg Hist Archaeobot 18:341–349
McPherson MA, Yang R-C, Good AG, Nielson RL, Hall LM (2009) Potential for seed-mediated gene flow in agroecosystems from transgenic safflower (Carthamus tinctorius L.) intended for plant molecular farming. Transgenic Res 18:281–299
Mokhtari N, Rahimmalek M, Talebi M, Khorrami M (2013) Assessment of genetic diversity among and within Carthamus species using sequence-related amplified polymorphism (SRAP) markers. Plant Syst Evol 299:1285–1294
Nimbkar N (2008) Issues in safflower production in India. In: Proceedings of the Seventh International Safflower Conference, Wagga Wagga.
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pearl SA, Bowers JE, Reyes-Chin-Wo S, Michelmore RW, Burke JM (2014) Genetic analysis of safflower domestication. BMC Plant Biol 14(1):43
Peng S, Feng N, Guo M, Chen Y, Guo Q (2008) Genetic variation of Carthamus tinctorius L. and related species revealed by SRAP analysis. Biochem Syst Ecol 36:531–538
Perrier X, Jacquemoud-Collet J (2006) DARwin software http://darwin.cirad.fr/darwin.
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ramu P, Billot C, Rami J, Senthilvel S, Upadhyaya H, Reddy LA, Hash C (2013) Assessment of genetic diversity in the sorghum reference set using EST-SSR markers. Theor Appl Genet 126:2051–2064
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385
Sehgal D, Raina SN (2005) Genotyping safflower (Carthamus tinctorius L.) cultivars by DNA fingerprints. Euphytica 146:67–76
Sehgal D, Rajpal VR, Raina SN, Sasanuma T, Sasakuma T (2009) Assaying polymorphism at DNA level for genetic diversity diagnostics of the safflower (Carthamus tinctorius L.) world germplasm resources. Genetica 135:457–470
Smith JR (1996) History, Chapter 1. In: Safflower. The American Oil Chemists Society Press, Champaign, pp 1–15
Smýkal P, Kenicer G, Flavell AJ, Corander J, Kosterin O, Redden RJ, Ford R, Coyne CJ, Maxted N, Ambrose MJ (2011) Phylogeny, phylogeography and genetic diversity of the Pisum genus. Plant Genet Resour 9:4–18
Sujatha M (2008) Biotechnological interventions for genetic improvement of safflower. In: Proceedings of the Seventh International Safflower Conference, Wagga Wagga, New South Wales pp. 3–6.
Tollefsrud MM, Sønstebø JH, Brochmann C, Johnsen Ø, Skrøppa T, Vendramin GG (2009) Combined analysis of nuclear and mitochondrial markers provide new insight into the genetic structure of North European Picea abies. Heredity 102:549–562
Van Zeist W, Waterbolk-Van Rooijen W (1992) Two interesting floral finds from third millennium BC Tell Hammam et-Turkman, northern Syria. Veg Hist Archaeobot 1(3):157–161
Vavilov NI (1951) The origin, variation, immunity and breeding of cultivated plants. Soil Sci 72:482
Velasco L, Pérez‐Vich B, Fernández‐Martínez J (2005) Identification and genetic characterization of a safflower mutant with a modified tocopherol profile. Plant Breed 124:459–463
Vos P, Hogers R, Bleeker M, Reijans M, van De Lee T, Hornes M, Friters A, Pot J, Paleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Weiss E (1983) Oilseed crops. In: Safflower. Longman Group Limited, Longman House, London
Yang YX, Wu W, Zheng YL, Chen L, Liu RJ, Huang CY (2007) Genetic diversity and relationships among safflower (Carthamus tinctorius L.) analyzed by inter-simple sequence repeats (ISSRs). Genet Resour Crop Evol 54:1043–1051
Yeh Francis C, Yang R, Boyle Timothy B, Ye Z, Mao Judy X (1999) POPGENE version 1.32, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Canada (http://www.ualbertaca/∼fyeh/).
Acknowledgments
This work was funded by the DST-PURSE grant of the Department of Science and Technology, Government of India, provided to University of Delhi. HA was supported by a research fellowship from University Grants Commission, India. The authors thank Professors R. Geeta and Deepak Pental of University of Delhi for their critical comments and suggestions. The authors would also like to extend their sincere thanks to anonymous reviewers whose critical comments have further improved the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
S. Kumar and H. Ambreen contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLSX 23 kb)
Rights and permissions
About this article
Cite this article
Kumar, S., Ambreen, H., Murali, T.V. et al. Assessment of Genetic Diversity and Population Structure in a Global Reference Collection of 531 Accessions of Carthamus tinctorius L. (Safflower) Using AFLP Markers. Plant Mol Biol Rep 33, 1299–1313 (2015). https://doi.org/10.1007/s11105-014-0828-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0828-8