Abstract
Cytoplasmic genetic male-sterility is used to produce hybrid onion (Allium cepa L.) seeds worldwide. In this paper, we present the results of research aimed toward identifying PCR-based markers linked to the Ms locus through amplified fragment length polymorphism (AFLP). After screening 512 AFLP primer combinations, only one AFLP fragment was identified as being flanking linked to the dominant Ms allele. Subsequently, the AFLP marker was converted into a sequence-characterized amplified region (SCAR) marker, designated as DNF-566, co-segregated with the dominant Ms allele in first backcross (BC1) segregated populations. Furthermore, we designed another molecular marker (RNS-357) co-segregated with the ms allele to identify different genotypes (i.e., MsMs, Msms, or msms). Both markers could be used for evaluating onion lines with different genetic backgrounds (including male-sterile lines, maintainer lines, male-fertile lines, and commercial based F1 hybrid cultivars). The results of this study indicate that maintainer plants could be directly selected by using these 2 SCAR markers in the onion breeding process, and this may contribute significantly toward breeding onion F1 hybrid cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytoplasmic genetic male-sterility (CMS) is used to produce hybrid onion (Allium cepa L.) seeds worldwide. The first description of male-sterile plants (S) was provided by Jones and Emsweller (1936). These plants contain the S cytoplasm, and are homozygous recessive at the Ms locus. Male-fertility in such plants is restored by a dominant allele at the nuclear male-fertility restoration locus Ms (Jones and Clarke 1943). In addition to S type CMS, onions have several other cytoplasm types, including CMS-T, which is caused by three independently segregated loci in the nuclear genome. Furthermore, some undefined cytoplasm types have also been reported (Berninger 1965; Pathak and Gowda 1993; Havey 2000). However, F1 hybrid cultivars primarily use just two types of cytoplasmic genetic male-sterility (CMS-S and CMS-T) systems. Of the two systems, the CMS-S system is the most widely used in the majority of hybrid onion cultivars, because of its stability in various environments (Havey 1995; Havey 2000). In CMS-S system, cytoplasmic male-sterile lines are seed propagated by crossing them with a maintainer line that possesses normal (N) male-fertile cytoplasm and a homozygous recessive genotype at the restorer locus N (msms). However, the extraction of maintainer lines from some onion populations is difficult and time-consuming, due to the biennial generation time of onions, high frequency of the dominant allele at Ms (Little and Jones 1944; Davis 1957; Havey and Randle 1996), and prevalence of the S cytoplasm (Satoh et al. 1993; Havey 1993; Havey and Bark 1994).
Marker-assisted selection (MAS) could improve the efficiency of traditional breeding programs, serving as a powerful tool that facilitates the transfer important agricultural economic genes into cultivars, such as disease resistance genes, high quality characteristic genes and selecting maintainer lines (Tanksley et al. 1989; Havey 1995; Kanzaki et al. 2010; Hayashi et al. 2011; Kuraparthy et al. 2011). Sato (1998) and Kim et al. (2009) developed the molecular markers via polymerase chain reaction (PCR) to identify the cytoplasm of a single onion plant. Consequently, this identification now requires just hours as opposed to years and allows breeders to distinguish the type of cytoplasm from a single onion plant without test crossing. However, despite progress in studying the polymorphism of mitochondrial or chloroplast cytoplasm, relatively limited information about the nuclear restorer genes has been reported. For instance, AOB272 is the closest restriction fragment length polymorphism (RFLP) marker to the Ms locus in the onion genome (Gökçe et al. 2002). OPT and PsaO markers derived separately from AOB272 and AGF136 are linked in opposite directions to the Ms locus, at distances of 1.5 and 6.4 cM, respectively (Bang et al. 2011). These markers are useful for the selection of specific alleles at the Ms locus in segregating families at linkage disequilibrium. In a previous study by our research group, a cDNA-based marker (WHR240) was successfully identified that was able to distinguish the nuclear restorer genes using cDNA-SRAP analysis (Huo et al. 2012). Although this marker is useful for the selection of maintainer lines in the MAS breeding program, it has limited application for large-scale selection because of its origin from RNA.
Amplified fragment length polymorphism (AFLP) combining the advantages of both RFLP and RAPD is a powerful marker technique (Savelkoul and Aarts 1999). This technique may be used to generate linkage maps (Tanksley et al. 1992; van Heusden et al. 2000) or screen markers around the loci of interest (Zhang et al. 2007). However, AFLP markers are relatively costly and technologically demanding, which limits their application for large-scale screening in the MAS program. Therefore, AFLP markers must be converted into easy-to-use markers, such as sequence-characterized amplified region (SCAR) markers.
In this study, we specifically focused on the onion with S and N type cytoplasm, and successfully obtained two useful SCAR markers tightly linked to the both alleles at Ms locus. The acquisition of these two markers will allow breeders to directly select maintainer lines in different background onions and contribute towards improving the speed of selecting hybrid F1 cultivars in onion.
Materials and methods
Plant materials and evaluation of onion fertility
Two male-sterile lines [(118 and 110), S (msms)], and one male-fertility-restored line [12–12, S (MsMs)], whose genotype were identified from the testcross progeny between male sterile plant and 12–12 (>20 individuals), and self-cross progeny (>20 individuals), were used to construct the BC1 segregating population. Another 11 male-sterile lines (8 intermediate day type and 3 long day type), 11 maintainer lines (10 intermediate day type and 1 long day type), and four sets of paternal lines (146, 149, 153, and 156), for which the MsMs genotype was identified by test cross (Huo et al. 2012), were used to validate the practical application of the molecular marker. All of these lines were selected by the Vegetable Research Institute of Shandong Academy of Agricultural Sciences (SAAS). Seven commercial F1 hybrid cultivars were purchased from the seed markets released from Japanese companies Takii Seed and Shippo Seed.
The sterility of the onion plants was determined based on the presence or absence of pollen during flowering. Flowers were checked daily for pollen production by rubbing mature anthers with the back of the hand. Once pollen was detected, the scape was tagged, and not checked again. Flowers of untagged umbels were repeatedly scored during the entire flowering period. Multiple umbels on an individual plant were independently scored. After flowering, plants were scored as male-fertile if one umbel was tagged.
DNA extraction and genetic pool construction
Total DNA was extracted from the young leaves of each individual onion plant using a plant DNA extraction kit (Tiangen Biotech, Beijing, China). DNA concentrations were measured using a spectrophotometer at a wavelength of 260 versus 280 nm, while DNA quality was assessed on 0.8 % agarose gel. Bulked segregant analysis (BSA) was used to identify AFLP markers linked to the target gene. An equal quantity of DNA from 10 fertile plants and 10 sterile plants was selected at random to form a male-fertile pool and male-sterile pool, respectively. The two BSA pools were then subjected to AFLP analysis.
DNA-AFLP analysis
DNA-AFLP analysis was performed according to the procedure described by Vos et al. (1995), with minor modifications. For each sample, 300 ng DNA was completely digested using two enzyme combinations, EcoRI/MseI and PstI/MseI, and then linked to adapters (EcoRI adapter: 5′-CTCGTAGACTGCGTACC-3′ and 5′-AATTGGTACGCAGTCTAC-3′; MseI adapter: 5′-GACGAT GAGTCCTGAG-3′ and 5′-TACTCAGGACTCAT-3′; PstI adapter: 5′-CTCGTAGACTGCGTACATGCA-3′ and 5′-TGTACGCAGTCTAC-3′). After dilution of the pre-amplified PCR products with deionized water (10 times), 16 EcoRI primers (AAC, AAG, ACA, ACC, ACG, ACT, AGC, AGG, AAT, AAA, AGT, AGA, ATC, ATG, ATT, ATA) and 16 PstI primers (GAA, GAC, GAG, GAT, GTA, GTC, GTG, GTT, GCA, GCC, GCG, GCT, GGA, GGC, GGG, GGT) were combined with 16 MseI primers (CAA, CAC, CAG, CAT, CTA, CTC, CTG, CTT, CCA, CCC, CCG, CCT, CGA, CGC, CGG, CGT) for selective amplification. The selective amplification products were separated on 6 % polyacrylamide denaturing sequencing gels, and stained with silver nitrate (Lu et al. 2001).
Polymorphic fragment isolation and sequencing
Differential fragments (TYAGG/CGT) from male-sterile pool and male-fertile pool by AFLP analysis were excised from polyacrylamide gels and used as templates for PCR re-amplification. The PCR products were checked on a 2.5 % agarose gel, and purified using a Tian Gel midi purification kit (Tiangen Biotech, Beijing, China). The purified PCR product was cloned into the pGEM®-T Easy Vector Systems (Promega, Madison, WI, USA), and was then sequenced by Shanghai Biosune Biological Engineering Technology and Service Co., Ltd.
Conversion of SCAR markers
Based on the TYAGG/CGT sequence, three forward primers (TF1, TF2, and TF3) and three reverse primers (TR1, TR2, and TR3) were designed by Primer premier 5.0 software, and then used to genomic walking according to the technology of high-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR) (Liu and Chen 2007) (Table 1). Two primers (FS-F, FS-R) were applied to amplify the fragment from genomic walking. Four primers (FN1, RN1, F3S2 and R3S2) were designed to develop SCAR marker based on the extended sequence (Table 1). PCR was performed using different combinations of primers in a total volume of 25 μl, containing 0.2 mM of each dNTP, 1 unit Ex Taq polymerase (TaKaRa Biotechnology, Dalian, China), 0.4 μM of each primer, 1× Ex Taq reaction buffer, and 50 ng of template DNA. The PCR procedure was as follows: initial denaturation at 95 °C for 6 min, followed by 35 cycles of 95 °C for 30 s, 58 °C for 45 s, 72 °C for 45 s, and then an additional extension was carried out at 72 °C for 5 min. The amplified products of the PCR were analyzed on 1.0 % agarose gels, and visualized by ethidium bromide staining.
Results
Construction of the segregating population
Two male-sterile lines (118 and 110), and one male-fertility-restored line [12–12, S (MsMs)] were sown in September 2004, and their bulbs were harvested in May 2005. These F1 populations obtained from two cross combinations [(118 × 12–12) and (110 × 12–12)] were sown in September 2006, and crossed with 118 and 110 in May 2008. Two BC1 combinations [118 × (118 × 12–12) and 110 × (110 × 12–12)] harvested in July 2008 were sown in a greenhouse in September 2008, surrounded by a 30-mesh net, and with the top covered by a plastic film. Plant density was 10 × 20 and 30 × 50 cm2 for seedlings and bulbs, respectively. The segregated BC1 populations were harvested in May 2010, and individuals were identified as being male-sterile or male-fertile during the flowering stages, according to the method described in materials and methods (plant materials and evaluation of onion fertility). All F1 phenotype of two cross combinations between male-sterile lines (118 and 110) and a male-fertility-restored line (12–12) exhibited a normal type (male-fertile) (Table 2). In the BC1 segregation generation of these crosses [118 × (118 × 12–12)] and [110 × (110 × 12–12)], two different phenotypes were shown, male-fertile and male-sterile. There were 122 male-fertile and 117 male-sterile individuals in [118 × (118 × 12–12)] population, and 51 male-fertile and 42 male-sterile individuals in [110 × (110 × 12–12)] population. Both segregation patterns fitted to the expected 1:1 ratio for a single gene model (Table 2).
Identification of AFLP markers linked to the nuclear male-fertility restoration locus (Ms)
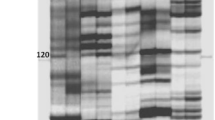
Ten fertile plants and ten sterile plants being selected randomly mixed in equal quantities to construct the male-fertile pool and male-sterile pool based on BSA method, respectively. Sixteen EcoRI primers and 16 PstI primers were combined with 16 MseI primers, resulting in 512 AFLP primer combinations to use the AFLP analysis. Approximately 23,000 bands were obtained that ranged in size from 50 to 1,000 bp. The majority of the primer combinations generated approximately 40–80 bands, with just a few primer combinations producing 30–50 bands. In the screen of 512 primer pairs, only one polymorphic fragment was obtained from the primer combination E-AGG/M-CGT. This fragment was only amplified in the male-fertile pool and male fertile individuals (Fig. 1). This result indicated that the AFLP marker was putatively linked to the dominant Ms allele, this marker was designated as TYAGG/CGT. For more precise and easy identification of their linkage relationship, and to validate the practical application, we converted an AFLP marker to SCAR markers.
AFLP pattern with primer combination E-AGG/M-CGT from 2 DNA pools. Lanes 1–10 10 different individuals from the sterile pool; lane 11 male sterile pool; lane 12 male fertile pool; lanes 13–22 10 different individuals from the fertile pool. The arrow indicates the product that is present in male fertile pool and individuals, and not in male sterile pool and individuals
Conversion of the AFLP marker to a SCAR marker and its linkage analysis
The polymorphism fragment TYAGG/CGT was purified, cloned and sequenced. The sequence of TYAGG/CGT had 176-bp lengths, and showed no match to any existing sequence in GenBank database by BLAST searches. Subsequently, the genomic walking was performed according to hiTAIL-PCR (Liu and Chen 2007) and a 1,527-bp fragment in male-fertile pool was amplified. Based on the 1,527-bp sequence, we designed a pair of primers (FS-F, FS-R) (Table 1) to amplify male-fertile pool and male-sterile pool in BC1 populations. Two type sequences (1,527 and 1,546-bp) were acquired in male-fertile pool, only 1,546-bp sequence in male-sterile pool. These results coincided with the genotype at Ms locus which were heterozygous and homozygous recessive in male-fertile pool and male-sterile pool, respectively. The sequences of 1,527 and 1,546-bp which were named as F1527 and S1546 were putatively linked to both dominant and recessive Ms alleles, respectively. Alignment of two genomic sequences (F1527 and S1546) revealed 53 SNP sites and eleven InDel events (27 InDel sites) (Fig. 2). Both of F1527 and S1546 were no significant similarity to any exiting sequence in GenBank database by megablast, and also the coding sequence of the F1527 and S1546 were not predicted by GENSCAN (http://genes.mit.edu/GENSCAN.html). Depending on the alignment results of the two genomic sequences (F1527 and S1546) we designed two primers (FN1 and RN1) of SCAR marker (Fig. 2) and amplified a 566-bp band in male-fertile pool and fertile individuals (Fig. 3). This marker showed a dominant character linked to the Ms locus, was designated as a dominant nuclear male-fertile allele (DNF-566).
Alignment of nucleotide sequences from AFLP fragment and genomic walking between male fertile pool and male sterile pool. F1527 and S1546 are amplified in male fertile pool, only S1546 in male sterile pool. The gray box is for the AFLP sequence. The italic nucleotides indicate the restriction endonuclease site of MseI and EcoRI, respectively. The asterisk indicates the consensus sequence and the dash indicates a nucleotide deletion. The arrows indicate the positions of primer-binding sites
Evaluation of the DNF-566 marker in individual plants from 2 DNA pools. M DL2000 ladder molecular marker. Lanes 1–10 10 different individuals from the fertile pool. Lane 11 male fertile pool. Lanes 12–21 10 different individuals from the sterile pool. Lane 22 male sterile pool. The arrow indicates that the 566 bp fragment is only present in fertile individuals, and not sterile individuals
DNF-566 marker was applied to two segregated populations [118 × (118 × 12–12) and 110 × (110 × 12–12)]. The 122 fertile individuals were amplified the 566-bp band, while 117 sterile individuals were negative in 239 BC1 individuals from [118 × (118 × 12–12)] population (Table 3). Fifty-one fertile individuals amplified the 566-bp band, while 42 sterile individuals showed no amplification (Table 3) in 93 BC1 individuals from the [110 × (110 × 12–12)] population which had a different background. The amplification results of the two different background populations supported their phenotypes. These results strongly indicate that the 566-bp fragment linked to the dominant Ms allele.
The genotype at Ms locus without amplifying the 566-bp fragment was considered to homozygous recessive genotype, because the 566-bp fragment of DNF-566 is a marker linked to the dominant Ms allele. In practical breeding programs, to avoid misclassification caused by the failure of PCR and identify different genotypes (MsMs, Msms, or msms), we designed a marker RNS-357 (recessive nuclear male-sterile allele, forward primer F3S2 and reverse primer R3S2) expectedly to link to the recessive ms allele (Fig. 2). Two segregated populations [118 × (118 × 12–12) and 110 × (110 × 12–12)] and their paternal line (12–12) were used to evaluate the reliability of the RNS-357 marker. The results showed the 357-bp fragment was amplified in each fertile and sterile individual of the two BC1 individuals possessing the recessive ms allele (Table 3) and was not amplified in paternal line (12–12) with homozygous dominant Ms allele (Fig. 4). Thus, we speculated that marker RNS-357 was linked to the recessive ms allele.
Evaluation of the RNS-357 marker in individual plants from 2 DNA pools. M DL2000 ladder molecular marker. Lanes 1 and 12 2 individuals from the paternal line (12–12). Lanes 2–11 10 different individuals from the sterile pool. Lanes 13–22 10 different individuals from the fertile pool. The arrow indicates that the 357-bp fragment accompanied the ms allele
Evaluating the applicability of SCAR markers
To test the applicability of the 2 SCAR markers in onion breeding programs, we validated the markers DNF-566 and RNS-357 using 13 male-sterile lines and 11 maintainer lines included two different ecotypes of intermediate and long day type, respectively, in addition to four paternal lines. Their genotypes were identified by test cross (Table 4). In all of these sterile and maintainer lines, only one 357-bp band was clearly related to the ms allele. In comparison, the four paternal lines showed one 566-bp band related to the Ms allele. Although these sterile lines, maintainer lines, and four paternal lines were derived from at least five different genetic backgrounds, clear 357-bp and 566-bp bands were observed in the genotype msms and MsMs, respectively. These results indicate that SCAR markers DNF-566 and RNS-357 were co-segregated with the dominant and recessive Ms allele, respectively. To test whether these markers were available in different F1 hybrid cultivars, seven commercial F1 hybrid cultivars were selected in this experiment (Table 5). All seven F1 hybrid cultivars contained 566 and 357-bp bands, despite these cultivars originating from two different companies and showing various different phenotypes, such as maturation time, plant height, leaf shape, and bulb color.
Discussion
In this study, we succeeded in identifying 2 SCAR markers co-segregated with dominant and recessive Ms alleles using AFLP and hiTAIL-PCR techniques in the onion. The 512 primer combinations were screened, but only one polymorphic fragment was amplified by EcoRI/MseI primer combinations. In most cases, the background of the electrophoresis was slightly tanned, which was probably caused by the huge genome and highly repetitive sequences. Efficient polymorphism was not found using PstI/MseI primer combinations. One reason might be that the onion genome has duplicated loci and methylation sensitivity to the PstI restriction enzyme (King et al. 1998; van Heusden et al. 2000). Because the size range of the molecular weight of fingerprints is concentrated at 50–500 bp (Vos et al. 1995), some AFLP markers could not be successfully converted into a SCAR marker (Ke et al. 2004). The genomic walking based on hiTAIL-PCR method (Liu and Chen 2007) was used to amplify unknown sequences flanking the AFLP marker. We obtained two types sequences (F1527 and S1546) respectively in BC1 populations through hiTAIL-PCR method. F1527 and S1546 existed in fertile individuals and S1546 in sterile individuals. Therefore, we deduced F1527 and S1546 were linked to the dominant Ms allele and recessive ms allele, respectively. Four specific primers of haplotype marker were designed in continuous polymorphic sites (continuous nucleotides differential sites, except for the primer R3S2) according to the alignment of F1527 and S1546 (Fig. 2). Consequently, the obtained AFLP marker TYAGG/CGT was converted to two SCAR markers, DNF-566 and RNS-357, linked to the dominant and recessive Ms alleles, respectively, by using hiTAIL-PCR method.
Onion is a biennial plant, with 4–8 years being required for maintainer lines to develop from an uncharacterized population or segregated family (Havey 1995). Therefore, the identification of molecular markers that are flanking the nuclear Ms locus would greatly facilitate the selection of maintainer lines. However, the onion has a huge nuclear genome that is more than 36 times larger than that of rice and 6 times larger than that of maize (Arumuganathan and Earle 1991). At present, several DNA markers, AOB272, OPT, PsaO etc. have been reported and the Ms locus was located in between the OPT and PsaO (Gökçe et al. 2002; Bang et al. 2011). These markers (AOB272, OPT and PsaO) are useful for segregating families because of their linkage disequilibrium. However, these may not be possible to identify genotypes of maintainer lines among plants from open-pollinated populations (Gökçe and Havey 2002; Bang et al. 2011). Therefore, the selection of maintainer lines from open-pollinated populations has been limited to their use. Huo et al. (2012) developed a cDNA marker, WHR240, which was combined with a previously reported cytoplasm marker to classify the cytoplasm into S or N type (Havey 1995; Sato 1998); it can be used to select the maintainer line through the expression of the pectin methylesterase gene in onion flower buds. This marker is based on RNA, which is not practical for large-scale selection. Compared to the conventional test-cross method, the marker techniques have significant utility for onion breeding, by economizing on the costs of labor and trials, as it reduces the number of testcrosses that are required to identify maintainer plants. However, they are not widely used in onion breeding because of their own limitations as mentioned above.
In the present researches, the DNF-566 and RNS-357 were tightly linked to the restorer locus (putatively Ms locus) without recombinant value, and both markers were derived from the lines of S-type-cytoplasm (Sato 1998; Kim et al. 2009). Moreover, Jones and Clarke (1943) reported that male-fertility in CMS-S system was restored by only one dominant allele at the nuclear male-fertility restoration Ms locus. These results were strongly suggested that our new markers (DNF-566 and RNS-357) should be located in between OPT and PsaO, and considering two restorer loci from our research and previous report was the same (Gökçe et al. 2002; Bang et al. 2011).
Two SCAR markers were linked to the dominant and recessive Ms alleles, respectively, which could precisely identify three genotypes (MsMs, Msms, and msms) at the Ms locus, and it effectively avoid the risk of misclassification caused by failed PCR in practical breeding. Furthermore, the utility of these two markers were confirmed in different origins of onion, including male-sterile lines, maintainer lines, and male-fertile lines (Table 4). In the evaluation of the seven commercial F1 hybrid cultivars, all of tested individuals amplified 357 bp fragments (Table 5), because the genotype at Ms locus of all individuals in F1 hybrid should be one of msms or Msms except for MsMs and each genotype at least possess recessive ms allele. On the other hand, we also obtained 566-bp fragment in all of F1 individuals. These results demonstrated that both markers were also available in these cultivars for clarifying the genotype of Ms locus, and the genotype of the paternal line in these cultivars were presumed as S/N (MsMs).
Melgar and Havey (2010) reported that the dominant Ms allele shows reduced penetrance in onion plant. It makes us have to considerate about the possibility of misclassification some male-fertile plants to male-sterile plants whether happened or not. However, both phenotypes of male-fertile and male-sterile in the segregation populations fitted to the expected 1:1 ratio for a single gene model in this experiment, which indicate that there was no segregation distortion in those BC1 populations. Further more, the recombinant individual was not observed between marker and male-sterile characters in BC1 populations, and both markers are suitable in all the validated populations. These demonstrated that the environmental effects were not strong enough to express low penetrance at dominant allele in this experiment. Melgar and Havey (2010) also reported some testcross families from male parents heterozygous at Ms that showed average proportions of male-fertile plants with lower variation across years for male-fertility restoration. Hence, from the breeding point of view, these markers might be involved in the Ms/ms alleles.
These results showed the two SCAR markers could be suitable for the usage in open-pollinated populations with various backgrounds, including long-day and short-day types and it is likely that we develop universal SCAR markers in the S and N cytoplasm, which are used for most of the F1 hybrid onion cultivars worldwide. These markers presented in the current study are considered to be one of the most powerful tools for MAS to select maintainer lines directly, and would make a significant contribution to onion F1 hybrid cultivars. Additionally, the alignment of the two genomic sequences (F1527 and S1546) revealed 53 SNP sites and eleven InDel events (27 InDel sites) (Fig. 2); both F1527 and S1546 were no significant similarity to any exiting sequence in GenBank database by megablast; the coding sequences of the F1527 and S1546 were not predicted by GENSCAN (http://genes.mit.edu/GENSCAN.html). Thus we are conducting further research on the verification of other markers (SNP, CAPS, SSR, InDel), allelic variation, haplotype analysis and development of the co-dominant marker based on the alignment of genomic sequence around the polymorphic fragment from genomic walking in diverse onion. And in the near future, we will focus our efforts on isolating the onion genomic region that carries the Ms locus, to determine the physical map and to obtain more information on the mechanism of male-sterility in the onion plant.
References
Arumuganathan K, Earle E (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Bang H, Cho DY, Yoo KS, Yoon MK, Patil BS, Kim S (2011) Development of simple PCR-based markers linked to the Ms locus, a restorer-of-fertility gene in onion (Allium cepa L.). Euphytica 179:439–449
Berninger E (1965) Contribution a l’etude de la sterilite male de l’oignon (Allium cepa L.). Ann Amélior Plant 15:183–199
Davis EW (1957) The distribution of the male sterility gene in onion. HortScience 70:316–318
Gökçe AF, Havey MJ (2002) Linkage equilibrium among tightly linked RFLPs and the Ms locus in open-pollinated onion populations. J Am Soc Hortic Sci 127:944–946
Gökçe AF, Sato Y, Michael J, Havey MJ (2002) Molecular tagging of the Ms locus in onion. J Am Soc Hortic Sci 127(4):576–582
Havey MJ (1993) A putative donor of S-cytoplasm and its distribution among open-pollinated populations of onion. Theor Appl Genet 86:128–134
Havey MJ (1995) Identification of cytoplasm using the polymerase chain reaction to aid in the extraction of maintainer lines from open-pollinated populations of onion. Theor Appl Genet 90:263–268
Havey MJ (2000) Diversity among male-sterility-inducing and male-fertile cytoplasms of onion. Theor Appl Genet 101:778–782
Havey MJ, Bark O (1994) Molecular confirmation that sterile cytoplasm has been introduced into open-pollinated cultivars of grano onions. J Am Soc Hortic Sci 119:90–93
Havey MJ, Randle WM (1996) Combining abilities for yield and bulb quality among long- and intermediate-day open-pollinated onion populations. J Am Soc Hortic Sci 121:604–608
Hayashi M, Ujiie A, Serizawa H, Sassa H, Kakui H, Oda T, Koba T (2011) Development of SCAR and CAPS markers linked to a recessive male sterility gene in lettuce (Lactuca sativa L.). Euphytica 180:429–436
Huo YM, Miao J, Liu BJ, Yang YY, ZHang YH, Wu X (2012) The expression of pectin methylesterase in onion flower buds is associated with the dominant male-fertility restoration allele. Plant Breed 131:211–216
Jones HA, Clarke AE (1943) Inheritance of male sterility in the onion and the production of hybrid seed. HortScience 43:189–194
Jones HA, Emsweller SL (1936) A male-sterile onion. HortScience 34:582–585
Kanzaki S, Akagi T, Masuko T, Kimura M, Yamada M, Sato A, Mitani N, Ustunomiya N, Yonemori K (2010) SCAR markers for practical application of marker-assisted selection in persimmon (Diospyros kaki Thunb.) breeding. J Japan Soc Hort Sci 79(2):150–155
Ke LP, Sun YQ, Liu PW, Yang GS (2004) Identification of AFLP fragments linked to one recessive genic male sterility (RGMS) in rapeseed (Brassica napus L.) and conversion to SCAR markers for marker-aided selection. Euphytica 138:163–168
Kim S, Lee ET, Cho DY (2009) Identification of a novel chimeric gene, orf725, and its use in development of a molecular marker for distinguishing among three cytoplasm type in onion (Allium cepa L.). Theor Appl Genet 118:433–441
King JJ, Bradeen JM, Bark O, McCallum JA, Havey MJ (1998) A low-density genetic map of onion reveals a role for tandem duplication in the evolution of an extremely large diploid genome. Theor Appl Genet 96:52–56
Kuraparthy V, Sood S, Guedira GB, Gill BS (2011) Development of a PCR assay and marker-assisted transfer of leaf rust resistance gene Lr58 into adapted winter wheats. Euphytica 180:227–234
Little T, Jones HA (1944) The distribution of the male sterility gene in varieties of onion. Herbertia 11:310–312
Liu YG, Chen YL (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43:649–656
Lu GY, Yang GS, Fu TD (2001) Silver stained AFLP-a novel assay for DNA fingerprinting in Brassica napus. J Huazhong Agric Univ 20:413–415
Melgar S, Havey MJ (2010) The dominant Ms allele in onion shows reduced penetrance. J Am Soc Hortic Sci 135:49–52
Pathak C, Gowda R (1993) Breeding for the development of onion hybrids in India: problems and prospects. Acta Hortic 358:239–242
Sato Y (1998) PCR amplification of CMS-specific mitochondrial nucleotide sequences to identify cytoplasmic genotypes of onion (Allium cepa L.). Theor Appl Genet 96:367–370
Satoh Y, Nagai M, Mikami T, Kinoshita T (1993) The use of mitochondrial DNA polymorphism in the classification of the individual onion plants by cytoplasmic genotypes. Theor Appl Genet 86:345–348
Savelkoul PHM, Aarts HJM (1999) Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol 37:3083–3091
Tanksley SD, Young ND, Paterson AH (1989) RFLP mapping in plant breeding: new tools for an old science. Biotechnology 7:257–264
Tanksley SD, Ganal MW, Prince JP (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160
Van Heusden AW, Van Ooijen JW, Vrielink-van GR, Verbeek WHJ, Wietsma WA, Kik C (2000) A genetic map of an inter-specific cross in Allium based on amplified fragment length polymorphism (AFLP™) markers. Theor Appl Genet 100:118–126
Vos P, Hogers R, Reijans M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Zhang D, Zhang Z, Yang K (2007) Identification of AFLP markers associated with embryonic root development in Populus tomentosa Carr. Silvae Genet 56:27–32
Acknowledgments
This project was supported by the National Natural Science Foundation of China (30871706, 31201635), Special Fund for Agro-scientific Research in the Public Interest (200903018), National Key Technology R&D Program (2012BADO2B04).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yan Yan Yang and Yu Meng Huo contributed equally to this study.
Rights and permissions
About this article
Cite this article
Yang, Y.Y., Huo, Y.M., Miao, J. et al. Identification of two SCAR markers co-segregated with the dominant Ms and recessive ms alleles in onion (Allium cepa L.). Euphytica 190, 267–277 (2013). https://doi.org/10.1007/s10681-012-0842-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-012-0842-6