Abstract
Foxtail millet downy mildew caused by obligate parasitic Oomycetes Sclerospora graminicola (Sac.) Schrot greatly reduces the foxtail millet yield. The purpose of this study was to find the specific bacterial and fungal populations that might be involved in the suppression of foxtail millet downy mildew. Bacterial and fungal communities in foxtail millet rhizosphere soils under two different cropping systems were compared, with high-throughput Illumina sequencing technology. Compared with foxtail millet continuous cropping system, the foxtail millet–soybean–potato (Si–Gm–St) rotational cropping system showed the lower foxtail millet disease incidence (3.00%) and higher yield (3892.00 kg ha–1). The Bacteria/fungi value of foxtail millet rhizosphere soil in Si–Gm–St rotational cropping system was higher than that in foxtail millet continuous cropping system. The enzyme activities of phosphatase, urease and sucrase showed the same trend. Lower abundance of pathogenic microorganisms and higher abundance of plant growth-promoting microorganisms were observed under the Si–Gm–St rotational cropping system. There was a significant negative correlation between the relative abundances of Nocardioides, Devosia, Neorhizobium and Pseudomonas and foxtail millet downy mildew incidence, but an extremely significant positive correlation between the relative abundances of Fusarium, Cylindrocarpon, Ilyonectria, Thermoascus, Myrothecium and the disease incidence (P < 0.01). Different foxtail millet cropping systems were found to occupy different rhizosphere bacterial and fungal communities, and high bacterial diversity and low fungal diversity might be associated with the effective mitigation of downy mildew.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Foxtail millet (Setaria italica L.) originating in China, is mainly grown in arid and semi-arid regions of the world (Xu et al. 2018). Downy mildew is the most destructive disease in foxtail millet production, caused by Sclerospora graminicola (Sac.) Schrot, and classified as Oomycetes (Michie et al. 2017). The pathogen oospores of foxtail millet downy mildew can be easily blown into the field by the wind after they are dry at the hair of the infected plant and on the head of the hedgehog, and then overwinter in the soil. After the seeds are sown in the next year, it will infect the plant as the foxtail millet germinates and grows. Therefore, under the millet continuous cropping system, the amount of pathogenic oospore in the soil will multiply year by year, and the downy mildew incidence will correspondingly increase. However, few reports on the prevention of this destructive disease.
Microbes in plant rhizosphere soils form an interactional mechanism that greatly improves soil quality and plant adaptability (Berendsen et al. 2012). For example, soil microbial communities secreting antibacterial properties can protect plants from soil-borne pathogens (Lugtenberg and Kamilova 2009). In addition, specific microorganisms can enhance crop yield by improving soil nutrient availability, degrading soil toxic compounds, and inhibiting the reproduction of soil pathogens (Bais et al. 2006; Wu et al. 2017). In recent years, the relationship between plant disease resistance and rhizosphere microbial diversity has been extensively studied in plant-pathogen interaction system, such as Candidatus Liberibacter asiaticus of Citrus (Wu et al. 2014), Arbuscular mycorrhizal of maize-wheat rotational system (Hu et al. 2015), Ustilago crameri Korn of foxtail millet (Han et al. 2016), and Pseudomonas fluorescens of cucumber (Zhou et al. 2017a). However, in light of foxtail millet downy mildew, little information is known on the role of rhizosphere microbial community in pathogen suppression.
Plant genotypes are closely related to their rhizosphere soil microbial composition. Plants provide nutrients and energy for the growth of soil microorganisms by producing root exudates. It is found that different crop rotational systems can form different rhizosphere microbial communities in the same plot (Frindte et al. 2020). Therefore, the diversity and richness of plant rhizosphere soil microorganisms are closely related to crop genotypes. Gu et al. finds that with the continuous planting of Chinese chives, the abundance of pathogenic fungi Fusarium has increased, while the abundance of beneficial bacteria Pseudomonas significantly has decreased in the rhizosphere soil (Gu et al. 2020).
Likewise, the abundance of Fusarium, Thelephora, Hortaea and Penicillium in the rhizosphere soil of the three-year continuous cropping system of Casuarina equisetifolia increased year by year (Zhou et al. 2019). Under the strawberry continuous cropping system, the increase of Fusarium, Humicola and Arthrobotrys in the strawberry rhizosphere soil has caused the accumulation of phenolic substances, which in turn inhibit the growth of soil-beneficial bacteria Bacillus, Sphingomonas and Sphingopyxis (Chen et al. 2020a). Other studies have also shown that the rotational cropping system can alleviate continuous cropping obstacles by changing the diversity and richness of microorganism and inhibiting the proliferation of pathogenic bacteria (Chen et al. 2020a). An example is that the tomato-celery-cucumber-Chinese cabbage rotational cropping system increases the Flavobacterium and Ohtaekwangia in the cucumber rhizosphere soil, which effectively alleviate the Fusarium wilt of cucumbers infected by Fusarium oxysporum f.sp. cucumerinum Owen (Zhou et al. 2017a). Soybean-oilseed rape rotation system increases the abundances of Sphingomonas, Bacillus, Streptomyces and Trichoderma in oilseed rape rhizosphere soil, and inhibits the clubroot disease caused by Plasmodiophora brassicae (Yang et al. 2020). However, for foxtail millet downy mildew, little information is known for the role of rhizosphere microbial community in pathogen suppression. Research on the composition of rhizosphere microbial community under the Si–Gm–St rotational cropping system can contribute to the discovery of potential microorganisms related to the downy mildew suppression. Therefore, the relationship between the composition of rhizosphere microbial community and downy mildew suppression under foxtail millet and different crop rotational cropping systems deserves further study.
In addition to microbial communities, soil enzymes also act as pivotal parts in the transformation of nitrogen, phosphorus, potassium and organic matter in soil (Pignataro et al. 2012; Burns et al. 2013). The studies of soil enzyme activity can indirectly characterize the physical and chemical properties of soil. Soil enzyme activities can be used as an important indicator in the evaluation of soil quality and fertility. Activities of polyphenol oxidase and urease are significantly positively correlated with the resistance of eggplant to Verticillium wilt (Zhou et al. 2012). And soil microbes greatly influence soil enzyme activities (Han et al. 2016). Previous reports have shown that the enhancement of bacterial diversity increased the enzyme activity in rhizosphere soil of apple trees (Sun et al. 2014a, b). All these indicate that high soil quality and good pathogen-inhibiting ability are closely related to high enzyme activity (Chen et al. 2021).
There are few reports on rotational cropping systems alleviating the obstacles to foxtail millet continuous cropping system, and even fewer studies have reported on the relationship between the changes of soil microbial community and foxtail millet downy mildew susceptibility. In the present study, we used high throughput Illumina sequencing method to compare the microbial communities of foxtail millet rhizosphere soils under the continuous and rotational cropping systems. The purpose of the study is to reveal the relationship between the rhizosphere microbial community and downy mildew disease incidence, and to explore specific fungi and bacterial taxa which can be used for suppressing this disease in the future.
Methods and materials
Experimental design and sample collection
Research plots were established at the southeast of Changzhi City, Shanxi Province, China (E 113°06′ N 36°11′), for the purpose of developing a long-term site for studying the 7-year continuous and rotational cropping systems. Soil type at the site is sandy clay loam. This site was planted with maize prior to establish our study. The basic soil properties were pH 7.24, soil organic matter (SOC) 17.63 g kg−1, available nitrogen (AN), available phosphorus (AP) and available potassium (AK) 26.98, 16.21, and 95.60 mg kg−1, respectively. The varieties of crops in this study were Changnong 35 foxtail millet, Zhonghuang 13 soybean (Glycine max (Linn.) Merr.) and Jinshu 16 potato (Solanum tuberosum L.). Two different cropping systems were foxtail millet continuous system and foxtail millet-soybean-potato (Si–Gm–St) rotational cropping system during growing season from 2014 to 2020. The area of the plot is 60 m2 (15 m × 4 m). There were three replicates for each experimental treatment.
Different crops were planted according to the following densities: plant spacing and row spacing of foxtail millet, soybean and potato were 10 cm × 20 cm, 20 cm × 40 cm and 25 cm × 65 cm, respectively. In 2020, seeds of foxtail millet were randomly sown in field plots under foxtail millet continuous and Si–Gm–St rotational cropping systems. Twenty healthy plants in each plot were randomly selected and sampled on July 7, 2020. First, loose soil adhered to the foxtail millet roots was gently shaken off. Then, the soil tightly adhered to the roots was brushed down and considered as the rhizosphere soil. The rhizosphere soils collected from twenty foxtail millet plants were evenly mixed as a sample. After passing through a 0.5 mm sieve, the mixed sample was divided into three parts for DNA extraction, enzyme activity determination and chemical property analysis, respectively.
Determination of soil chemical properties
Soil pH was measured by an IQ150/pH meter (Spectrum1 Technologies Inc., Aurora, IL, USA) at soil/water 1:5. 10 g of soil was treated with 2 M potassium chloride solution, and then a continuous flow analyzer (Skalar Sanþþ, Skalar Co., The Netherlands) was used to determine the available nitrogen (AN) (Mulvaney 1996). AP and AK were determined by Olsen’s method (Olsen and Dean 1982) and flame atomic absorption spectrophotometry, respectively. The dichromic acid oxidation method of Walkley and Black (1934) was used to measure SOC. Soil microbial biomass carbon (MBC) was extracted by chloroform-K2SO4 fumigation with a correction factor of 0.33 (Doan et al. 2013; Lipiec et al. 2015). Soil microbial biomass nitrogen (MBN) was measured using the method of Brookes et al. with a correction factor of 0.54 (Brookes et al. 1985).
Foxtail millet downy mildew incidence survey
The determination of foxtail millet downy mildew incidence was carried out during the ear formation period. The healthy or diseased foxtail millet plants were identified based on their visual symptoms. The diseased foxtail millet showed 2–3 pieces of heart leaf tissue were split into filaments that were curved like hair without heading, or the heading was deformed into a hedgehog shape. A large amount of yellow–brown powder (pathogenic oospores) was scattered in the diseased tissue. The foxtail millet downy mildew incidence in each plot was calculated according to the following formula: number of infected foxtail millet/ number of total foxtail millet × 100%.
Leaf photosynthesis and antioxidant enzymes activity measurement
During August, the net photosynthetic rate of the foxtail millet flag leaves was measured using the LI-6400 photosynthesis assay system (Li-Cor Inc., Lincoln, NE, USA). Then, the foxtail millet flag leaves were collected for enzyme activity determination. Superoxide dismutase (POD) activity was assayed based on the oxidation of guaiacol using H2O2 as described by the method of Hammerschmidt et al. (1982). Polyphenol oxidase (PPO) activity was assayed with the method of Yan et al. (2014).
Soil enzyme activity measurement
The activity of urease (UR) and sucrase (SUC) was measured as described by Yao and Huang (Yao and Huang 2006). Phosphatase (PP) activity was carried out as described by Tabatabai and Bremner (Tabatabai and Bremner 1969). Polyphenol oxidase (PPO) activity was determined using the method of Guan (Guan 2020).
Soil DNA extraction, Illumina sequencing and data processing
Soil microbial genomic DNA was extracted from each sample using an E.Z.N.A.®Soil DNA Kit (Omega, Carlsbad, CA, USA). Soil bacterial and fungal communities were sequenced with Illumina Miseq technology. Primer sets of F338 (5′-ACTCCTACGGGAGGCAGCAG-3′)/R806 (5′-GGACTACHVGGGTWTCTAAT-3′) and ITSF ((5′ GGAAGTAAAAGTCGTAACAAGG-3′)/ITSR (5′-GCTGCGTTCTTCATCGATGC-3′) were applied to amplify the bacterial 16S rDNA gene (V3–V4) and the fungal ITS gene, respectively. The PCR protocol was 4 min denaturation at 94 °C 30 s, followed by 25 cycles of 94 °C 30 s, 50 °C annealing 45 s, 70 °C elongation 30 s, and an extension at 70 °C for 4 min.
The PCR amplification products were purified by a rapid Gel Extraction Kit (Qiagen, Valencia, CA, USA). Barcoded V3-V4 of bacteria and ITS of fungi amplicons were sequenced with 7 cycle index reads on the Illumina Miseq platform. Sequence reads including ambiguous bases, or average phred score less than 25, or homopolymer run more than 6, or sequence lengths less than 150 bp, or mismatches in primers were eliminated. Only sequences without any mismatches and overlap more than 10 bp in length were assembled. The effective sequences from each sample were uploaded to QIIME for further analysis at 97% similarity of operational taxonomic units (OTU).
The Ribosomal Database Project (RDP) classifier was used to conduct the taxon-dependent analysis (Lan et al. 2012). USEARCH 6.1 in QIIME was used to identify and remove chimeric sequences (Caporaso et al. 2010). Singleton OTUs was eliminated after quality control filtered reads, resulting in an average of 50,531 quality bacterial sequences and 47,038 quality fungal sequences (Table 2). The average read lengths of bacterial 16S rDNA genes and fungal ITS regions were 438 bp and 252 bp, respectively. The accession numbers for bacterial and fungal sequencing data entered at NCBI are PRJNA825072 SRP368639 and PRJNA825114 SRP368847, respectively.
Quantitative PCR
A StepOnePlus™ real-time PCR system (Thermo) was used to estimate the abundance of total bacteria, fungi, Pseudomonas and nifH in foxtail millet rhizosphere soil. For total bacteria and fungi, forward primer (5′-GGGTTGCGCTCGTTGC-3′)/reverse primer (5′-ATGGYTGTCGTCAGCTCGTG-3′) and forward primer (5′-GGGTTGCGCTCGTTGC-3′)/reverse primer (5′-ATGGYTGTCGTCAGCTCGTG-3′) were used for amplification, respectively. The abundance of Pseudomonas spp. and nifH were amplified using the primers PsF (5′-ACTTTAAGTTGGGAGGAAGGG-3′) /PsR (5′-ACACAGGAAATTCCACCACCC-3′) and nifHF (5′-AAAGGYGGWATCGGYAARTCCACCAC-3′)/nifHR (5′-TTGTTSGCSGCRTACATSGCCATCAT-3′), respectively. Primer sets of total 16S rDNA, fungal ITS, Pseudomonas spp. and nifH gene were designed and synthesized by Wcgene Biotechnology Corporation (Shanghai, China). Each assay was repeated three times. Assay results were expressed as log10 values (target copy number g−1 soil) for statistics and analysis.
Taxonomy classification and statistical analysis
Abundance counts at the genus level were log2 transformed and then normalized (Zhao et al. 2013). The Chao and Shannon diversity indices and rarefaction curves were evaluated according to the method of Yu et al. (2015). OTU comparisons of soil samples were presented using a Venn diagram. Column charts were drawn using Sigmaplot, and * indicates significant difference (P < 0.5). SPSS 22.0 software was used for independent sample t-test to analyse the significance of differences between two samples (Chicago, IL, USA). Redundancy analysis (RDA) was performed using Canoco 5.0 and the Monte Carlo permutations test (499 permutations) was used to confirm whether soil microbial community compositions were correlated with the soil enzyme activities and properties.
Results
Soil chemical properties, MBC, MBN, foxtail millet disease incidence and yield
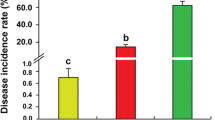
Soil chemical properties and microbial biomass were changed under the two different cropping systems (Table 1). Compared with foxtail millet continuous cropping system, AN, AP, AK, SOC, MBC and MBN were significantly increased under the Si–Gm–St rotational cropping system. There was little difference in pH between the two different cropping systems. Under Si–Gm–St rotational cropping system, the foxtail millet downy mildew incidence was 3.00%, which was significantly lower than that under foxtail millet continuous cropping system (15.33%) (Table 1). Furthermore, foxtail millet yields of the two different cropping systems were determined. Table 1 showed that foxtail millet continuous cropping system greatly decreased foxtail millet yield. The foxtail millet downy mildew disease resistance under Si–Gm–St rotational cropping system was significantly higher than that under foxtail millet continuous cropping system.
Physiological characteristics of the foxtail millet flag leaves
Furthermore, foxtail millet plant responses to different cropping systems were assessed by Pn and activities of POD and PPO. As shown in Fig. 1, Si–Gm–St rotational cropping system significantly increased Pn. The activity of PPO also showed the same trend (Fig. 1). It was worth noting that after Si–Gm–St rotational cropping, the PPO activity of foxtail millet increased by 68.80% (Fig. 1). Compared with foxtail millet continuous cropping system, POD activity decreased by 28.92% under Si–Gm–St rotational cropping system.
Soil enzyme activity
Compared with foxtail millet continuous cropping system, the activities of UR, PP, SUC and PPO were significantly increased by 100.65%, 40.36%, 71.97% and 65.48% under Si–Gm–St rotational cropping system, respectively (Fig. 2).
Soil microbial community richness and diversity
High-throughput Illumina sequencing technology of 16S rDNA/ITS was employed to characterize differences of the bacterial and fungal community compositions in the foxtail millet rhizosphere soils under two different cropping systems. The rarefaction curve was nearly saturated in all soil samples, implying a reasonable depth and coverage of sequencing (Fig. 3a, b). After quality-filtered and chimera-check, we obtained 48,120 and 52,942 bacteria sequences from foxtail millet rhizosphere soil of foxtail millet continuous and Si–Gm–St rotational cropping system, respectively (Table 2). Fungal sequences were 45,158 and 48,918 under foxtail millet continuous and Si–Gm–St rotational cropping system, respectively (Table 2). Under foxtail millet continuous cropping system and Si–Gm–St rotational cropping systems, the average operation taxonomic unit (OTU) number of bacteria was 4335 and 4450, and the average OTU of fungi was 696 and 733, respectively (Fig. 3c, d). Meanwhile, the diversity indices of bacterial and fungal communities were compared between the two different treatments. As shown in Table 2, the alpha-diversity of both bacteria and fungi was higher under Si–Gm–St rotational cropping system than that under foxtail millet continuous cropping system.
Soil bacterial community composition and structure
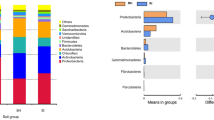
As shown in Fig. 4a, 17 phyla and subphylum were harboured across all samples with Alphaproteobacteria, Actinobacteria, Gemmatimonadetes, Chloroflexi, Bacteroidetes, Acidobacteria, Firmicutes, Gammaproteobacteria, Deltaproteobacteria, Betaproteobacteria Nitrospirae, Planctomycetes, Cyanobacteria, Tectomicrobia, Verrucomicrobia, Saccharibacteria and Armatimonadetes as the major taxa, which occupied more than 99% of the relative abundance of bacteria. However, there were still some variations in relative abundance of each phylum in different samples. Compared with foxtail millet continuous cropping system, the relative abundances of Chloroflexi, Gammaproteobacteria and Cyanobacteria were higher, and the relative abundances of Bacteroidetes and Firmicutes were lower under Si–Gm–St rotational cropping system (P < 0.05) (Fig. 4a). Genera with an average abundance greater than 0.10% in each soil sample were determined as major, and applied in the subsequent analysis. Compared with foxtail millet continuous cropping system, the relative abundances of Nocardioides, Solirubrobacter, Geodermatophilus, Devosia, Neorhizobium, Rubellimicrobium, Pseudomonas and Ensifer were higher, while the relative abundances of Aeromicrobium, Rubrobacter, Candidatus Entotheonella and Achromobacter were lower under Si–Gm–St rotational cropping system (P < 0.05) (Fig. 5a). Pearson’s correlation analysis found that the relative abundances of Gemmaproteobacteria and Chloroflexi phyla had a significant negative correlation with disease incidence, while an extremely significant positive correlation between Bacteroidetes and Firmicutes phyla with disease incidence (P < 0.01) (Table S1). The relative abundances of Nocardioides, Solirubrobacter, Geodermatophilus, Devosia, Neorhizobium, Rubellimicrobium and Pseudomonas had a negative association with disease incidence, while an extremely positive correlation between the relative abundance of Aeromicrobium, Rubrobacter, Candidatus Entotheonella and disease incidence was observed (Table S2).
Soil fungal community composition and structure
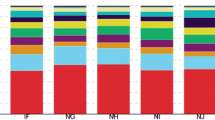
Ascomycota and Basidiomycota were the dominant phyla in all soil samples, and the relative abundances of these two phyla occupied more than 83% of the fungal sequences. Moreover, the relative abundances of Zygomycota and Chytridiomycota were more than 1% in at least one sample (Fig. 4b). The foxtail millet continuous cropping system has more relative abundance of Basidiomycota than the Si–Gm–St rotational cropping system (P < 0.05). Compared with foxtail millet continuous cropping system, the relative abundances of Mortierella, Ascobolus, Conocybe, Cyathus, Rhizophlyctis, Podospora and Acremonium were higher and the relative abundances of Myrothecium, Corynespora, Thermoascus, Pseudogymnoascu, Gymnoascus, Thermomyces, Schizothecium, Ilyonectria, Cylindrocarpon, Fusarium were lower under Si–Gm–St rotational cropping system (Fig. 5b). A significant positive correlation between the relative abundance of Basidiomycota and disease incidence was shown in a Pearson’s correlation analysis (P < 0.01) (Table. S1). The relative abundances of Mortierella, Acremonium, Podospora, Conocybe, Cyathus and Ascobolus genera had negative association with disease incidence, while an extremely positive correlation between disease incidence and Fusarium, Cylindrocarpon, Ilyonectria, Pseudogymnoascus, Thermoascus and Myrothecium was observed (P < 0.01) (Table S2).
Correlation analysis
The effects of soil chemical properties on microbial community composition were investigated using RDA analysis (Fig. 6). Figure 6a showed that the axe1 and axe2 accounted for 55.57% and 24.42% of the distribution of soil bacterial community structure at the phylum level. SOC, AN, AP and AK had the highest correlation with Betaproteobacteria, Gammaproteobacteria, Cyanobacteria and Chloroflex. Figure 6c showed that the axe1 and axe2 accounted for 70.26% and 12.57% of the distribution of soil bacterial community structure at the genus level. The Nocardioides, Solirubrobacter, Geodermatophilus, Devosia, Neorhizobium, Rubellimicrobium, Pseudomonas and Ensifer genera had a positive correlation with SOC, AN, AP and AK. Figure 6b showed that the axe1 and axe2 accounted for 80.72% and 3.85% of the distribution of fungal community structure at the phylum level. SOC, AN, AP and AK were positively correlated with Cercozoa and Chytridiomycota. Figure 6d showed that the axe1 and axe2 explained 75.59% and 16.75% of the distribution of fungal community structure at the genus level. The Fusarium, Acremonium, Cyphellophora, Ophioceras and Preussia genera had a positive correlation with SOC, AN, AP and AK (Fig. 6d). AN, AP, AK and SOC significantly affected the distribution of fungal communities (Fig. 6b, d). At the genus level, soil chemical properties significantly affected the distribution of bacterial communities (Fig. 6c). The effect of soil enzyme activities on soil chemical properties was shown in Fig. 6e. The axe1 and axe2 accounted for 96.30% and 0.55% of the changes of chemical properties (Fig. 6e). UR, PP, SUC and PPO were significantly correlated with SOC, AN, AP and AK.
Soil microbial abundances
Compared with foxtail millet continuous cropping system, the abundances of soil bacteria, Pseudomonas spp. and nifH were 2.71, 2.40 and 1.82 times than that under Si–Gm–St rotational cropping system, respectively (Table. 3). Bacteria/Fungi value was significantly higher under foxtail millet continuous cropping system than that under Si–Gm–St rotational cropping system (P < 0.05) (Table 3).
Discussion
Si–Gm–St rotational cropping system improves foxtail millet physiological indicators and rhizosphere microbial richness and diversity
Rhizosphere microbial community composition and structure of healthy plant can increase plant disease resistance and maintain plant health. Studying plant microbial community structure of rhizosphere soil can provide new ideas for exploring functional microorganisms that might inhibit soil-borne plant pathogens (Schweitzer et al. 2008). In agricultural production, different crop rotational systems can alleviate the spread of soil-borne diseases caused by crop continuous cropping systems. Qin et al. found that after potato rotation with oat or faba bean, the incidence of tuber black scurf decreased the abundances of beneficial microorganisms in potato rhizosphere soil and potato yield (Qin et al. 2022). Xu et al. showed that compared with foxtail millet-maize, foxtail millet-potato-maize and foxtail millet-maize-soybean rotational systems, the Si–Gm–St rotational system harbored the highest foxtail millet yield and lowest downy mildew incidence (Xu et al. 2022). Thus, we investigated the microbial community composition of foxtail millet rhizosphere soils under foxtail millet continuous and Si–Gm–St rotational cropping systems. Compared with foxtail millet continuous cropping system, Pn and antioxidant enzyme activities of foxtail millet flag leaves were changed under Si–Gm–St rotational cropping system (Fig. 1). The composition of nutrients and secondary metabolites excreted by plant roots may be affected by these physiological changes, which in turn alter plant rhizosphere microbial community composition and structure (Han et al., 2016). A previous report showed that the bacterial community composition in the rice rhizosphere soil changed after rice-maize rotation (Maarastawi et al. 2018), which is consistent with our findings under Si–Gm–St rotational cropping system (Table 3). Similarly, the richness and diversity of beneficial bacteria significantly increased in the tomato rhizosphere soil under tomato-celery rotational cropping system (Jin et al. 2021). Plant genotype has an important effect on the microbial community compositions of rhizosphere soil. Plants can produce root exudates and exfoliants, which provide nutrients and energy for the growth of soil microorganisms. Different plant genotypes can stimulate or inhibit the growth of specific microorganisms in their rhizosphere soils by secreting different compounds (Han et al. 2016). At the same time, rhizosphere soil microbes can also affect plant growth by changing the soil micro-ecological environment. Our result showed that the composition of rhizosphere bacterial and fungal communities varied greatly under different cropping systems (Figs. 4, 5). These differences may be caused by the rotation of soybean and potato with foxtail millet. Previous research found that the bacterial OUT of cucumber rhizosphere soil increased under tomato-celery-cucumber-Chinese cabbage rotational cropping system, which also proved our findings (Zhou et al. 2017a). Many studies have shown that the distribution of high abundance and diversity of functional bacteria is closely related to disease suppression (Rosenzweig et al. 2012; Wu et al. 2015). The results of high-throughput sequencing showed that the Chao and Shannon values of bacteria in foxtail millet rhizosphere soil were significantly increased under Si–Gm–St rotational cropping system, and the number of effective sequences also significantly increased (Table 2). The higher diversity of bacterial community composition, the more stable rhizosphere ecosystem can be provided for plants, which will be beneficial to promote crop growth and resist the invasion of plant soil-borne pathogens (Chen et al. 2020b). Moreover, the enrichment of antagonistic microorganisms in plant rhizosphere soil usually helps to improve plant disease resistance. Therefore, we conclude that higher rhizosphere antagonistic bacterial diversity in our study might be beneficial for suppressing foxtail millet downy mildew suppression.
Si–Gm–St rotational cropping system improves diversity of functional bacteria in foxtail millet rhizosphere soil
We also conducted an in-depth study on the species of bacterial community composition of foxtail millet rhizosphere soil under foxtail millet continuous and Si–Gm–St rotational cropping systems. Compared with foxtail millet continuous cropping system, large amounts of Cyanobacteria and Gemmaproteobacteria were observed under Si-Gm-St rotational cropping system(Fig. 3). Species of Cyanobacteria phylum were mainly involved in the fixation of free nitrogen in agricultural soils (Zhou et al. 2017b). The relative abundance of Gammaproteobacteria in foxtail millet rhizosphere soil increased under Si–Gm–St rotational cropping system (Fig. 4). Other results showed that Gammaproteobacteria was copiotrophic bacteria and suitable for proliferation in soil with high content of SOC and AN (Fierer et al. 2007; Fierer et al. 2012). In our results, there were higher levels of AN, nifH and SOC under Si–Gm–St rotational cropping system (Tables1, 3), which supported this conclusion. Likewise, Gammaproteobacteria was enriched in corn-rice rotational cropping soil than in corn or rice continuous cropping soil in the Philippine (Maarastawi et al. 2018). Moreover, the relative abundances of Firmicutes and Bacteroidetes were increased in foxtail millet rhizosphere soil under foxtail millet continuous cropping system compared with Si–Gm–St rotational cropping system. The difference in relative abundances of Cyanobacteria, Gammaproteobacteria, Bacteroidetes and Firmicutes indicated that these four phyla were closely related to the occurrence of foxtail millet downy mildew incidence (Table S1). To further study on the relationship between bacterial taxa and disease occurrence, we performed a genus level analysis. We speculate that the increase of bacterial diversity under Si–Gm–St rotational cropping system might be related to the higher nutrient level in soil and foxtail millet downy mildew suppression. Xu et al. found that higher AN and SOC were conducive to the distribution of bacterial diversity in the soil (Xu et al. 2018). Neorhizobium is a non-symbiotic neorhizobia isolated from dryland soils (Soenens et al. 2018). These non-nodular endophytes colonize root nodules can maintain a good micro-ecological environment between plants and soil microorganisms. It has been reported that Devosia netuniae contains the nifH gene and can form a unique nitrogen-fixing foot nodule symbionts with the aquatic legume Neptunia natans (L.f.) Druce. (Rivas et al. 2002). Chen et al. found that intercropping of peanut and maize increased the abundance of nifH compared with monoculture maize (Chen et al. 2017). Combining these reports with our results, we conclude that Si–Gm–St rotational cropping system may be more suitable for the proliferation of nitrogen-fixing bacteria. Other studies have shown that some species of the genera Neorhizobium and Devosia may participate in the nitrogen cycle in the soil, increasing the level of AN. However, in order to better demonstrate the nitrogen fixation mechanism of Neorhizobium and Devosia, related in-vitro experiments are still needed. Nocardioides degraded carbendazim and other organic compounds containing benzene ring (Busato et al. 2016). Our study found an enrichment of Pseudomonas spp., members of which are known to be endophytic bacteria that promote plant growth. For example, Pseudomonas fuorescens has been used to promote the plant growth by producing 2, 4-diacetylphloroglucinol (Landa et al. 2003). Pseudomonas koreensis group strains were dominated in the cocoyam rhizosphere and can suppress Pythium root rot by producing cyclic lipopeptide (Qin et al. 2022). Other results showed that Pseudomonas spp. was involved in the decomposition of alcohols, ethers and phorate in soil, and could antagonize soil pathogens such as Rhizoctonia solani Ag3 (Garbeva et al. 2004). In addition, q-PCR results found that the abundance of Pseudomonas spp. was increased under Si–Gm–St rotational cropping system, which further verified the conclusion of soil microbial diversity analysis (Table 3). Meanwhile, the correlation analysis showed that Pseudomonas spp. had a negative correlation with the disease incidence (Table S1). Aeromicrobium isolated from ginseng rhizosphere soil can produce erythromycin, a macrolide antibiotic with antibacterial function. Therefore, our results combined with earlier studies suggested that Pseudomonas spp. and Aeromicrobium might play important roles in the suppression of downy mildew pathogen. However, future studies such as in vitro antifungal tests using isolated Pseudomonas spp. and Aeromicrobium bacteria, are still needed to identify their antagonistic activities against smut disease pathogen and better understand the relative mechanisms.
Si–Gm–St rotational cropping system improves diversity of functional fungi in foxtail millet rhizosphere soil
In addition, studies on the fungal community composition showed that Basidiomycota was increased in foxtail millet continuous cropping system. This is consistent with Wu’s research results in the rhizosphere soil of ginseng continuous cropping system (Wu et al. 2019). Leite et al. found that Basidiomycota has the ability to degrade cellulose (Leite et al. 2017). Some species of Basidiomycota phylum cultures secrete exopolysaccharides with strong antioxidant properties and immunity, good antibacterial properties and antitumor activity (Osińska-Jaroszuk et al. 2015). RDA analysis showed that SOC, AN and AP were significantly positively correlated with the distribution of soil fungal diversity at the genus level (Fig. 6d). Some Mortierella spp. could promote plant growth by producing antibiotics against various plant pathogens (Xiong et al. 2017; Liu et al. 2020a). Fusarium graminearum could infect the roots of maize seedlings (Zhao et al. 2013). Fusarium oxysporum f.sp. niveum. was the pathogen of watermelon rhizosphere Fusarium wilt. Further studies showed that Fusarium spp. could colonize asparagus rhizosphere and increase the disease incidence (Gilardi et al. 2013). In this study, the increased abundance of Fusarium may be related to the enhancement of soil borne diseases in the rhizosphere of Rehmannia glutinosa. Ophioceras was suitable for living in soils with high nitrogen levels and had the function of breaking down carbohydrates (Garrett et al. 2010). Cyathus striatus could produce highly active extracellular enzymes that promoted the cycling of carbon, nitrogen and phosphorus in soil (Wang et al. 2014). Acremonium coenophialum could increase the productivity and stress tolerance of tall fescue (Belesky and Fedders 1995; Martínez-culebras et al. 2004). Thermostable hemicellulase produced by Thermomyces lanuginosus may contribute to hemicellulose degradation during composting (Zhang et al. 2015). The main functions of the above microbial groups were concentrated in the decomposition and denitrification of cellulose, thereby promoting the cycle of carbon and nitrogen. It was also the reason for the decreasing of AN and SOC in foxtail millet continuous cropping system. It was found that the number of fungi could participate in soil nutrient cycling increased, while some pathogenic fungi decreased under Si–Gm–St rotational cropping system. This indicated that Si–Gm–St rotational cropping system could reduce the spread of soil borne diseases.
Si–Gm–St rotational cropping system increases the activity of nutrient cycling enzymes in foxtail millet rhizosphere soil
The Si–Gm–St rotational cropping system increased aboveground plant diversity, and the residues and root exudates of different plant genotypes enhanced soil nutrients and microbial diversities (Liu et al. 2020b). The abundant soil microbes can increase the enzyme activities of rhizosphere soil. Otherwise, the increased enzyme activities promoted the decomposition of nutrients in the soil, thereby increasing the microbial diversities in the soil (Ndabankulu et al. 2022). For example, soil alkaline phosphatase plays an important role in the mineralization of organic matter, which can convert phosphorus components in soil into available inorganic phosphorus, thereby increasing the diversity of soil microorganisms (Ullah et al. 2019). In addition, the other studies also show that PP activity in the rhizosphere soil of resistant banana varieties is higher than that of susceptible varieties (Sun et al. 2013). Compared with foxtail millet continuous cropping, higher nitrogen levels increased soil microbial diversity under crop rotation treatments, which in turn promotes the activities phosphatase and urease (Treseder and Vitousek 2001). The hydrolysis of sucrose by sucrase can reflect the conversion ability of SOC, and the catabolism of sucrose can provide a carbon source for the growth and reproduction of microorganisms (Cantarella et al. 2018). PPO activity is positively correlated with plant disease resistance. PPO is an important oxidoreductase, which plays an important role in soil antibacterial defense system (Zhou 2012). Furthermore, millet rhizosphere soil of the Si–Gm–St rotational system harbored higher enzyme activities as compared to the millet continuous cropping system (Fig. 2). The reason may be that the increase of rhizosphere microbial community diversity is closely related to higher soil enzyme activity (Sun et al. 2014a, b). Some specific bacterial and fungal taxa may support effective nutrient cycling, and can resist soil diseases to a certain extent, and thus to provide better nutritional conditions for foxtail millet growth under Si–Gm–St rotational cropping system.
Conclusions
Compared with foxtail millet continuous cropping system, the diversities and abundances of bacteria and fungi were increased, the foxtail millet downy mildew incidence was reduced, and the yield was increased under Si–Gm–St rotational cropping system. Downy mildew suppression may be associated with high bacterial diversity and enzyme activity in the rhizosphere soil. Furthermore, the specific rhizosphere microbial community was composed, especially with the high abundance of beneficial bacteria Pseudomonas and Aeromicrobium and low abundance of pathogenic fungi Fusarium. The two genera Pseudomonas and Aeromicrobium could be used as the potential bacteria to suppress downy mildew in the future, although their antagonistic activities against the obligate parasitic oomycetes pathogen still need to be further identified. In conclusion, the changes of soil microbial community structure and function are jointly restricted and influenced by Si–Gm–St rotational cropping and soil environmental factors. Our findings provide a basis for further development of sustainable and environmentally compatible agricultural systems under Si–Gm–St rotational cropping system.
Availability of data and material
Not applicable.
Code availability
Not applicable.
References
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizoshpere interactions with plant sand other organisms. Annu Rev Plant Biol 57:233–266. https://doi.org/10.1146/annurev.arplant.57.032905.105159
Belesky DP, Fedders JM (1995) Tall fescue development in response to Acremonium coenophialum and soil acidity. Crop Sci 35(2):529–533
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486. https://doi.org/10.1016/j.tplants.2012.04.001
Brookes PC, Landman A, Pruden G, Jenkinson DS (1985) Chloroform fumigationand the realease of soilnitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol Biochem 17:837–842
Burns RG, Deforest JL, Marxsen J, Sinsabaugh RL, Stromberger ME, Wallenstein MD, Weintraub MN, Zoppini A (2013) Soil enzymes in a changing environment: current knowledge and future directions. Soil Biol Biochem 58:216–234. https://doi.org/10.1016/j.soilbio.2012.11.009
Busato JG, Papa G, Canellas LP, Adani F, de Oliveira AL, Leão TP (2016) Phosphatase activities and its relationship with physical and chemical parameters during vermicomposting of cake and cattle manure. J Sci Food Agric 96:1223–1230. https://doi.org/10.1002/jsfa.7210
Cantarella H, Otto R, Soares JR, Silva AGB (2018) Agronomic efficiency of NBPT as a urease inhibitor: a review. J Adv Res 13:19–27. https://doi.org/10.1016/j.jare.2018.05.008
Caporaso GJ, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich J, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Chen J, Arafat Y, Wu LK, Xiao ZG, Li QS, Khan MA, Khan MU, Lin S, Lin WX (2017) Shifts in soil microbial community, soil enzymes and crop yield under peanut/maize intercropping with reduced nitrogen levels. Appl Soil Ecol 124:327–334. https://doi.org/10.1016/j.apsoil.2017.11.010
Chen P, Wang YZ, Liu QZ, Zhang YT, Li WH (2020) Phase changes of continuous cropping obstacles in strawberry (Fragaria×ana assa Duch.) production. Appl Soil Ecol 155:103626
Chen J, Gong J, Xu M (2020b) Implications of continuous and rotational cropping practices on soil bacterial communities in pineapple cultivation. Eur J Soil Biol 97:103172. https://doi.org/10.1016/j.ejsobi.2020.103172
Chen YP, Tsai CF, Rekha PD, Ghate SD, Huang HY, Hsu YH, Liaw LL, Young CC (2021) Agricultural management practices influence the soil enzyme activity and bacterial community structure in tea plantations. Bot Stud 62:8. https://doi.org/10.1186/s40529-021-00314-9
Doan TT, Jusselme MD, Lata JC, van Nguyen B, Jouquet P (2013) The earthworm species Metaphire posthuma modulates the effect of organic amendments (compost vs. vermicompost from buffalo manure) on soil microbial properties. A laboratory experiment. Eur J Soil Biol 59:15–21. https://doi.org/10.1016/j.ejsobi.2013.08.005
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. https://doi.org/10.1890/05-1839
Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R (2012) Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6:1007–1017. https://doi.org/10.1038/ismej.2011.159
Frindte K, Zoche SA, Knief C (2020) Development of a distinct microbial community upon first season crop change in soils of long-term managed maize and rice fields. Front Microbiol. https://doi.org/10.3389/fmicb.2020.588198
Gilardi G, Garibaldi A, Gullino ML (2013) Seed transmission of Plectosphaerella cucumerina, causal agent of leaf spot of Diplotaxis tenuifolia in Italy. Phytoparasitica 41(4):411–416. https://doi.org/10.1007/s12600-013-0302-4
Garbeva P, Veen JAV, Dirk VEJ (2004) Assessment of the diversity, and antagonism towards Rhizoctonia solani AG3, of Pseudomonas species in soil from different agricultural regimes. FEMS Microbiol Ecol 47:51–64. https://doi.org/10.1016/S0168-6496(03)00234-4
Garrett SD (2010) Soil conditions and the take-all disease of wheat: The relation between soil reaction and soil aeration. Ann Appl Biol 24(4):747–751
Gu Y, Wang Y, Wang P, Wang C, Li M (2020) Study on the diversity of fungal and bacteria communities in continuous cropping fields of Chinese chives (Allium tuberosum). Biomed Res Int 5:1–14. https://doi.org/10.1155/2020/3589758
Hammerschmidt R, Nuckles EM, Kuc JA (1982) Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol Plant Path 20:73–82
Han Y, Xu L, Liu L, Yi M (2016) Illumina sequencing reveals a rhizosphere bacterial community associated with foxtail foxtail millet smut disease suppression. Plant Soil 410:1–11. https://doi.org/10.1007/s11104-016-3042-7
Hu J, Yang A, Zhu A, Wang J, Dai J, Wong MH, Lin X (2015) Arbuscular mycorrhizal fungal diversity, root colonization, and soil alkaline phosphatase activity in response to maize-wheat rotation and no tillage in North China. J Microbiol 53(7):454. https://doi.org/10.1007/s12275-015-5108-2
Jin L, Yu J, Jin N, Xie J, Lyu J (2021) Effects of different vegetable rotations on the bacterial community structure and tomato growth in a continuous tomato cropping substrate. PLoS ONE 16(9):e025743. https://doi.org/10.1371/journal.pone.0257432
Lan YM, Wang Q, James RC, Rosen GL (2012) Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS ONE 7(3):e32491. https://doi.org/10.1371/journal.pone.0032491
Leite MFA, Pan Y, Bloem J, Berge H, Kuramae EE (2017) Organic nitrogen rearranges both structure and activity of the soil-borne microbial seedbank. Sci Rep 7:42634. https://doi.org/10.1038/srep42634
Landa BB, Mavrodi DM, Tomashow LS, Weller DM (2003) Interactions between strains of 2,4-Diacetylphloroglucinolproducing Pseudomonas fuorescens in the rhizosphere of wheat. Phytopathology 93:982–994. https://doi.org/10.1094/PHYTO.2003.93.8.982
Lipiec J, Brzezinska M, Turski M, Szarlip P, Frac M (2015) Wettability and biogeochemical properties of the drilosphere and casts of endogeic earthworms in pear orchard. Soil Tillage Res 145:55–61. https://doi.org/10.1016/j.still.2014.08.010
Liu K, Bandara M, Hamel C, Knight JD, Gan Y (2020a) Intensifying crop rotations with pulse crops enhances system productivity and soil organic carbon in semi-arid environments. Field Crop Res 248:107657
Liu L, Huang X, Zhang J, Cai Z, Chang Y (2020b) Deciphering the combined effect and relative importance of soil and plant traits on the development of rhizosphere microbial communities. Soil Biol Biochem 148:107909
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556. https://doi.org/10.1146/annurev.micro.62.081307.162918
Maarastawi SA, Katharina F, Marius L, Claudia K (2018) Crop rotation and straw application impact microbial communities in Italian and Philippine soils and the rhizosphere of zea mays. Front Microbiol 9:1295. https://doi.org/10.3389/fmicb.2018.01295
Michie K, Yukie H, Akira A, Hiroki Y, Satoshi N, Hideko K, Hiroki T, Hiromasa S, Joe W, Sophien K, Ryohei T (2017) Genome analysis of the foxtail foxtail millet pathogen Sclerospora graminicola reveals the complex effector repertoire of graminicolous downy mildews. BMC Genom 18(1):897. https://doi.org/10.1186/s12864-017-4296-z
Martínez-culebras PV, Abad-Campos P, García-Jiménez J (2004) Molecular characterization and PCR detection of the melon pathogen Acremonium cucurbitacearum. Eur J Plant Pathol 110(8):801–809. https://doi.org/10.1007/s10658-004-2490-8
Mulvaney RL (1996) NitrogenInorganic forms. In: Sparks DL (ed) Methods of soil analysis. Part 3 Chemical methods. SSSA and ASA, Madison, pp 1123–1200
Ndabankulu K, Egbewale SO, Tsvuura Z (2022) Magadlela A (2022) Soil microbes and associated extracellular enzymes largely impact nutrient bioavailability in acidic and nutrient poor grassland ecosystem soils. Sci Rep. https://doi.org/10.1038/s41598-022-16949-y
Oni F, Geudens N, Onyeka J, Olorunleke O, Salami A, Omoboye O, Arias A, Adiobo A, Neve S, Ongena M, Martins J, Höfte M (2020) Cyclic lipopeptide-producing Pseudomonas koreensis group strains dominate the cocoyam rhizosphere of a Pythium root rot suppressive soil contrasting with P. putida prominence in conducive soils. Environ Microbiol 22(12):5137–5155. https://doi.org/10.1111/1462-2920.15127
Olsen SR, Sommers LE (1982) Phosphorus. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis. Part 2. Chemical and microbiological properties. SSSA, Madison, pp 403–430
Osińska-Jaroszuk M, Jarosz-Wilkołazka A (2015) Extracellular polysaccharides from Ascomycota and Basidiomycota: production conditions, biochemical characteristics, and biological properties. World J Microbiol Biotechnol 31:1823–1844. https://doi.org/10.1007/s11274-015-1937-8
Pignataro A, Moscatelli MC, Mocali S, Grego S, Benedetti A (2012) Assessment of soil microbial functional diversity in a coppiced forest system. Appl Soil Ecol 62:115–123. https://doi.org/10.1016/j.apsoil.2012.07.007
Qin J, Bian C, Duan S, Wang W, Li G, Jin L (2022) Effects of different rotation cropping systems on potato yield, rhizosphere microbial community and soil biochemical properties. Front Plant Sci 13:999730. https://doi.org/10.3389/fpls.2022.999730
Rosenzweig N, Tiedje JM, Quensen JFIII, Meng Q, Hao JJ (2012) Microbial communities associated with potato common scabsuppressive soil determined by pyrosequencing analyses. Plant Dis 96:718–725. https://doi.org/10.1094/PDIS-07-11-0571
Rivas R, Velázquez E, Willems A, Vizcaíno N, Subba-Rao NS, Mateos PF, Gillis M, Dazzo FB, Martínez Molina E (2002) A new species of Devosia that forms a unique nitrogen-fxing root-nodule symbiosis with the aquatic legume Neptunia natans (L.f.) Druce. Appl Environ Microbiol 68:5217–5222. https://doi.org/10.1128/AEM.68.11.5217-5222.2002
Schweitzer JA, Bailey JK, Fischer DG, LeRoy CJ, Lonsdorf EV, Whitham TG, Hart SC (2008) Plant-soil-microorganism interactions: heritable relationship between plant genotype and associated soil microorganisms. Ecology 89:773–781. https://doi.org/10.1890/07-0337.1
Soenens A, Imperial J (2018) Novel, non-symbiotic isolates of Neorhizobium from a dryland agricultural soil. Peer J 6:e4776. https://doi.org/10.7717/peerj.4776
Sun J, Peng M, Wang Y, Li W, Xia Q (2013) The effects of different disease-resistant cultivars of banana on rhizosphere microbial communities and enzyme activities. FEMS Microbiol Lett 345:121–126
Sun J, Zhang Q, Zhou J, Wei Q (2014a) Pyrosequencing technology reveals the impact of different manure doses on the bacterial community in apple rhizosphere soil. Appl Soil Ecol 78:28–36. https://doi.org/10.1016/j.apsoil.2014.02.004
Sun J, Zhang Q, Zhou J, Wei Q (2014b) Illumina amplicon sequencing of 16S rRNA tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLoS ONE 9:e111744. https://doi.org/10.1371/journal.pone.0111744
Tabatabai MA, Bremner JM (1969) Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol Biochem 1:301–307. https://doi.org/10.1016/0038-0717(69)90012-1
Treseder KK, Vitousek PM (2001) Effects of soil nutrient availability on investment in acquisition of N and P in Hawaiian rain forests. Ecology 82(4):946–54
Trivedi P, He Z, Van Nostrand JD, Albrigo G, Zhou J, Wang N (2013) Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. Isme J 6(2):363–383. https://doi.org/10.1038/ismej.2011.100
Ullah S, Chao A, Huang S, Jia JZ, Ping H (2019) The responses of extracellular enzyme activities and microbial community composition under nitrogen addition in an upland soil. PLoS ONE 14(9):e0223026. https://doi.org/10.1371/journal.pone.0223026
Walkley A, Black CA (1934) An estimation of the degtjareff method for determining soil organic matter and a proposed modifcation of the chromic acid titration method. Soil Sci 37:29–38
Wang W, Li Y, Wang H, Zu Y, Singer AC (2014) Differences in the activities of eight enzymes from ten soil fungi and their possible influences on the surface structure, functional groups, and element composition of soil colloids. PLoS ONE 9(11):e111740. https://doi.org/10.1371/journal.pone.0111740
Wu K, Yuan S, Wang L, Shi J, Zhao J, Shen B, Shen Q (2014) Effects of bio-organic fertilizer plus soil amendment on the control of tobacco bacterial wilt and composition of soil bacterial communities. Biol Fertil Soils 50:961–971. https://doi.org/10.1007/s00374-014-0916-9
Wu F, Chen S, Chang C, An M, Zhou X, Xu W (2015) Rhizosphere soil microorganism populations andcommunity structures of different watermelon cultivars with differing resistance to Fusarium oxysporum f. sp. Niveum Inoculation. Pak J Bot 47:1535–1546. https://doi.org/10.1139/w11-015
Wu J, Jiao Z, Zhou J, Guo F, Ding Z, Qiu Z (2017) Analysis of bacterial communities in rhizosphere soil of continuously cropped healthy and diseased konjac. World J Microbiol Biotechnol 33:134. https://doi.org/10.1007/s11274-017-2287-5
Wu A-L, Jiao X-Y, Fan F-F, Wang J-S, Guo J, Dong E-W, Wang L-G, Shen X-M (2019) Effect of continuous sorghum cropping on the rhizosphere microbial community and the role of Bacillus amyloliquefaciens in altering the microbial composition. Plant Growth Regul 89:299–308. https://doi.org/10.1007/s10725-019-00533-y
Xiong W, Li R, Ren Y, Liu C, Zhao Q, Wu H, Jousset A, Shen Q (2017) Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol Biochem 107:198–207. https://doi.org/10.1016/j.soilbio.2017.01.010
Xu W, Wu F, Chang C, Liu S (2013) Effects of wheat as companion cropping on growth, soil enzymes and disease resistance of watermelon. Allelopath J 32:267–278
Xu L, Yi M, Yi H, Guo E, Zhang A (2018) Manure and mineral fertilization change enzyme activity and bacterial community in foxtail millet rhizosphere soils. World J Microb Biot 34(1):8. https://doi.org/10.1007/s11274-017-2394-3
Xu L, Dong X, Zhang A, Guo E, Li S (2022) Effects of the different rotation patterns on physiological and biochemical indicators and yield of millet’s. Acta Agric Boreal Sin 37(3):68–76. https://doi.org/10.7668/hbnxb.20192822
Yan YH, Li JL, Zhang XQ, Yang WY, Wan Y, Ma YM, Zhu YQ, Peng Y, Huang LK (2014) Effect of naphthalene acetic acid on adventitious root development and associated physiological changes in stem cutting of Hemarthria compressa. PLoS ONE 9:e90700. https://doi.org/10.1371/journal.pone.0090700
Yang X, Huang X, Wu W, Xiang Y, Zhang L, Liu Y (2020) Effects of different rotation patterns on the occurrence of clubroot disease and diversity of rhizosphere microbes. J Integr Agric 19(9):2265–2273. https://doi.org/10.1016/S2095-3119(20)63186-0
Yao KY, Huang CY (2006) Soil microbial ecology and experimental techniques. Science Press, Beijing, p 201
Yu B, Lou Z, Zhang D, Shan AD, Yuan HP, Zhu NW, Zhang KH (2015) Variations of organic matters and microbial community in thermophilic anaerobic digestion of waste activated sludge with the addition of ferric salts. Bioresour Technol 179:291–298. https://doi.org/10.1016/j.biortech.2014.12.011
Zhang L, Ma H, Zhang H, Xun L, Chen G, Wang L (2015) Thermomyces lanuginosus is the dominant fungus in maize straw composts. Bioresour Technol 197:266–275. https://doi.org/10.1016/j.biortech.2015.08.089
Zhang MM, Wang N, Hu YB, Sun GY (2018) Changes in soil physicochemical properties and soil bacterial community in mulberry (Morus alba L.)/alfalfa (Medicago sativa L.) intercropping system. Microbiol Open 12:e0189781. https://doi.org/10.1002/mbo3.555
Zhou L, Li J, Luo Y, Liu S, Chen J, Wang J, Bai Y, Lin W, Wu Z (2019) Variation in soil fungal community structure during successive rotations of Casuarina equisetifolia plantations as determined by highthroughput sequencing analysis. Plant Growth Regul 87:445–453. https://doi.org/10.1007/s10725-019-00483-5
Zhao L, Wang G, Siegel P, He C, Wang H, Zhao W, Zhai Z, Tian F, Zhao J, Zhang H, Sun Z, Chen W, Zhang Y, Meng H (2013) Quantitative genetic background of the host influences gut microbiomes in chickens. Sci Rep 3(5):1163. https://doi.org/10.1038/srep01163
Zhou B (2012) Resistance of eggplant (Solanum melongena L.) to verticillium wilt correlates to microbial abundance and soil enzyme activities. Am J Exp Agric 2:557–572
Zhou X, Liu J, Wu F (2017a) Soil microbial communities in cucumber monoculture and rotation systems and their feedback effects on cucumber seedling growth. Plant Soil 415(1–2):507–520. https://doi.org/10.1007/s11104-017-3181-5
Zhou X, Smith H, Ana GS, Jayne B, Ferran GP, Daniele D (2017b) Differential responses of dinitrogen fixation, diazotrophic Cyanobacteria and ammonia oxidation reveal a potential warming-induced imbalance of the N-Cycle in biological soil crusts. PLoS ONE 11(10):e0164932. https://doi.org/10.1371/journal.pone.0164932
Acknowledgements
We thank Shanghai Sangon Biotechnology Co., Ltd. for the Illumina sequencing work.
Funding
The study was supported by The National Natural Science Foundation of China (31371868, 31500504); National Special Fund for the Construction of Modern Agricultural Technology System (CARS-06-13.5-A10); Chinese Academy of Sciences Key Project Sub-project (2020YFD1000803-2); Shanxi Provincial Department of Education Science and Technology Innovation Project (201804038).
Author information
Authors and Affiliations
Contributions
LX and HY design and supervise the conduct of experiments. Experiment implementation and data by AZ, EG and LX, XL prepared the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that there is no conflict of interest.
Ethical approval
All authors have read and agreed to the published version of the manuscript.
Consent for publication
Not applicable.
Additional information
Communicated by Xingchun Wang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, L., Yi, H., Zhang, A. et al. Rhizosphere soil microbial communities under foxtail millet continuous and rotational cropping systems and their feedback effects on foxtail millet downy mildew suppression. Plant Growth Regul 99, 161–175 (2023). https://doi.org/10.1007/s10725-022-00936-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-022-00936-4