Abstract
Regeneration failure and productivity decline, which is collectively known as consecutive monoculture problem (CMP), were observed during long-term monoculture Casuarina equisetifolia plantations. In this study, the high-throughput sequencing method was applied to determine whether the rhizospheric microbial community composition would be significantly degenerated by consecutive monoculture in C. equisetifolia plantations. The results showed that the soil fungal community structure exhibited obvious differences among the first rotation plantation (FCP), the second rotation plantation (SCP), and the third rotation plantation (TCP). Both the Shannon and Simpson diversity indices of the soil fungal community in the FCP were significantly higher than in the SCP (P < 0.05). Additionally, the relative abundance of Fusarium, Thelephora, Hortaea and Penicillium were significantly higher in the SCP and TCP soils than in the FCP soils, suggesting that certain fungi gradually became predominant in the continuous monoculture plantation soils. Conversely, the relative abundance of Tolypocladium and Trichoderma were significantly lower in the SCP and TCP soils than in the FCP soils, suggesting that some microbes gradually decreased in the continuous monoculture plantation soils. Overall, the results demonstrated that the long-term pure plantation pattern exacerbated the microecological imbalance in rhizospheric soils of C. equisetifolia and markedly decreased soil microbial community diversity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Casuarina equisetifolia is a she-oak species of the genus Casuarina, which is native to the east coast of Australia. C. equisetifolia has been widely planted all around the world because of its characteristics of high yield and fast growth (Zhong et al. 2013). This species has adapted to harsh environmental conditions, including high salinity, poor soils, and heavy metal pollution (Scotti-Campos et al. 2016; Vijayabhama et al. 2018). More importantly, C. equisetifolia plays significant economic and ecological roles by offering shelter belts for numerous animal species, stabilizing coastal sand dunes, and protecting against storms (Karthikeyan et al. 2013). Indeed, C. equisetifolia has been introduced to China’s southeastern coastal areas as windbreaks to improve the environment for more than half a century (Liu et al. 2015; Li et al. 2018).
However, studies during the past few decades have shown regeneration failure and productivity decline in long-term monoculture C. equisetifolia plantations in a phenomenon referred to as CMP (Wardle et al. 2004). CMP results in slow growth, low output, and aggravation of disease and insect pests of C. equisetifolia (Long et al. 2018). This phenomenon has long been a complex problem in forest management; therefore, its formation mechanism and control have been the focus of many studies. Soil nutrients deficiency has been considered one of the reasons for regeneration failure of C. equisetifolia, but CMP cannot be solved by application of chemical fertilizer (Karthikeyan 2016). In addition, previous studies have suggested that some root exudates may be responsible for the C. equisetifolia replanting problem, such as 12, 13-dihydromicromeric acid, betulinic acid, and catechins (Veluthakkal and Dasgupta 2012; Kelderer et al. 2012). Although much work has been conducted to investigate methods of controlling CMP in C. equisetifolia plantations, such as the use of biological organic fertilizer, plant growth hormone, and mycorrhizal inoculation, there has been little success (Hata et al. 2015; Zhang et al. 2016).
Investigations of CMP have gradually come to focus on rhizospheric biological processes. Rhizospheric microorganisms are crucial to forest ecosystems because of their critical role in regulating ecosystem balance via fundamental ecological processes, such as mineralization and decomposition (Bennett et al. 2012). In recent years, studies have suggested that the imbalance of rhizospheric microbial community structure was one of the main reasons for CMP (Shi et al. 2011; Li et al. 2014a, b). Moreover, some studies have shown that root exudates could stimulate harmful fungal pathogens or inhibit beneficial microorganisms within long-term monoculture rotations (Wu et al. 2016a, b). However, few studies have investigated variations in the rhizospheric microbial community in successive rotations of C. equisetifolia. In the present study, we investigated whether the soil microbial community composition and diversity would significantly degenerate in response to consecutive monoculture in C. equisetifolia plantations. The specific goal of this study was to illuminate shifts in the rhizospheric microbial composition and diversity in successive rotations of C. equisetifolia plantations by using high-throughput sequencing technology.
Materials and methods
Field description and soil sampling
The sampling area was located in Hui’an National Forest Farm, on the western ChongWu Peninsula (24°54′N, 118°55′E), which is a barrier island that protects against typhoons in Fujian Province, China. The arid climate consists of scarce precipitation, with a mean of 1000–1500 mm annually, and an annual mean evaporation of 2000 mm. The sandy soils of the sampling site are infertile and vulnerable to erosion.
Rhizospheric soil samples were randomly collected from three sampling positions (20 m × 20 m) on November 10, 2017. Among the three sampling sites, soil samples were collected from 0 to 20 cm depths by a core sampler (diameter of 3.5 cm), with 20 random repetitions samples from each positions at a least distance of 4 m. The 20 soil cores from each sampling positions were mixed into one composite soil sample for analysis. The composite samples were subsequently sieved using 2-mm mesh, after which the DNA was immediately extracted. The remaining sieved soil samples which from each sampling location were divided into two portions, one part stored at − 80 °C for further analysis, and another part were air-dried at room temperature for soil physicochemical properties.
Measurement of soil nutrient
Soil pH was determined by the glass electrode pH meter (1:2.5 soil–water suspensions) (Thomas et al. 1996). The total and available counts of nitrogen, phosphorus and potassium were worked out referring to the methods by Zhao et al. (2017) and Kaur and Garg (2017).
DNA extraction and high-throughput sequencing
The total DNA from individual rhizospheric soil samples was extracted in three replicates using a SoilGen DNA kit (CWBIO, Beijing, China) according to the manufacturer’s instructions. The DNA quality was tested on 1% agarose gels, and the DNA concentration was determined using a Nanodrop 2000C Spectrophotometer (Thermo Scientific, Waltham, MA, USA). The DNA was then diluted with sterile water to 1 ng/µL, after which the internal transcribed spacer (ITS1) was expanded with the designated primers ITS5F and ITS2R (Coyer et al. 2013). The PCR amplification experiments were accomplished in the 30 µL volume involving 15 µL of 2 × Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Beverly, MA, USA), 3 µL of each primer (6 µM), 10 ng DNA and sterile water. The PCR amplification conditions were shown in the supplementary information.
The PCR products of individual rhizospheric soil samples were co-mixed in equal proportions and then purified with a Qiagen Gel Extraction Kit (Qiagen GmbH, Hilden, Germany). Sequencing libraries were developed from a TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, USA) after which high-throughput sequencing was carried out on the Illumina HiSeq 2500 platform.
Operational taxonomic units (OTUs) based sequence analysis
After sequencing, the sequences reads were allocated to samples based on their unique barcodes. Paired-end reads were incorporated applying FLASH (V1.2.7) (Tanja and Salzberg 2011). After chimera removal and quality filtering, the remaining sequences named as effective tags ultimately acquired for performing OTU clustering at 97% sequence similarity through Uparse software (Uparse v7.0.1001) (Sun et al. 2014; Edgar 2013). Species annotations were implemented through the Unite Database according to the BLAST algorithm, which was performed with the QIIME software (Version 1.7.0) (Koljalg et al. 2013).
Statistical analyses
Based on the abundances of normalized OTUs, alpha and beta diversity analyses were conducted according to the standardized data. Alpha analyses consisted of determination of the diversity of individual species based on the observed-species, chao1 (http://www.mothur.org/wiki/Chao), abundance-based coverage estimator (ACE) (http://www.mothur.org/wiki/Ace), and Shannon (http://www.mothur.org/wiki/Shannon) and Simpson (http://www.mothur.org/wiki/Simpson) diversity indices. Alpha diversity indices were was determined by QIIME (version 1.7.0) (Caporaso et al. 2010) and displayed with R software (version 2.15.3). Beta diversity analysis consisted of an unweighted UniFrac distance grounded on the abundance of lineages to estimate the diversity between different samples, including principal coordinate analysis (PCoA) and unweighted pair-group method with arithmetic means (UPGMA) clustering. PCoA analysis was displayed by WGCNA package, stat packages and ggplot2 package in R software (version 2.15.3). UPGMA clustering was conducted by QIIME software (version 1.7.0). Statistical analysis was processed by DPS software (version 7.51), and analysis of variance (ANOVA) was used to conclude significance of difference by Tukey’s test (P < 0.05).
Results
Soil physicochemical properties
The experimental results showed that the SCP and TCP had a higher pH under the successive rotations of C. equisetifolia. The total nitrogen (TN) in rhizospheric soils of FCP was significantly higher than in SCP and TCP (P < 0.05). In the FCP, alkaline nitrogen (AN) was significantly higher than in the TCP (P < 0.05), while it was higher in the SCP (not significant, P > 0.05). Total phosphorus (TP) was significantly higher in the FCP and SCP than in the TCP. The available phosphorus (AP), total potassium (TK), and available potassium (AK) did not differ significantly among treatments (P > 0.05) (Table S1).
OTUs cluster and species annotation
The impact of successive rotations of C. equisetifolia was assessed by using ITS1 deep pyrosequencing to investigate the soil fungal community is concerned. Following filtration analysis, a total of 359,640 effective sequences were found from nine rhizospheric soil samples with a mean of 39,960 effective tags. The sequences from the nine soil samples were clustered into 5399 OTUs at a 97% similarity cut-off level. The average number of OTUs in the FCP, SCP, and TCP was 654, 600, and 545, respectively (Fig. S1). On average, more than 90% of the effective tags could be classified at the class level, but only 15.6% at the genus level (Fig. S2). Rarefaction curve analyses indicated that the observed species number trended to be stable in 30,000 sequences (Fig. S3).
Alpha diversity indices
To explain the species diversity, alpha diversity indices were evaluated. The richness and diversity indices of the fungal communities in individual rhizospheric soil samples were obtained with a cutoff of 35,692 sequences. The observed species, Chao1 and ACE were significantly higher in the FCP than in the TCP (P < 0.05), and higher than in the SCP (not significant, P > 0.05). In the FCP, the fungal community showed significantly higher Shannon and Simpson diversity indices than in the SCP (P < 0.05). There was no significant difference in the Shannon and Simpson diversity indices between the FCP and TCP (Table 1).
Beta diversity indices
The results of the unweighted Unifrac distances can reveal differences in community composition and structure of total fungal groups between different plantation treatments. The unweighted Unifrac distance between the FCP and SCP was 0.393, while it was 0.513 between the FCP and TCP. When compared with the FCP, the unweighted Unifrac distance increased with the aggravation of successive rotations.
PCoA and UPGMA clustering
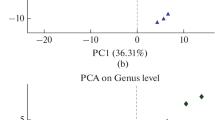
According to the unweighted UniFrac distance, both PCoA analysis and the UPGMA clustering were conducted to compare fungal community structures across all rhizospheric soil samples. The goal of PCoA and UPGMA was to identify distinct differences among generations and similarities within the same generation of rhizospheric soil samples. The first two principal components of identified by PCoA accounted 42.95% (PC1) and 26.67% (PC2) of the overall rhizospheric soil fungal community alterations, respectively (Fig. 1a). Additionally, PCoA revealed the fungal community in FCP and SCP were divided from that in TCP by PC1, and the community in FCP was divided from that in SCP by PC2 (Fig. 1a). Furthermore, the results of hierarchical clustering showed that fungal community structure from FCP and SCP were gathered together, and then clustered together as a single group with TCP (Fig. 1b).
Venn diagram analysis
The exclusive and shared OTUs among different sets were observed based on Venn diagram analysis. The results revealed that the percentage of OTUs exclusively constructed in the FCP was 17.9% at the species level (201 species), while the percentage of OTUs exclusively constructed in the SCP was 10.2% (114 species). The proportion of species-level OTUs exclusively constructed in the TCP was 9.7% (109 species). Furthermore, the number of OTUs shared in FCP and SCP was 158 (14.1%), revealing a similar community structure in the FCP and SCP. The number of OTUs shared in the FCP, TCP, and SCP was 35.0% (393 species) (Fig. 2a).
Alterations in rhizospheric soil fungal community composition and structure under successive rotations
According to the Ribosomal Database Project classifier through the Unite Database, all of the OTUs in individual rhizospheric soil samples were classified into different clusters. The results showed that the predominant fungal phyla in three different treatments, listed from most to least common, were Ascomycota, Basidiomycota, Zygomycota, Glomeromycota, Chytridiomycota, and Neocallimastigomycota. Of these phyla, Ascomycota was the dominant phylum, accounting for 40.9%, 85.6%, and 73.8% of the populations in the FCP, SCP, and TCP, respectively (Fig. 2b).
At the genus level, the relative abundance of Fusarium, Thelephora, Hortaea, and Penicillium significantly increased, while the relative abundance of Tolypocladium, Trichoderma, Melanconiella, and Polyporus decreased significantly. Among these, Fusarium was considered one of the dominant plant pathogens for many crucial plantations. The top fungal genera, which are listed in Table 2, included Fusarium, Tolypocladium, Trichoderma, and Penicillium.
As shown in the heat map of the 35 most abundant fungi (relative abundance > 1%) at the genus level, there were distinct alterations in the fungal community composition and structure in replanted soils at three time scales. Moreover, when compared with FCP, the dissimilarity of the community structure of fungi was enhanced under the extended monoculture regime of C. equisetifolia, implying that the rhizosphere fungal community gradually changed with increasing years of successive rotations (Fig. 3).
Discussion
Consecutive monoculture problem, also known as soil disease or replant failure, is a pivotal factor threatening sustainable and stable development of a variety of vegetations. The rapid growth and high yield of cultivated tree species, including C. equisetifolia, Eucalyptus robusta, and Chinese fir plantations, suffer from a significant decline under successive rotations. Studies have shown that successive rotations culture problem is caused by many factors, including soil nutrient deficiency, the autotoxicity of root exudates, and the imbalance of soil microorganisms (Wu et al. 2016a, b). In this study, the results of soil chemical properties also showed that TN, AN, and TP were higher in the FCP than in the SCP and TCP (Table S1). Previous studies suggested that successive rotations could lead to the consumption of soil available nutrients in C. equisetifolia stands, but the application of fertilizer does not prevent replant disease, indicating that CMP is more closely associated with autotoxicity and soil microbes (Berendsen et al. 2012).
Moreover, many scholars have revealed that allelochemicals released by roots did not possess sufficient conditions to directly impact the health of adjacent plants or host plants. Therefore, the belowground microbial community structure and functional diversity has attracted increasing attention (Haney and Ausubel 2015; Xiong et al. 2016). Numerous species of microorganisms come into contact with plant roots, resulting in a complicated plant-related microbial community regarded as the second genome of plants, which is essential to their growth and productivity. Fungi, the major decomposers, play a role in biogeochemical cycles in the soil ecosystems (Li et al. 2014a, b). In this study, the results of deep pyrosequencing demonstrated that the composition and diversity of the soil fungal community was significantly altered in the rooting zone after successive rotations of C. equisetifolia. Moreover, both PCoA and UPGMA clustering revealed an obvious partition among the three rhizospheric soil samples of FCP, SCP, and TCP. Additionally, the fungal community diversity indices of the FCP were significantly higher than those of the SCP. This results showed that the richness and diversity of soil fungal community in C. equisetifolia decreased significantly with the increase of CMP. Generally, bacteria will increase when soil nutrients are enough. Conversely, fungi will increase when soil nutrient decline, because fungi are more able to adapt to the bad soil environment. In this study, we found that in FCP soil, adequate soil nutrition resulted in the reproduction of bacteria. However, in SCP and TCP soil, nutrient content decrease resulted in the reproduction of fungi. Similar result was also reported by Zhao et al. (2013) in which soil fungal diversity increased in Eucalyptus monocultures.
Our findings exhibited that successive rotations of C. equisetifolia resulted in a significant increase in the relative abundance of Fusarium, Penicillium, Thelephora, and Hortaea, while Trichoderma, Polyporus, Tolypocladium, and Melanconiella decreased significantly. It is well known that most Fusarium are causative agents of root rot and wilt in a number of important plants (Van Wees et al. 2008), such as Chinese fir, Eucalyptus robusta, cucumber and strawberry (Bashir et al. 2012; Zhao et al. 2017). In this study, Fusarium showed a significantly higher relative abundance in SCP or TCP than in FCP. The increase in genus Fusarium from the FCP to SCP and TCP was primarily a result of the increase of Fusarium oxysporum by approximately 682% and 830%, respectively. It was also reported that many Penicillium could strengthen protection against pathogenic agents and facilitate the growth of plants, but some could also induce plant sickness. For example, Leelasuphakul et al. (2008) revealed that inoculation of a suspension of the Penicillium digitatum conidia into injured citrus fruit resulted in signs and symptoms at day 3 and corrupted at day 5. However, the relative abundance of Trichoderma showed a reduction from 1.44% in the FCP to 0.35% in the TCP. According to previous reports, some Trichoderma have been exploited as biocontrol agents against fungal diseases of the soil (Harman 2006).
Based on the above discussion, regeneration failure and productivity decline of C. equisetifolia could be because of the rapid reproduction of possible pathogenic agents and the consumption of plant beneficial fungi in replant failure soils. Many studies have shown that the quantity of fungal pathogens gradually increased under monoculture regime (Zhou et al. 2012). Moreover, studies have demonstrated that alterations in the rhizospheric soil microbial community play a far more important role in the successive rotation problem of plants than allelopathic effects (Haichar et al. 2008; Chen et al. 2018). Additionally, there is increasing evidence that plant root exudates can significantly reshape rhizospheric soil microbial community structure and microflora could then affect plant healthy growth in plant-soil ecosystems (Huang et al. 2014). Some studies demonstrated that the phenolic compounds that were released during dissolution and then accumulated in soils around the rhizosphere had a negative impact on the growth of Chinese fir plantations (Kõljalg et al. 2013; Peiffer et al. 2013). Additionally, many researchers detected that the inhibition or promotion of certain rhizospheric microorganisms induced by root exudates might be the dominant factor influencing soil disease within peanut crops, particularly F. oxysporum.
Conclusion
The successive rotations of C. equisetifolia can alter rhizosphere fungal communities by reducing plant beneficial fungi and enriching host-specific pathogens, which might be the key factor triggering soil disease of this tree species. Nevertheless, the impact of pathogens and beneficial microbes mentioned in this study on C. equisetifolia need to be verified in subsequent experiments. In addition, further studies are required to detect the roles of root exudations in the rhizosphere to address the consecutive productivity problem associated with C. equisetifolia, as well as the mechanisms underlying the rhizospheric mutual impact between plant beneficial microbes and pathogenic microbes.
Abbreviations
- CMP:
-
Consecutive monoculture problem
- FCP:
-
First rotation plantation
- SCP:
-
Second rotation plantation
- TCP:
-
Third rotation plantation
- TN:
-
Total nitrogen
- AN:
-
Alkaline nitrogen
- TP:
-
Total phosphorus
- AP:
-
Available phosphorus
- TK:
-
Total potassium
- OTU:
-
Operational taxonomic unit
- AK:
-
Available potassium
- ACE:
-
Abundance-based coverage estimator
- PCoA:
-
Principal coordinate analysis
- UPGMA:
-
Unweighted pair-group method with arithmetic means
- OTUs:
-
Operational taxonomic units
References
Bashir K, Ishimaru Y, Nishizawa NK (2012) Molecular mechanisms of zinc uptake and translocation in rice. Plant Soil 361:189–201. https://doi.org/10.1007/s11104-012-1240-5
Bennett AJ, Bending GD, Chandler D, Hilton S, Mills P (2012) Meeting the demand for crop production: the challenge of yield decline in crops grown in short rotations. Biol Rev Camb Philos Soc 87:52–71. https://doi.org/10.1111/j.1469-185x.2011.00184.x
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486. https://doi.org/10.1016/j.tplants.2012.04.001
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Chen J, Arafat Y, Wu LK, Xiao ZG, Li QS, Khan MA, Khan MU, Lin S, Lin WX (2018) Shifts in soil microbial community, soil enzymes and crop yield under peanut/maize intercropping with reduced nitrogen levels. Appl Soil Ecol 124:327–334. https://doi.org/10.1016/j.apsoil.2017.11.010
Coyer JA, Hoarau G, Kuo J, Tronholm A, Veldsink J, Olsen JL (2013) Phylogeny and temporal divergence of the seagrass family Zosteraceae using one nuclear and three chloroplast loci. Syst Biodivers 11:271–284. https://doi.org/10.1080/14772000.2013.821187
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996. https://doi.org/10.1371/journal.pone.0078866
Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230. https://doi.org/10.1038/ismej.2008.80
Haney CH, Ausubel FM (2015) Plant microbiome blueprints. Science 349:788–789. https://doi.org/10.1126/science.aad0092
Harman DG, Blanksby SJ (2006) Trapping of a tert-adamantyl peroxyl radical in the gas phase. Chem Commun 8: 859-861. https://doi.org/10.1039/b516408g
Hata K, Kawakami K, Kachi N (2015) Higher soil water availability after removal of a dominant, nonnative tree (Casuarina equisetifolia Forst.) from a subtropical forest. Pac Sci 9:445–460. https://doi.org/10.2984/69.4.2
Huang XF, Chaparro JM, Reardon KF, Zhang RF, Shen QR, Vivanco JM (2014) Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92:267–275. https://doi.org/10.1139/cjb-2013-0225
Karthikeyan A (2016) Frankia strains for improving growth, biomass and nitrogen fixation in Casuarina equisetifolia seedlings. J Trop For Sci 28:235–242. https://doi.org/10.1007/bf00336081
Karthikeyan A, Chandrasekaran K, Geetha M, Kalaiselvi R (2013) Growth response of Casuarina equisetifolia Forst. rooted stem cuttings to Frankia in nursery and field conditions. J Biosci 38:741–747. https://doi.org/10.1007/s12038-013-9362-3
Kaur H, Garg N (2017) Recent perspectives on cross talk between cadmium, zinc, and arbuscular mycorrhizal fungi in plants. J Plant Growth Regul 8:1–14. https://doi.org/10.1007/s00344-017-9750-2
Kelderer M, Manici LM, Caputo F, Thalheimer M (2012) Planting in the ‘inter-row’ to overcome replant disease in apple orchards: a study on the effectiveness of the practice based on microbial indicators. Plant Soil 57:381–393. https://doi.org/10.1007/s11104-012-1172-0
Kõljalg U, Nilsson RH, Abarenkov K, Larsson KH (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. https://doi.org/10.1111/mec.12481
Leelasuphakul W, Hemmanee P, Chuenchitt S (2008) Growth inhibitory properties of Bacillus subtilis, strains and their metabolites against the green mold pathogen (Penicillium digitatum, Sacc.) of citrus fruit. Postharvest Biol Tec 48:113–121. https://doi.org/10.1016/j.postharvbio.2007.09.024
Li N, Zheng YQ, Ding HM, Li HP, Peng HZ, Jiang B, Li HB (2018) Development and validation of SSR markers based on transcriptome sequencing of Casuarina equisetifolia. Trees-Struct Funct 32: 41-49. https://doi.org/10.1007/s00468-017-1607-6
Li XG, Ding CF, Hua K, Zhang TL, Zhang YN, Zhao L, Yang YR, Liu JG, Wang XX (2014a) Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol Biochem 78:149–159. https://doi.org/10.1016/j.soilbio.2014.07.019
Li XG, Ding CF, Zhang TL, Wang XX (2014b) Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol Biochem 72:11–18. https://doi.org/10.1016/j.soilbio.2014.01.019
Liu XZ, Lu YC, Xie YS, Xue Y (2015) The positive interaction between two nonindigenous species, Casuarina (Casuarina equisetifolia) and Acacia (Acacia mangium), in the tropical coastal zone of south China: stand dynamics and soil nutrients. Trop Conserv Sci 8:598–609. https://doi.org/10.1177/194008291500800302
Long F, Xie BB, Liang AJ, Li J (2018) Replant problem in Casuarina equisetifolia L.: isolation and identification of allelochemicals from its roots. Allelopathy J 43:73–82. https://doi.org/10.26651/allelo.j./2018-43-1-1131
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110:6548–6553. https://doi.org/10.1073/pnas.1302837110
Scotti-Campos P, Duro N, Costa MD, Pais IP, Rodrigues AP, Batista-Santos P, Semedo JN, Leitão AE, Lidon FC, Pawlowski K (2016) Antioxidative ability and membrane integrity in salt-induced responses of Casuarina glauca Sieber ex Spreng. in symbiosis with N2-fixing Frankia Thr or supplemented with mineral nitrogen. J Plant Physiol 196–197:60–69. https://doi.org/10.1016/j.jplph.2016.03.012
Shi SJ, Richardson AE, O’Callaghan M, DeAngelis KM, Jones EE, Stewart A, Firestone MK, Condron LM (2011) Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol 77:600–610. https://doi.org/10.1111/j.1574-6941.2011.01150.x
Sun J, Zhang Q, Zhou J, Wei QP (2014) Pyrosequencing technology reveals the impact of different manure doses on the bacterial community in apple rhizosphere soil. Appl Soil Ecol 78:28–36. https://doi.org/10.1016/j.apsoil.2014.02.004
Tanja M, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Thomas JC, Berger F, Jacquier M, Bernillon D, Baud-Grasset F, Truffaut N, Normand P, Vogel T M, Simonet P (1996) Isolation and characterization of a novel gamma-hexachlorocyclohexane-degrading bacterium. J Bacteriol 178: 6049-6055. https://doi.org/10.1128/jb.178.20.6049-6055.1996
Van Wees SC, Van der Ent S, Pieterse CM (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11:443–448. https://doi.org/10.1016/j.pbi.2008.05.005
Veluthakkal R, Dasgupta MG (2012) Isolation and characterization of pathogen defence-related class I chitinase from the actinorhizal tree Casuarina equisetifolia. For Pathol 42:467–480. https://doi.org/10.1111/j.1439-0329.2012.00781.x
Vijayabhama M, Jaisankar R, Raj SV, Baranidharan K (2018) Spatial–temporal variation of casuarina spread in Cauvery delta and north eastern zone of Tamil Nadu, India: a spatial autoregressive model. J Appl Stat 45:1–7. https://doi.org/10.1080/02664763.2016.1247786
Wardle DA, Bardgett RD, Klironomos JN, Setälä H, van der Putten WH, Wall DH (2004) Ecological linkages between aboveground and belowground biota. Science 304:1629–1633. https://doi.org/10.1126/science.1094875
Wu LK, Chen J, Wu HM, Qin XJ, Wang JY, Wu HM, Khan MU, Lin S, Xiao ZG, Luo XM (2016a) Insights into the regulation of rhizosphere bacterial communities by application of bio-organic fertilizer in Pseudostellaria heterophylla monoculture regime. Front Microbiol 7:1788. https://doi.org/10.3389/fmicb.2016.01788
Wu LK, Chen J, Wu HM, Wang JY, Wu YH, Lin S, Khan MU, Zhang ZY, Lin WX (2016b) Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci Rep UK 6:26601. https://doi.org/10.1038/srep26601
Xiong W, Zhao Q, Xue C, Xun W, Zhao J, Wu H, Li R, Shen Q (2016) Comparison of fungal community in black pepper-vanilla and vanilla monoculture systems associated with vanilla Fusarium wilt disease. Front Microbiol 7:117. https://doi.org/10.3389/fmicb.2016.00117
Zhang Y, Zhong CL, Han Q, Jiang QB, Chen Y, Chen Z, Pinyopusarerk K, Bush D (2016) Reproductive biology and breeding system in Casuarina equisetifolia (Casuarinaceae)—implication for genetic improvement. Aust J Bot 64:120–128. https://doi.org/10.1071/bt15184
Zhao J, Wan SZ, Zhang CL, Liu ZF, Zhou LX, Fu SL (2013) Contributions of understory and/or overstory vegetations to soil microbial PLFA and nematode diversities in Eucalyptus monocultures. PLoS ONE 9:e85513. https://doi.org/10.1371/journal.pone.0085513
Zhao J, Mei Z, Zhang X, Xue C, Zhang CZ, Ma TF, Zhang SS (2017) Suppression of Fusarium wilt of cucumber by ammonia gas fumigation via reduction of Fusarium population in the field. Sci Rep-UK 7:43103. https://doi.org/10.1038/srep43103
Zhong CL, Mansour S, Nambiar-Veetil M, Franche C (2013) Casuarina glauca: a model tree for basic research in actinorhizal symbiosis. J Biosci 38:815–823. https://doi.org/10.1007/s12038-013-9370-3
Zhou X, Yu G, Wu F (2012) Soil phenolics in a continuously mono-cropped cucumber (Cucumis sativus L.) system and their effects on cucumber seedling growth and soil microbial communities. Eur J Soil Sci 63:332–340. https://doi.org/10.1111/j.1365-2389.2012.01442
Acknowledgements
We thank LetPub (http://www.letpub.com) for providing linguistic assistance during the preparation of this manuscript. This work was supported by the Chinese National Natural Science Foundation (Grant No. 31500443), Natural Science Foundation of Fujian Province, China (Grant No. 2018J01617), the Scientific Research Foundation of Fujian Agriculture and Forestry University (Grant No. XJQ201718), and the Fujian-Taiwan Joint Innovative Centre for Germplasm Resources and Cultivation of Crops (Grant No. 2015-75. FJ 2011 Program, China).
Author information
Authors and Affiliations
Contributions
WZY and LWX conceived and directed the project. WZY, ZLT and LWX designed all experiments. LY, LJJ, WJY, CJ, LSY and BY did all of experiments. ZLT and LJJ performed the integrated data analysis. ZLT and WZY wrote the manuscript with the assistance and approval of all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing financial interests.
Ethical approval
This study has been approved by the Hui’an National Forest Farm Management Committee, which takes care of the planning and protection of Hui’an National Forest Farm. The study did not involve any endangered or protected species. All of the data in this study can be published and shared.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liuting, Z., Jianjuan, L., Yang, L. et al. Variation in soil fungal community structure during successive rotations of Casuarina equisetifolia plantations as determined by high-throughput sequencing analysis. Plant Growth Regul 87, 445–453 (2019). https://doi.org/10.1007/s10725-019-00483-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-019-00483-5