Abstract
Cytoplasmic male-sterility (CMS) is the primary source for the production of commercially viable onion F1 hybrids. The molecular interplay of the cytoplasmic-nuclear genes propels the restoration of fertility in the CMS system, making it cost-effective and stable. The use of the molecular markers that can determine the cytoplasm type and nuclear genotype reduces the amount of time and labour required. This study characterized the morphology of male sterile and fertile plants based on anther colour and pollen viability. Additionally, the molecular characterization of the organellar DNA differentiating cytoplasm and nuclear genotypes in 35 commercially grown open-pollinated varieties of Indian short-day onion was attempted. Our results revealed that morphological and microscopic observations for the identification of male sterile and fertile plants were not 100% corroborative. Markers located in the chloroplast (accD) and mitochondrial DNA (MKFR) revealed that Indian cultivars exhibited a greater frequency of N (normal) cytoplasm, lower frequency of S (sterile) cytoplasm and no occurrence of T (sterile) cytoplasm. All three markers viz., AcPMS1, AcSKP1, and jnurf13 revealed that 93 to 99% of the plants of all the varieties had homozygous recessive (msms) alleles at the Ms locus. The OPT marker classified the plants as having 38% msms, 39% Msms and 21% MsMs genotypes and needs further investigation. This underscores the pressing need for additional markers to precisely discern the Ms locus, facilitating the identification of male sterile and maintainer plants within open-pollinated populations of Indian short-day onions. Notably, male sterile cytoplasm was identified in eight commercial varieties, marking a pioneering revelation in Indian onion cultivation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The bulb onion (Allium cepa L., 2n = 2x = 16), which belongs to the Amaryllidaceae family, is one of the most important crops and has been domesticated and cultivated in the Old World for more than 5000 years. Due to their peculiar properties as food and their therapeutic and ethnopharmacological value, onion plants are preferred for consumption across the globe and are cultivated under all climates (Brewster 2008). Although it is an important vegetable crop, breeders have not focused much on its genetics, breeding and genomic research. The main reason for this limited research is due to its biennial nature, highly cross-pollinating nature, high degree of inbreeding depression and giant genome size (Khar 2022).
In India, the phenomenal increase in onion production from 3.23 to 26.74 million tonnes reflects the enhanced awareness of onion consumption over the last three decades i.e., from 1990 to 2020. As the leading country in terms of harvested acreage and total production, India harvested 26.7 MMTs from an area of 1.62 million hectares. However, Indian farmers are getting experiencing comparatively lower productivity (16.4 t/ha) than farmers in other regions such as Korea (79.62 t/ha), the USA (71.10 t/ha), Japan (49.32 t/ha) and China (21.86 t/ha) (FAOSTAT 2023). The foremost reasons for the lower productivity are the cultivation of open-pollinated varieties (OPVs) or locally produced seeds by farmers who have no or less quality control or regulation over seed production. Several scientific reports have demonstrated that compared with open-pollinated varieties, hybrid cultivars exhibit great heterosis and display greater uniformity in terms of their horticultural and morphological characteristics (Nunes et al. 2014; Netrapal and Singh 1999). In India, Shashikanth et al. (2007) observed higher heterosis of 45.31% over better parent and 27.40% over the standard check in one of its hybrid whereas Singh and Bhonde (2011) observed higher gross yield in hybrids ‘Mercedes’ (72.5 t/ha), Linda Vista (71.6 t/ha), Cougar (67.5 t/ha) compared to the open pollinated variety ‘ALR’ (42.7 t/ha). Henceforth, the bulb productivity of Indian onions might also be improved by selecting and developing well-adopted high-yielding F1 hybrids.
Onion hybrid breeding started after the reports of cytoplasmic male sterility induced by S-cytoplasm (Jones and Emsweller 1936) and complete restoration of fertility through the single nuclear dominant (Ms) restorer gene (Jones and Clarke 1943). The development of onion hybrids using this system is the only feasible and economical method for increasing onion productivity and bulb uniformity while maintaining desirable quality and disease resistance. The S-cytotype is popular for hybrid development worldwide due to its complete stability under diverse and varying environmental conditions and further this is inherited simply by a single dominant nuclear gene (Havey 2000). Nevertheless, the hindrances in developing onion hybrids remain because of their biennial habit and male sterility mechanism, which requires 4 to 8 years for identification of the cytoplasm and genotype of nuclear Ms alleles through progeny testing (Havey 2000). Henceforth, specific molecular markers were supposed to be the ideal tool to speed up this time-consuming and laborious procedure of identifying maintainers for hybrid development. Specific PCR-based molecular markers used to distinguish N and S types of cytoplasm (Havey 1995; Sato 1998; Von Kohn et al. 2013) and all three N, S and T cytotypes (Kim et al. 2009) have been tested. Identification of male sterile onion lines based on visual and/or microscopic examination is easy. However, the isolation of maintainer lines requires progeny testing for genotyping of the nuclear Ms locus. Molecular markers closely linked with the Ms locus were initially reported by Gokce and Havey (2002) followed by other marker types (Bang et al. 2011; Kim 2014; Kim et al. 2015; Huo et al. 2015) and single nucleotide polymorphisms (SNPs) (Havey 2013) tightly associated with the Ms locus. To date, no commercial Indian onion hybrids from the public sector are available at the national scale. Despite the fact that Indian onion researchers have been working for the last 60 years, the development of onion hybrid cultivars has not gained momentum. A positive step in this direction are some reports of deployment of PCR based markers for cytoplasm and Ms locus identification in Indian onions (Chaurasia et al. 2010; Saini et al. 2015; Khar and Saini 2016; Khar et al. 2022). However, these reports are not comprehensive enough to provide an overall picture of the cytoplasmic and Ms locus distributions in Indian onion populations. To accelerate hybrid breeding programme, the first step is to identify male sterile and maintainer lines in different genetic backgrounds to serve as base materials. Hence, this work aimed to identify cytoplasm types and nuclear fertility restorer (Ms) locus in populations of commercially grown short-day Indian onion open-pollinated varieties.
Materials and methods
Plant materials
The plant material comprised 35 open-pollinated commercially cultivated short-day Indian varieties (OPVs) representing more than 10 states. The varieties represent diversity present in Indian onions and bulb colour varied from white to dark red (Table 1). These varieties are being maintained at the Division of Vegetable Science, ICAR-Indian Agricultural Research Institute, New Delhi.
Morphological assessment of anthers
A visual assessment of flowers on the basis of anther colour could lead to the use of morphological markers for the identification of male sterile and fertile plants. Phenotypically, male sterile flowers and fertile plants were distinguished based on anther colour. The initial hypothesis was that light green anthers would be considered as male sterile and that dark green anthers would be considered as male fertile plants (Pathak 1997).
Microscopic assessment of pollens
For pollen viability, flowers were collected in the morning from umbels which had 60% dehisced flowers. Anthers were collected and pollens were dispersed on a microscope slide and stained with 0.5% acetocarmine solution (Khar and Saini 2016). Pollens were observed under a microscope and classified as viable if the pollens were elliptical in shape with two stained nuclei and if the cytoplasm was pink. Pollen that was clumped, and transparent with an unusual shape was identified as non-viable.
Collection of samples for DNA extraction
Twenty-four plants from each variety were tagged and assigned serial numbers ranging from 1 to 24. After three to four weeks of bulb planting, tender leaf tips of the 1–2 cm portion were cut with a sterilized pair of scissors, immediately wrapped in aluminum foil and put in liquid nitrogen for extraction of DNA. A total of 840 samples were taken from 35 OPVs.
Genomic DNA extraction and PCR amplification
Genomic DNA was isolated, from 840 individual plants of 35 OPVs, using the CTAB method with minor modifications (Murray and Thompson 1980). For the determination of cytoplasm, two specific PCR based molecular markers were explored. The mitochondrial orf725 gene mentioned as a MKFR marker (Kim et al. 2009) and an indel in the accD gene in chloroplast genome denoted as the accD marker (Von Kohn et al. 2013) were used (Supplementary Table 1). To identify the male fertility (Ms) locus, four identified PCR markers, namely, OPT (Bang et al. 2011), AcSKP1 (Huo et al. 2015), jnurf13 (Kim 2014) and AcPMS1 (Kim et al. 2015), were used (Supplementary Table 2).
Results and discussion
The CMS mechanism is an excellent model for examining the nuclear and cytoplasmic interactions since fertility restoration depends upon nuclear genes that govern the cytoplasmic dysfunction in terms of the viability of pollen. This genetic system is the most preferred approach for the commercial and wide exploitation of heterosis in various field and horticultural crops and is characterized by the genetic expression of developmental differences among hybrids and their respective parents. Onion hybrids exhibit higher heterosis over open pollinated varieties in terms of agronomical characteristics (Netrapal and Singh 1999; Nunes et al. 2014). Hybrids are quite popular among onion growers worldwide due to their ability to yield higher quantities of onions with uniform bulb, morphological and maturity traits. This crop is strictly cross-pollinated and is classified as a crop showing steep inbreeding depression.
Morphological assessment and pollen sterility status
The fertility and sterility status of the flower buds were assessed by examining anther colour visually and examining pollen dust by touching the flowers. It was observed that visual examination led to the identification of 789 (93.93%) plants as fertile whereas 51 (6.07%) were observed to be sterile. A wide range of anther and pollen colour was also observed and it varied from light creamish yellow to dark green colour (Fig. 1).
It was observed that the colour of anther or pollen could not decide the sterility status of the pollen. In Pusa Sona, anthers were light green and visually sterile but upon microscopic examination, they were not sterile. Similar discrepancies were observed in PWF, L-819, Bhima Safed, Bhima Shweta, RO252, Punjab Naroya and KRR (Supplementary Table 3). Earlier, it has been documented that light green coloured anthers possess sterile pollen in onion flowers (Pathak 1997; Santos et al. 2010; Saini et al. 2015). Further, Santos et al. (2010) elicited that light green anthers in onion flowers were male-sterile. The present investigation revealed that anther and pollen colour alone cannot decide the sterility level of pollen, since dark green and other coloured anthers contain sterile pollen. Khar and Saini (2016) also hypothesized that male sterility has no correlation with the anther colour, and our results confirm their findings.
Microscopic assessment of pollen
In plant species, the use of staining chemicals to separate viable and nonviable pollen grains has been exploited for decades (Peterson et al. 2010). Acetocarmine staining for onion ideally works to assess aborted or non-aborted pollens (Khar and Saini 2016; Lee and Havey 2020). In the present study, considerable variation in anther and pollen colour was observed (Supplementary Table 4) and it was cumbersome to determine the fertility versus sterility level of plants on the basis of colour visually. Hence, acetocarmine staining solution (0.5%) was used to distinguish the sterility status of the pollen grains. The scoring of viable pollen grains was based on the strongly stained nucleus and cytoplasm, whereas, no or lightly stained pollen grains were scored as non-viable (Fig. 2). Apart from stain, abnormal or misshapen pollens were also recorded in the aborted category. A total of 24 plants per variety were tagged for assessing viability via visual and microscopic studies. There was discrepancy between the visual and microscopic studies for most of the varieties. e.g., In Pusa Shobha, 21 fertile and 3 sterile plants were observed visually but microscopic observation led to the detection of 23 fertile and 1 sterile plant. This was the case for most of the varieties. This implies that the selection of plants based on visual examination for sterility is not advisable. Microscopic studies are important for a valid assessment of sterility. Male sterility was also detected in at least eleven varieties that can be used for the development of male sterile and maintainer lines in different genetic backgrounds. Based on the visual examination, 93.9% (789) were fertile and 6.1% (51) were sterile whereas microscopic studies revealed that out of 840 plants, 804 (95.71%) were fertile, and 36 (4.29%) were sterile plants. The highest percentage (62.5%) of sterile plants was observed in Arka Bheem followed by KRR (25.0%) and Pusa Red (20.8%). The colour of anthers and pollen might be influenced by the prevailing environmental conditions of the specific region. With increasing temperature and plant age, continuous variation in colour was observed. Another observation was that the fertile pollen possessed some adhesive traits whereas, the sterile pollen grains showed dryness and dullness. It might be due to the manipulation in the synthesis in the glucose, fatty acids, pectin and cellulose as described in the cotton by Wu et al. (2015). Further in potato, mature sterile pollens were recorded with significant decline in the carbohydrate pool and augmentation in the amino acid concentration. Various alterations in lipophilic compounds and fatty acid pool led to defective cell wall morphology of pollen grain depending upon the deposition of callose and sporopollenin quality (Shishova et al. 2019).
Cytoplasm identification
In onion, the sterile (S) type of cytoplasm is extensively exploited and preferred globally for the development of F1 hybrids in the onion crop owing to its greater stability under diverse climatic and environmental conditions. The polymorphism in mtDNA for differentiation of N and S type of cytoplasm are significantly faster than test crosses and saves abundant time to accelerate hybrid development. Identification of the plants that possess male sterile cytoplasm and the apt fertility restorer nuclear gene(s) is the critical step for the initiation of a heterosis breeding program.
In the present investigations, all 840 individual plants of thirty-five genotypes were analyzed using two cytoplasmic specific markers viz., accD and MKFR. Based on accD marker, out of 840 plants, 803 (95.6%) were having normal (N) and 37 (4.4%) had sterile (S) cytoplasm. None of the plants was recorded with T-cytoplasm. The same results were observed by using MKFR markers for identification of the N and S cytoplasm. The chloroplast based InDel primer accD determined 37 (4.4%) individuals possessing S-cytoplasm in the eight commercial cultivars that included Pusa Shobha (1), Pusa Red (10), GJWO-11 (1), Bhima Shweta (1), Arka Bheem (15), Kalyanpur Red Round, PRO-6 (1) and Punjab Naroya (1). Similarly, the mitochondrial marker MKFR followed the same trend and validated the results obtained from accD. The MKFR too couldn’t predict any plant possessing T-cytoplasm. T cytoplasm was reported by Khar et al. (2022) in the hybrid ‘T821’ being grown under Indian conditions. Recently, the ‘T’ like cytoplasm has been designated as ‘R’ since the sterility is restored by single dominant Ms allele (Havey and Kim 2021). The OPVs popularly grown in Northern Indian conditions for the last 30 years namely Pusa Red and Kalyanpur Red Round exhibited 17 (35.41%) plants having S-cytoplasm. Pusa Red has earlier been reported to have S cytoplasm (Khar and Saini 2016; Khar et al. 2022). On the other hand, Arka Bheem, a tri-parental synthetic variety having pinkish red and elongated globe bulbs possessed a maximum number (15/24) of plants containing S-cytoplasm. This can be expected since the initial work on onion male sterility started at IIHR, Bengaluru (Pathak et al. 1980) from where Arka Bheem has been released. One of the parents used in synthetic development may be male sterile. Except for two (GJWO-11 and Bhima Shweta), all S-cytoplasm-containing varieties belong to the red bulb category. This implies that various institutes are actively engaged in breeding efforts to introduce S cytoplasm into Indian varieties where these cultivars have been released. However, the absence of the use of molecular markers might have led to oversight, potentially causing breeders to miss identifying the sterility status. Khar and Saini (2016) reported that 31 (93.9%) of the Indian OPVs had N cytoplasm and only 2 (5.8%) had S cytoplasm. Similarly, Khar et al. (2022) reported that 91.3% of Indian OPVs had N cytoplasm whereas 8.7% had S cytoplasm. These reports were based on one plant per OPV whereas we used 24 plants per OPV and the results are a true indicator of the sterility status in Indian onion.
Genotyping of the Ms locus
In general, aberrant mitochondrial genes induce male sterility in the higher plants which can be reverted by the interaction of nuclear gene(s) known as fertility restorer genes (Rf). In onion, one dominant Rf gene (Ms locus) was reported to govern the restoration of fertility in the S-cytoplasm (Jones and Clarke 1943). However, three independent loci were observed to be involved in the restoration of T-cytoplasm fertility status (Schweisguth 1973).
In this study, four PCR-based molecular markers (AcPMS1, AcSKP1, jnurf13 and OPT) were exploited for genotyping the Ms locus in diverse Indian onion populations. Among 840 individual plants, AcPMS1, AcSKP1, jnurf13, and the OPT primer predicted 98.33% (826/840), 99.52% (836/840), 99.40% (835 /840) and 38.81% (326/840) homozygous recessive genotypes (msms), respectively. Whereas, these markers predicted 0.12% (1/840), 0.0% (0/840), 0.12% (1/840) and 21.67% (182/840) homozygous dominant alleles (MsMs), respectively. The frequencies of observed heterozygous dominant (Msms) allele were 1.55% (13), 0.48% (4), 0.48% (4) and 39.52% (332), respectively (Table 2).
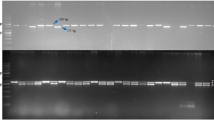
For the Ms locus, the AcPMS1 marker identified 13 heterozygous (Msms) plants from 840 plants including 4 each from Early Grano (Fig. 3) and Bhima Safed, 3 from Pusa Red and one from Pusa White Round and Bhima Dark Red.
Except for AcSKP1, all the other three Ms specific markers predicted homozygous dominant alleles in the Indian short-day onion population. Maximum homozygous recessive alleles were observed in all the plants of all the accessions. The marker jnurf13 identified 99.40% of the plants with the msms genotype, and AcSKP1 predicted 98.33% of the plants with homozygous recessive Ms alleles. The marker OPT showed considerably unpredictable results compared to the other markers (Fig. 4).
This marker detected 332 plants out of 840 (39.52%) with heterozygous (Msms) and 21.67% homozygous dominant (MsMs) alleles. However, the AcSKP1 marker exhibited no dominant homozygous genotype. A total of 836 out of the 840 plants possessed a recessive homozygous Ms locus (msms) according to the AcSKP1 marker. One plant had a homozygous dominant genotype at the Ms locus (MsMs) according to AcPMS1 and jnurf13 markers (Table 3).
Khar and Saini (2016) predicted that the primer AcPMS1 gave more accurate results than AcSKP1 based on visual observations of 25 commercially released varieties of Indian onion and some exotic lines grown under short-day conditions. The findings of this study suggest that all the primers (except OPT) predicted OPVs with similar frequencies. The efficacy of the use of the AcPMS1 and jnurf13 markers for correctly classifying onion populations was contested by Khar and Saini (2016) as they did not always predict the correct type of Ms genotype in the Indian population. However, the same markers were used on the North American onion population (Havey and Von Kohn 2017). AcPMS1 was recommended as a better marker as it displayed no recombination events with Ms locus whereas jnurf13 showed 3 events of recombination. The frequency of the Ms locus predicted by the OPT marker is completely different from that predicted by other three markers and needs further study. The results obtained from PCR markers need to be correlated with parents and their selfed progenies to arrive at a meaningful conclusion. Till then, we can conclude that the PCR marker AcPMS1 can be used for Ms locus determination in OPVs. Based on our findings, the homozygous recessive (msms) allele was the predominant Ms locus type with N cytoplasm, which contradicts the findings of previous studies (Pike 1986; Pathak 1997) in which the frequency of Nmsms plants in a population was found to be less than 5%. This identification of A, B and R/C (restorer line) lines from OPVs will aid in the development for high-yielding F1 hybrids with the low production cost of hybrid seed.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author upon reasonable request.
References
Bang H, Cho DY, Yoo KS, Yoon MK, Patil BS, Kim S (2011) Development of simple PCR-based markers linked to the Ms locus, a restorer-of-fertility gene in onion (Allium cepa L.). Euphytica 179:439–449
Brewster JL (2008) Onions and other vegetable alliums (No. 15). CABI
Chaurasia AK, Adsul GG, Nair D, Subramaniam VR, Krishna B, Sane PV (2010) Diversity in Indian and some exotic onion cultivars as revealed by genomic and mitochondrial DNA. Acta Hortic 859:207–220
FAOSTAT (2023) Onion production, area and productivity. https://www.fao.org/faostat/en/#data/QCL.Accessed on 13 Feb 2024
Gokce AF, Havey MJ (2002) Linkage equilibrium among tightly linked RFLPs and the Ms locus in open pollinated onion populations. J Am Soc Hortic Sci 127:944–946
Havey MJ (1995) Identification of cytoplasms using the polymerase chain reaction to aid in the extraction of maintainer lines from open-pollinated populations of onion. Theor Appl Genet 90:263–268
Havey MJ (2000) Diversity among male-sterility-inducing and male-fertile cytoplasms of onion. Theor Appl Genet 101:778–782
Havey MJ (2013) Single nucleotide polymorphisms in linkage disequilibrium with the male-fertility restoration (Ms) locus in open-pollinated and inbred populations of onion. J Am Soc Hortic Sci 138:306–309
Havey MJ, Kim S (2021) Molecular marker characterization of commercially used cytoplasmic male sterilities in onion. J Am Soc Hortic Sci 146:351–355
Havey MJ, Von Kohn C (2017) Efficacy of molecular markers jnurf13 and AcPms1 for prediction of genotypes at the nuclear Ms locus in North American open pollinated populations of onion. Hortic Sci 52:1052–1053
Huo YM, Liu BJ, Yang YY, Miao J, Gao LM, Kong SP, Wang ZB, Kitano H, Wu X (2015) AcSKP1, a multiplex PCR based co-dominant marker in complete linkage disequilibrium with the male-fertility restoration (Ms) locus, and its application in open pollinated populations of onion. Euphytica 204:711–722
Jones HA, Clarke A (1943) Inheritance of male sterility in the onion and the production of hybrid seed. J Am Soc Hortic Sci 43:189–194
Jones HA, Emsweller SL (1936) A male-sterile onion. J Am Soc Hortic Sci 34:582–585
Khar A (2022) Molecular breeding in allium. Edible alliums-botany, production and uses (Rabinowitch HD and Thomas B). CABI Publishing, UK, pp 284–295
Khar A, Saini N (2016) Limitations of PCR-based molecular markers to identify male-sterile and maintainer plants from Indian onion (Allium cepa L.) populations. Plant Breed 135:519–324
Khar A, Zimik M, Verma P, Singh H, Mangal M, Singh MC, Gupta AJ (2022) Molecular marker-based characterization of cytoplasm and restorer of male sterility (Ms) locus in commercially grown onions in India. Mol Biol Rep 49:5535–5545
Kim S (2014) A codominant molecular marker in linkage disequilibrium with a restorer-of-fertility gene (Ms) and its application in re-evaluation of inheritance of fertility restoration in onions. Mol Breed 34:768–778
Kim S, Lee E, Cho DY, Han T, Bang H, Patil BS, Ahn YK, Yoon M (2009) Identification of a novel chimeric gene, orf725, and its use in development of a molecular marker for distinguishing among three cytoplasm types in onion (Allium cepa L.). Theor Appl Genet 118:433–441
Kim S, Kim CW, Park M, Choi D (2015) Identification of candidate genes associated with fertility restoration of cytoplasmic male-sterility in onion (Allium cepa L.) using a combination of bulked segregant analysis and RNA-seq. Theor Appl Genet 128:2289–2299
Lee HI, Havey MJ (2020) Variable penetrance among different sources of the male fertility restoration allele of onion. Hortic Sci 55:543–546
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Netrapal N, Narendra Singh NS (1999) Heterosis for yield and storage parameters in onion (Allium cepa L.). Indian J Agric Sci 69:826–829
Nunes RLC, Oliveira ABD, Dutra AS (2014) Agronomic performance of onion hybrids in Baraúna, in the semi-arid region of Brazil. Rev Ciênc Agron 45:606–611
Pathak CS (1997) A possible new source of male sterility in onion. Acta Hortic 433:313–316
Pathak CS, Singh DP, Deshpande AA (1980) Annual Report, IIHR, Bangalore, pp 34–36
Peterson R, Janet P, Slovin CC (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. Int J Plant Biol 1:e13
Pike LM (1986) Onion Breeding. In: Publishing AVI (ed) Breeding vegetable crops. Basset MJ. Co., Inc., Westport, Connecticut, pp 357–394
Saini N, Hedau NK, Khar A, Yadav S, Bhatt JC, Agrawal PK (2015) Successful deployment of marker assisted selection (MAS) for inbred and hybrid development in long-day onion (Allium cepa L.). Indian J Genet Plant Breed 75:93–98
Santos CAF, Leite DL, Oliveira VR, Rodrigues MA (2010) Marker-assisted selection of maintainer lines within an onion tropical population. Sci Agric 67:223–227
Sato Y (1998) PCR amplification of CMS-specific mitochondrial nucleotide sequences to identify cytoplasmic genotypes of onion (Allium cepa L.). Theor Appl Genet 96:367–370
Schweisguth B (1973) E ́ tude d’un nouveau type de sterilite ́ male chez l’oignon, Allium cepa L. Ann Ame ́lior Plant 23:221–233
Shashikanth Evoor SE, Gowda RV, Gangappa E, Monohar RK (2007) Heterosis for yield, yield components and quality traits in onion (Allium cepa L.). Karnataka J Agric Sci 20:813–815
Shishova M, Puzanskiy R, Gavrilova O, Kurbanniazov S, Demchenko K, Yemelyanov V, Pendinen G, Shavarda A, Gavrilenko T (2019) Metabolic alterations in male-sterile potato as compared to male-fertile. Metabolites 9(2):24. https://doi.org/10.3390/metabo9020024
Singh RK, Bhonde SR (2011) Performance studies of exotic onion (Allium cepa L.) hybrids in the Nasik region of Maharashtra. Indian J Hill Farming 24:29–31
Von Kohn C, Kiełkowska A, Havey MJ (2013) Sequencing and annotation of the chloroplast DNAs and identification of polymorphisms distinguishing normal male-fertile and male-sterile cytoplasm of onion. Genome 56:737–742
Wu Y, Min L, Wu Z et al (2015) Defective pollen wall contributes to male sterility in the male sterile line 1355A of cotton. Sci Rep 5:9608. https://doi.org/10.1038/srep09608
Funding
This work was financially supported by a research grant awarded under the Core Research Grant-Science and Engineering Research Board (CRG-SERB) Scheme, Government of India (CRG/2019/006525) awarded to the corresponding author.
Author information
Authors and Affiliations
Contributions
HS was responsible for the layout of the trial, field work, data collection and writing of the first draft. Dr MZ was responsible for DNA isolation, marker validation and molecular analysis. Dr MM, KG, SS contributed to the formulation and guidance of the research and manuscript editing. Dr AK is the supervisor of this research paper, and he contributed to the development of the concept, guided the research, wrote and edited the manuscript critically for publication. Finally, all the authors contributed to the final approval of the version to be published. Hence, before submission for publication, all the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article. The authors have no financial or proprietary interests in any material discussed in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, H., Zimik, M., Mangal, M. et al. Distribution pattern of cytoplasm and restoration of male fertility (Ms) locus in short-day tropical Indian onion populations. Euphytica 220, 70 (2024). https://doi.org/10.1007/s10681-024-03334-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-024-03334-1