Abstract
Controlled environments are pivotal in all bioconversion processes, influencing the efficacy of biocatalysts. In this study, we designed a batch bioreactor system with a packed immobilization column and a decontamination chamber to enhance phenol and 2,4-dichlorophenol degradation using the hyper-tolerant bacterium Pseudomonas aeruginosa STV1713. When free cells were employed to degrade phenol and 2,4-DCP at a concentration of 1000 mg/L, the cells completely removed the pollutants within 28 h and 66 h, respectively. Simultaneous reductions in chemical oxygen demand and biological oxygen demand were observed (phenol: 30.21 mg/L/h and 16.92 mg/L/h, respectively; 2,4-dichlorophenol: 12.85 mg/L/h and 7.21 mg/L/h, respectively). After assessing the degradation capabilities, the bacterium was immobilized on various matrices (sodium alginate, alginate-chitosan-alginate and polyvinyl alcohol-alginate) to enhance pollutant removal. Hybrid immobilized cells exhibited greater tolerance and degradation capabilities than those immobilized in a single matrix. Among them, polyvinyl alcohol-alginate immobilized cells displayed the highest degradation capacities (up to 2000 mg/L for phenol and 2500 mg/L for 2,4-dichlorophenol). Morphological analysis of the immobilized cells revealed enhanced cell preservation in hybrid matrices. Furthermore, the elucidation of the metabolic pathway through the catechol dioxygenase enzyme assay indicated higher activity of the catechol 1,2-dioxygenase enzyme, suggesting that the bacterium employed an ortho-degradation mechanism for pollutant removal. Additionally, enzyme zymography confirmed the presence of catechol 1,2-dioxygenase, with the molecular weight of the enzyme determined as 245 kDa.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Phenol and 2,4-dichlorophenol (2,4-DCP) are widely distributed in the environment, especially in water and soil, causing various health hazards to living organisms while being a major reason for groundwater pollution (Panigrahy et al. 2022). Major sources of phenolic pollutants include industrial wastewater, agricultural run-off, domestic waste and natural causes (death and decay of organic matter) (Krastanov et al. 2013). Phenolic exposure can cause the production of various toxic metabolites within the organisms. These toxic metabolites can lead to mutation or malignancy (Barik et al. 2021). 2,4-DCP is one of the lethal derivatives of phenol. Both phenol and 2,4-DCP possess strong antimicrobial characteristics and have huge industrial and medicinal applications. Although many physical and chemical methods are available for the removal of phenolic pollutants from the environment, bioremediation always receives attention due to its eco-friendly nature and the fact that it does not produce any harmful byproducts (Bala et al. 2022).
Furthermore, bacteria are widely used for the degradation of phenolic pollutants due to their fast-doubling time and less specificity towards substrates. Many members of the genus Bacillus (Tripathi and Garg 2013) and Pseudomonas (Pinheiro et al. 2018) have been reported for phenolic pollutant degradation. However, the chemical stability of polychlorinated compounds, characterized by their resilient C–Cl bonds, poses challenges for microbial degradation processes (Oluwasanu 2018). Bacteria use two major mechanisms to effectively utilize these pollutants: ortho degradation and meta-cleavage pathways (Mishra 2017). In both pathways, the first step involves the conversion of phenol to catechol by the enzyme phenol monooxygenase. In the ortho-degradation pathway, catechol is cleaved by the catechol 1,2-dioxygenase enzyme to form cis-cis-muconic acid, which is further degraded into Krebs cycle intermediates. In the meta pathway, catechol is first converted to 2-hydroxymuconic semialdehyde by the enzyme catechol 2,3-dioxygenase, which is then degraded into Krebs cycle intermediates. Overall, both pathways lead to the complete degradation of phenolic pollutants into simpler compounds that bacteria can utilize for energy production and growth (Patel et al. 2017). Therefore, the detection of major enzymes can help to determine the metabolic pathway of pollutant removal in bacteria.
Very few Pseudomonas bacterial species have been reported to utilize 2,4-DCP effectively. However, most of them fail to survive under higher concentrations of phenol and 2,4-DCP due to their strong antimicrobial properties (Pinheiro et al. 2018). Therefore, removing higher concentrations of these phenolic pollutants from the environment has been a fascinating area of research for many years. Cell immobilization (single and hybrid) is considered as an appealing option for the degradation of higher concentrations of antimicrobial pollutants since it can significantly protect cells from the toxic effects of pollutants due to direct contact while increasing cell survivability and degradation efficiency (Lin and Cheng 2020). The hybrid immobilization method uses a combination of two or more immobilization matrices. It has several advantages over other immobilization techniques, including increased stability of the cells and enzymes, improved mass transfer of nutrients and waste products and increased resistance to harsh environmental conditions. The technique has potential applications in various fields, including biocatalysis, biosensors and bioremediation (Abarian et al. 2019).
All bioconversion processes require a controlled environment for the biocatalyst to perform well. Bioreactors play a crucial role in degradation studies, especially when investigating biological treatment methods for removing contaminants from wastewater (Sengupta et al. 2022). They provide a controlled environment for studying pollutant degradation. Parameters such as temperature, pH, dissolved oxygen levels and nutrient concentrations can be carefully controlled and monitored, ensuring optimal conditions for the growth and activity of phenol-degrading microorganisms (Lin and Cheng 2020). Different types of batch reactor models are often preferred in degradation studies for specific applications due to their simplicity, cost-effectiveness, and suitability for particular research objectives (Lin and Gu 2023). Similarly, packed bed reactors are often used to treat phenolic wastewater using immobilized matrices (Basak et al. 2019).
The strong antimicrobial characteristics of phenol and 2,4-DCP present a significant challenge for researchers in this field. The search for bacteria capable of efficiently degrading and tolerating higher concentrations of these phenolic pollutants remains a pressing concern. Moreover, the identification of a suitable immobilization material has the potential to enhance bacteria tolerance capacities. In addition, while many bioreactor models typically support either free cells or immobilized cells, those accommodating both modalities can offer a more economical and effective solution for wastewater treatment. Considering these research gaps, the present study focused on exploring the degradation capabilities of the hyper-tolerant bacterium P. aeruginosa STV1713.
In the present study, the hyper-tolerant bacterium P. aeruginosa STV1713 was immobilized on various matrices and employed in the degradation of phenol and 2,4-DCP within a custom-designed batch biological reactor to assess its degradation capabilities. The study also aimed to identify the most suitable matrix for pollutant degradation. Furthermore, experiments were conducted to elucidate the pathways of pollutant degradation by the bacterium.
Materials and methods
Bacterium and cultural conditions
The bacterium used in the present study was a well-adapted phenol-tolerant strain of P. aeruginosa, STV1713, isolated from oil-contaminated soil and maintained at the microbiology laboratory of the National Institute of Technology Calicut (Kerala, India). The composition of the Bushnell Hass Mineral Salt Medium (BH-MSM; MgSO4: 0.2 g, CaCl2: 0.02 g, K2HPO4: 1.0 g, KH2PO4: 1.0 g, NH4NO3: 1.0 g and 0.05 g FeCl3/L; pH 7 ± 0.1) used in the current study was adopted from Subramaniam et al. (2019). All the experiments were conducted at optimized conditions of pH 7.0 and a temperature of 32.5 °C. Additionally, all the experiments were conducted in triplicates.
Batch reactor: designing and fabrication
A batch reactor was designed for the biological treatment of phenolic wastewater using free and immobilized cells of P. aeruginosa, STV1713. The reactor had two chambers for efficient pollutant removal: the bio-reaction chamber and the decontamination chamber. The bio-reaction chamber had a height of 450 mm and a diameter of 150 mm, resulting in a height-to-diameter ratio of 3:1. The chamber was constructed using borosil glass with a thickness of 4 mm, ensuring chemical resistance and structural integrity. The total volume of the cylinder was 8 L with a working volume of 6 L, which provided ample space for the bio-reaction to occur efficiently. For monitoring and maintaining the essential degradation parameters, a pH probe was integrated into the design to detect and regulate the pH levels in real-time.
Additionally, a temperature probe was employed to monitor and control the temperature within the bioreactor, supporting the optimal conditions for microbial activity. The probes were housed through a fabricated lid. Moreover, additional ports were provided in the lid for sample collection, UV light (ultraviolet light for chamber sterilization), and aeration (fitted with an air filter). A magnetic stirrer (with temperature and rpm control) was used to mix the reactor contents homogeneously and to maintain the biocatalyst in suspension. In the biological reactor, a specialized and removable perforated packed column (3:1) was constructed for using immobilized cells (stainless steel net chamber). The process volume while using immobilized cells was set to 4 L. The bioreactor featured a separate decontamination chamber (second chamber), which matched the size of the primary chamber to ensure environmental safety. This secondary chamber facilitated the thorough decontamination of wastewater after treatment. A UV lamp with a λmax of 254 nm (i.e., UV-C) was used for decontamination. It helped to ensure that the treated water met the required quality standards before releasing it into the environment (Fig. 1).
Degradation studies in batch mode
The biological reactor system was sterilized for 30 min by the UV irradiation method. Next, an additional steam sterilization for 20 min was performed to ensure the sterility of the valves and connections. Synthetic wastewater (Hosseini and Borghei 2005) separately containing phenol (1000 mg/L), 2,4-DCP (1000 mg/L), and mixed pollutants (2000 mg/L; containing equal concentrations of phenol and 2,4-DCP) were treated separately. Pollutant containing synthetic wastewater was carefully transferred to the reactor under aseptic conditions. Bacterial inoculum (10%) was added to the bio-reaction mixture. Once the reactor was filled (leaving proper head space) and ready, the lid was closed, and the pH and temperature sensors were activated. The reactor's temperature was set to 32.5 °C, and the stirring speed was adjusted to 125 rpm. This continuous agitation of the medium played a crucial role in ensuring constant contact between the pollutants and the cells. Aeration was facilitated through the filtered air from the air valve. The same procedure was applied to conduct subsequent degradation studies involving immobilized cells. During bacterial inoculation, the immobilized cells were loaded in the immobilization chamber. The samples taken at regular intervals (4–8 h) were simultaneously analyzed for the pollutant, biological oxygen demand (BOD) and chemical oxygen demand (COD) removals, as described previously (Samudro and Mangkoedihardjo 2010). Once the complete removal of the pollutant was confirmed, the water was drained into the second chamber, i.e., the decontamination chamber and subjected to UVC treatment for 15 min (Chatzisymeon et al. 2011) before its release into the environment. The viability of the bacterium in the outlet sample was checked through culturing methods.
Single and hybrid cell immobilization studies
The cells of P. aeruginosa STV1713 were immobilized on different immobilization matrices viz. sodium alginate (single immobilization), alginate-chitosan-alginate (ACA) and polyvinyl alcohol- alginate (PVA-alginate) to achieve maximum degradation of the selected pollutants, phenol, and 2,4-DCP. The immobilized beads were repeatedly used for degradation until they lost biocatalytic activity. Overnight cultures of P. aeruginosa STV1713 separately grown in 1000 mg/L phenol and 2,4-DCP were harvested by centrifugation (2508×g for 10 min at 4 °C), and the pellets were subsequently immobilized. The final cell concentration for all immobilization techniques was 1.5 × 108 CFU/mL, corresponding to a McFarland standard of 0.5. All the experiments were carried out in the bioreactor. The perforated immobilization column was packed with immobilized cells, and the biological chamber was filled with the required quantity of BH-MSM containing phenolic pollutants. The column was attached to the bioreactor’s lid and immersed in the phenolic water. Continuous agitation was provided to ensure that the immobilized cells were in constant contact with the phenolic pollutant water. Samples were taken at definite intervals (4–8 h) to check pollutant removal, as described previously (Subramaniam et al. 2020).
Sodium alginate immobilization
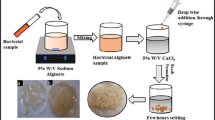
During alginate immobilization, sodium alginate was used in two different concentrations, 3% and 4%. The sodium alginate solutions were prepared, autoclaved, and cooled down to 40 °C, and 10 mL of bacterial suspension was added to each preparation. This resulted in final alginate concentrations of 3% and 4% (w/v). Similarly, 0.2 M calcium chloride (CaCl2) solution was prepared, sterilized and placed on a gel rocker with moderate agitation. The previously prepared bacterial-sodium alginate suspensions were added drop by drop into the CaCl2 solution from a height of 20 cm using a Pasteur pipette to facilitate the bead formation. The beads were kept for 1 h in the CaCl2 solution for gelling. Later, the beads were washed thrice with normal saline before being used for the experiments (Abarian et al. 2019).
ACA encapsulation
Bacterial cells (10 mL) were mixed with a 1.5% sterile sodium alginate solution. The resulting alginate-cell mixture was extruded into a solution of 0.2 M CaCl2 and left for 30 min in the solution for gelling. The beads were covered with a 0.5% chitosan solution (using a volume ratio of 1:5 beads to solution), and after 10 min, excess chitosan was removed by washing the beads with normal saline. Finally, the beads were immersed in a 0.15% sodium alginate solution to neutralize the residual chitosan. In order to create capsules with liquid cores, the beads were treated with a solution of 55 mM sodium citrate (Lu et al. 2012).
PVA-alginate entrapment
A solution of PVA (5%) and sodium alginate (0.8%) was made by dissolving them in distilled water at 80 °C. The resulting solution was autoclaved, cooled to 40 °C, and mixed with 10 mL of bacterial cell culture. The homogenous solution was then extruded in CaCl2 solution (0.2 M) and kept for 1 h to facilitate proper gelling of the beads. Afterward, the beads were rinsed with normal saline and used for degradation studies (Ruan et al. 2018).
Degradation studies using the immobilized cells were started with initial concentrations of 2000 mg/L phenol and 1400 mg/L 2,4-DCP (maximum concentrations of pollutants degraded by free cells of P. aeruginosa STV1713) and the results were compared with degradation capabilities of the free cells [shake flask culture method: Degradation studies started with an initial pollutant concentration of 400 mg/L (the concentration at which the bacterium was isolated).The bacterium was inoculated in to BH-MSM separately containing phenol and 2,4-DCP and incubated at 32.5 °C. After the complete removal of pollutants, the concentration was increased to 500 mg/L, and the experiments were repeated until it showed maximum degradation capabilities). Subsequently, they were used for the degradation of higher concentrations of pollutants.
Morphological analysis of immobilized cells
The alginate and PVA immobilized beads were sectioned, stained and analyzed to evaluate the morphological characteristics of immobilized bacteria. First, the beads were exposed to a 2.5% glutaraldehyde solution in 0.1 M sodium cacodylate buffer and left at 4 °C for 24 h to fix them. The fixative solution was removed, and the beads were washed thrice for 10 min with 0.1 M cacodylate washing buffer. Subsequently, the beads were incubated in a solution containing 1% osmium tetroxide (OsO4) and 1.5% potassium ferrocyanide (KFeCN) at 4 °C for 30 min. After the OsO4 treatment, another wash with 0.1 M cacodylate buffer was performed. The beads were then dehydrated with ethanol, gradually increasing from 10 to 90%, with 5 min intervals between each step. This was followed by two further dehydration steps with 100% ethanol, each lasting 10 min. Upon dehydration, the beads were infiltrated with epon by gradually adding increasing amounts of epon to ethanol (ranging from 1:1 to 3:1), with 30 min intervals between each addition. Next, pure epon, de-aerated under vacuum (constant pressure < 25 psi), was added to the beads, and they were left under vacuum for 1 h to remove any ethanol residue and air bubbles in the epon. Finally, the wells containing the beads were refilled with fresh epon and placed in an oven at 60 °C for 48 h to allow the epon to polymerize. Semi-thin sections with a thickness of 500 nm were prepared using an Ultracut-E ultramicrotome (Leica Reichert-Jung Ultra Microtome ULTRACUT E). These sections were then mounted on glass slides, stained with toluidine blue for 1 min, washed and observed under a light microscope (Labomed) (Mollaei et al. 2010).
Repeated batch degradations with immobilized cells
The immobilized beads were used repeatedly in a batch process to degrade phenolic pollutants (with two initial pollutant concentrations, 1000 mg/L and 1200 mg/L) to demonstrate their durability and stability. The degradation process was repeated with the same conditions until the degradation efficiency of the biocatalysts was no longer effective.
Biomass estimation
The biomass estimation method (based on the cell protein content in the gel beads) developed by Seung et al. (2005) was used to test the effectiveness of cells in the immobilized matrices. The method involved taking 1 g of gel beads and cutting them into small pieces on a glass plate using a surgical knife. The pieces were then placed in a test tube and treated with a 10% solution of sodium dodecyl sulfate (SDS) for 2 h to extract cell protein. After centrifugation, the protein content of the gel beads was assessed using the Lowry protein estimation method, as previously described (Lowry et al. 1951), with Bovine serum albumin (BSA; Sigma, USA) as the standard.
Metabolic pathway elucidation
After determining the degradation capabilities of the bacterium under immobilized conditions, efforts were undertaken to understand the metabolic mechanisms of phenol and 2,4-DCP degradation in P. aeruginosa STV1713 by enzyme assay and enzyme zymography.
Enzyme assay
The bacterial cells grown in BH-MSM separately containing 1000 mg/L phenol and 2,4-DCP were collected by centrifugation at 2508×g for 10 min. The pellets were washed twice using 0.33 M Tris–HCl buffer (pH 7.6). The cells were sonicated (Sonics Vibra-cell™) on ice for 4 min (30 s on and 30 s off cycle). After sonication, the cell extracts were centrifuged at 15,775×g for 20 min at 4 °C. The cell-free extracts were immediately used to test the activity of catechol dioxygenase enzymes as previously described (Lee et al. 2022).
Enzyme activity was calculated with the following formula;
where E is the molar extinction coefficient of the enzyme, L is the length of the cuvette, and △OD is the change in absorbance. “E” for catechol 1,2-dioxygenase (E260) is 16,800 mol/L/cm and catechol 2,3-dioxygenase (E375) is 14,700 mol/L/cm.
Lowry’s method (Lowry et al. 1951) determined the total protein concentration in the cell-free extract, with bovine serum albumin as the standard. The following equation calculated specific activities:
Enzyme zymography
Enzyme zymogram and SDS-PAGE analyses were conducted to confirm the presence of the degradation enzyme in the bacterium and its molecular weight. The crude enzyme extract (cell-free extract) from “Enzyme assay” section was electrophoresed under reducing conditions (SDS-PAGE- 4% stocking gel and 12% separating gel). After electrophoresis, the gels were soaked in a 1% catechol (substrate) solution to visualize the product bands formed due to the enzyme–substrate reaction. The zymogram band patterns were compared with normally stained SDS gels loaded with molecular markers to detect the molecular weight of the enzyme (Surendra et al. 2017).
Results
Batch reactor: designing and fabrication
A batch reactor was designed and constructed (Fig. 2) to facilitate the efficient treatment of phenolic wastewater by optimizing growth conditions conducive to the bacterium P. aeruginosa STV1713. Batch degradation studies started with an initial concentration of 1000 mg/L phenol and 2,4-DCP.
Biodegradation in batch mode: COD and BOD removal assays
While cultivated in the presence of 1000 mg/L phenol, P. aeruginosa STV1713 exhibited a reduction in COD and BOD within the culture medium, with removal rates recorded at 30.21 mg/L/h and 16.92 mg/L/h, respectively Fig. 3a. The bacterium achieved the reduction of COD and BOD values from their initial concentrations of 846.05 mg/L and 473.98 mg/L to significantly lower levels of 12.57 mg/L and 11.84 mg/L, respectively (over a duration of 28 h). Figure 3a illustrates the utilization of the adapted culture of P. aeruginosa STV1713 as the inoculum, which notably eliminated the lag phase in phenol degradation, consequently leading to direct reductions in both COD and BOD.
Graphs showing BOD, COD, and pollutant removal percentages during the degradation of phenol (a), (1000 mg/L) and 2,4-DCP (b), (1000 mg/L) by free cells of P. aeruginosa STV1713 in the batch bioreactor. Error bars were added to the graphs to show the experimental error observed among triplicate data points
When cultivated in the presence of 1000 mg/L 2,4-DCP, the bacterium reduced both the COD and BOD within the culture medium, with removal rates of 12.85 mg/L/h and 7.21 mg/L/h, respectively (Fig. 3b). The bacterium achieved the reduction of COD and BOD values from their initial concentrations of 848.72 mg/L and 476.45 mg/L to significantly lower levels of 18.92 mg/L and 12.17 mg/L, respectively, over a duration of 66 h. Although phenol and 2,4-DCP (1000 mg/L) were fully eliminated, traces of residual COD and BOD persisted to some extent. This retention could be attributed to the release of soluble byproducts (organic acids, exopolysaccharides etc.) into the synthetic wastewater during the degradation process.
After treating individual pollutants, the degradation performance of the bacterium in the presence of mixed pollutants was assessed. Remarkably, the bacterium reduced both the COD and BOD within the culture medium, with removal rates of 17.7 mg/L/h and 8.47 mg/L/h, respectively (Fig. 4). The bacterium caused the reduction of COD and BOD values from their initial concentrations of 1136.5 mg/L and 542.46 mg/L to significantly lower levels of 17.6 mg/L and 12.05 mg/L, respectively, over a duration of 65 h with complete removal of pollutants. These findings strongly indicated that the bacterium exhibited efficient degradation of higher concentrations of mixed pollutants, similar to its performance with individual pollutants. Furthermore, the study highlights that the presence of the pollutant mixture did not impede the degradation potential of P. aeruginosa STV1713.
Single and hybrid immobilization studies
The bacterium P. aeruginosa STV1713 immobilized on various matrices such as ACA, 3% alginate, 4% alginate and PVA-alginate showed improved degradation and biocatalytic efficiencies. The degradation studies started with an initial concentration of 2000 mg/L phenol and 1400 mg/L 2,4-DCP. After confirming the degradation efficiencies, they were subsequently employed for the degradation of higher concentrations of phenol and 2,4-DCP.
Sodium alginate immobilization
The total time the cells immobilized in alginate (3%) took for the complete degradation of 2000 mg/L phenol and 1400 mg/L 2,4-DCP was 46 h and 47.10 h, respectively. The results showed a considerable drop-in degradation time, around 23.33% and 46.54% less than freely suspended cells (shake flask culture method) required to degrade an equivalent initial concentration of phenol and 2,4-DCP, respectively (Fig. 5a–d). During the degradation of both pollutants, cells immobilized on 3% sodium alginate showed no significant difference in total time of degradation compared to 4% alginate immobilization (Fig. 5c–d). Therefore, 3% alginate was used in subsequent degradation studies.
Degradation pattern of a phenol and b 2,4-DCP by the free cells of P. aeruginosa STV1713. Degradation pattern of c phenol and d 2,4-DCP by P. aeruginosa STV1713 cells immobilized on different matrices such as 3% and 4% alginate, ACA, and PVA-alginate at initial concentrations of 2000 mg/L and 1400 mg/L, respectively. Error bars were added to the graphs to show the experimental error observed among triplicate data points
Degradation studies using ACA and PVA immobilized cells
Cells trapped in ACA showed the same level of proficiency as cells trapped in 3% alginate during phenol and 2,4-DCP degradation. The ACA immobilized cells could completely degrade phenol and 2,4-DCP (2000 mg/L and 1400 mg/L, respectively) within 45 h and 46 h, respectively (Fig. 5c and d).
Using a gelling solution consisting of CaCl2 in combination with PVA-alginate was believed to prevent the clustering. Also, it was expected to improve the surface properties of the beads and create beads with a spherical shape (Mollaei et al. 2010). Results showed that the agglomeration was prevented, and the PVA-alginate beads were equally capable of degradation and completely degraded phenol and 2,4-DCP within 44.5 h and 45 h, respectively (Fig. 5c and d).
Degradation of higher phenolic concentration by immobilized cells
After initial degradation experiments, cells immobilized in alginate (3%), ACA and PVA-alginate were used to degrade higher concentrations of phenolic pollutants (phenol—2200 to 2500 mg/L; 2,4-DCP- 1500 to 2000 mg/L) to access their pollutant tolerance and degradation efficiency. The results showed that compared to single immobilization (Sodium alginate 3%), hybrid immobilizations (PVA-alginate and ACA) showed better degradation performances in all pollutant concentrations. In the case of phenol degradation, both single and hybrid immobilized cells completely degraded phenol up to 2300 mg/L. In contrast, when the concentration was increased to 2500 mg/L, only PVA-alginate beads showed complete degradation within 100 h. During 2,4-DCP degradation, only hybrid immobilized cells showed complete degradation abilities till 1700 mg/L within 85 h. Sodium alginate (3%) immobilized cells had less tolerance under higher 2,4-DCP concentrations. Later, when the pollutant concentration was raised to 2000 mg/L, only PVA-alginate beads provided the best performance and completely degraded the pollutant within 102 h (Fig. 6a and b).
Morphological analysis of immobilized cells
Microscopic examination of the samples through light microscopy unveiled the excellent preservation of cell morphological characteristics following their embedding in epon resin. Clusters of bacteria were observed within the polymer matrix of beads, indicating that the bacterial activities were confined to the microenvironment of these colonies. The density and distribution of the bacteria varied extensively depending on the composition of the beads (Fig. 7a and b). The bacterial colonies were notably congested inside the sodium alginate beads, whereas in the PVA-alginate beads, individual colonies had ample space for their activities, indicating better preservation of cells in the hybrid matrices.
Repeated batch degradation with immobilized beads
The robustness and longevity of the trapped cells were analyzed by reusing the beads in various batches of degradations. Table 1 presents information on the efficiency of diverse immobilized biocatalysts when exposed to recurring degradation cycles at two different initial concentrations (1000 mg/L and 1200 mg/L) of phenol and 2,4-DCP. For phenol degradation with an initial concentration of 1000 mg/L, up to 16 batches of degradation reactions were performed over a period of 15.3 days using PVA-alginate. In comparison, ACA and 3% alginate beads were continuously reused 14 (14.5 days) and 15 (15 days) times, respectively. For the same initial concentration of 2,4-DCP, PVA-alginate beads could be reused for 9 (15 days) batches, while ACA and 3% alginate beads could be employed for 8 (13.3 days) and 7 times (12.2 days), respectively. Again, it was observed that PVA-alginate beads displayed the most efficient performance when the phenol concentration was raised to 1200 mg/L. It was able to operate for 9 uninterrupted batches, which spanned across 9 days. In comparison, ACA and alginate beads were reused for 8 (8.3 days) and 7 (7.5 days) batches, respectively. Similarly, PVA-alginate beads possessed higher activity under the same concentration of 2,4-DCP. It could be employed for 7 (13.1 days) continuous batches, while ACA and 3% alginate beads showed maximum bio-catalytic activity till 6 (11.5 days) and 5 (9.7 days) batches, respectively.
Biomass estimation
The long-term stability of the biocatalysts and the protein content of the cells trapped in the beads were measured as an indicator of changes in biomass during semi-continuous degradation of 1000 mg/L phenol and 2,4-DCP. Table 2 shows that the PVA-alginate hybrid beads had the maximum cell loading capacity in both the middle and final runs. The hybrid beads demonstrated outstanding stability and exceptional surface properties, resulting in the least cell leakage and the highest biomass content. Consequently, the protein content of the cells immobilized in 3% alginate was the lowest, consistent with the findings from the degradation and batch experiments.
Metabolic pathway elucidation
The metabolic mechanisms of pollutant removal in P. aeruginosa STV1713 were determined by enzyme assay and catechol zymography.
Enzyme assay
In the present study, the bacterium P. aeruginosa STV1713 was found to use catechol 1,2-dioxygenase enzyme to catabolize phenol and 2,4-DCP (Table 3). The enzyme analysis revealed that the organism uses the ortho cleavage pathway for pollutant degradation due to the low activity of catechol 2,3-dioxygenase compared to catechol 1,2-dioxygenase.
Enzyme zymography
An enzyme zymogram and SDS-PAGE were conducted to confirm the presence of catechol 1,2-dioxygenase in the bacterium during pollutant degradation and determine its molecular weight. The enzyme–substrate (catechol 1,2-dioxygenase and catechol) reaction product (cis-cis muconic acid) appeared as distinct bands in the zymogram gel [(b) L4- phenol grown sample and L5-2,4-DCP grown sample), corresponding to the protein bands in the SDS-PAGE gel at the molecular weight 245 kDa (Fig. 8a and b).
a SDS electrophoretic band pattern of proteins isolated from P. aeruginosa STV1713 during the degradation of phenol (L1) and 2,4-DCP (L2). Lane L1 shows the protein marker. b Zymogram gel showing catechol 1,2-dioxygenase enzyme–substrate reaction bands corresponding to the SDS gel bands at molecular weight 245 kDa. Lane L4 represents the phenol-grown sample, while L5 represents the 2,4-DCP-grown sample
Discussion
Phenolic compounds are ubiquitous environmental pollutants (Guo et al. 2020). In the environment, phenol is degraded by various microorganisms and they utilize phenol as their sole carbon and energy source. Bacteria, fungi, and algae are the common phenol-degrading microbes (Dell’ Anno et al. 2021), among which bacteria are the most widely used. The chemical stability of the C–Cl bond and the potent antimicrobial properties of polychlorinated compounds pose challenges for the degradation by most environmental microorganisms (Nikel et al. 2013).
Cell immobilization is an innovative and effective way to treat such toxic pollutants, as it can protect the cells by avoiding direct contact with them. Though cell immobilization is commonly employed, hybrid immobilization is not so common. The current study used 3% sodium alginate, ACA and PVA-alginate to immobilize P. aeruginosa STV1713. The results were very impressive, as immobilization conferred more protection to the cells and enhanced the rate of phenolic pollutant removal. Compared to single immobilization, hybrid immobilizations were found to be very effective in terms of degradation ability, consistency in performance and efficiency in maintaining the catalytic activity. The PVA-alginate immobilized cells could degrade higher concentrations of phenol and 2,4-DCP up to 2500 mg/L and 2000 mg/L, respectively. Hybrid immobilized cells had better stability in higher concentrations of phenolic pollutants. Similarly, Mollaei et al. (2010) immobilized Pseudomonas sp. SA01 cells on various matrices such as glycerol, pectin, PVA-alginate, and sodium alginate. The cells showed better degradation efficiency of phenol at different initial concentrations. In line with these investigations, Ahmad et al. (2012) immobilized phenol-degrading Acinetobacter sp. Strain AQ5NOL 1 in gellan gum and compared its degradation abilities with free cells. They reported that after immobilization, the tolerance level of the bacterium increased, and it grew up to 1900 mg/L phenol, whereas free cells grew only up to 1100 mg/L. Table 4 compares the degradation capabilities of the present immobilization system (PVA- alginate, ACA and sodium alginate) with other immobilized systems reported in the literature for degradation of phenol and 2,4-DCP.
Providing the best-suited conditions for the growth of bacterium is crucial in every biological treatment system as it can directly affect the functional capabilities of the biocatalyst. Bioreactors are specialized systems for providing the required optimized conditions for biological reactions. In the present study, we have introduced a batch biological reactor model for phenolic pollutant removal using free and immobilized cells of P. aeruginosa STV1713. The biological reactor featured a specialized and removable packed column for immobilized cells. Additionally, we have provided a separate decontamination chamber for the treatment of biologically treated water before its discharge into the environment. The decontamination chamber greatly helped while reusing the immobilized cells. When synthetic phenolic wastewater was used for the degradation in the bioreactor, we could observe a gradual reduction in both COD and BOD along with pollutant removal. While phenol and 2,4-DCP at a concentration of 1000 mg/L were completely eliminated, traces of residual COD and BOD persisted to some extent, which could be attributed to the release of soluble byproducts of bacterial metabolism into the synthetic wastewater during the degradation process (Afzal et al. 2007). Similarly, many batch reactor models have been proposed for the treatment of phenolic pollutants. For instance, Lin and Gu (2023) studied the kinetics of phenol biodegradation using custom-made batch and continuous reactors. In line with these investigations, Lin and Cheng (2020) studied phenol degradation by free and immobilized cells of Pseudomonas putida BCRC 14365 in a fabricated batch reactor.
In bacteria, after an initial breakdown into catechol, phenolic pollutants are further degraded via two metabolic pathways: the ortho-cleavage pathway and the meta-cleavage pathway (Sasi and Suchithra 2023). In the present study, the bacterium P. aeruginosa STV1713 was found to utilize catechol 1,2-dioxygenase enzyme for the catabolism of phenol and 2,4-DCP. Enzyme assay analysis suggested that the organism follows the ortho cleavage pathway due to the high activity of catechol 1,2-dioxygenase. However, the organism also possessed an inherent capability for the meta-cleavage pathway, which was demonstrated by the lower activity of the catechol 2,3-dioxygenase enzyme. In line with the current study, Asimakoula et al. (2023) studied the phenol degradation pathway in Pseudarthrobacter phenanthrenivorans Sphe3. They could find higher activities of catechol 1,2 dioxygenase compared to catechol 2,3 dioxygenase. The results suggested that the bacterium used the ortho-cleavage pathway for phenol utilization. In contrast to the current study, Mahiudddin et al. (2012) and Shourian et al. (2009) reported the meta-cleavage mechanism of phenol degradation in Pseudomonas fluorescens PU1 and Pseudomonas sp. SA01. Similarly, Ahmad et al. (2017) studied the pathways of phenol degradation in Acinetobacter sp. strain AQ5NOL 1 and they found that the bacterium utilized phenol via the meta-cleavage pathway.
In the present study, SDS PAGE and enzyme zymogram analysis further confirmed the presence of catechol 1,2-dioxygenase enzyme in the degradation samples. SDS PAGE analysis indicated that the enzyme has a molecular weight of 245 kDa. Corresponding to the SDS-PAGE banding pattern, there were enzyme–substrate (catechol) reaction bands (Cis-Cis muconic acid) on the zymogram gel, clearly indicating the presence of the degradation enzyme. In a similar study, native zymogram analysis revealed the presence of catechol 2,3-dioxygenase enzyme in Bacillus pumilus strain MG03 (Chris Felshia et al. 2014). This showed that enzyme zymography is a potentially reliable method for the detection of catabolic enzymes during degradation studies.
In summary, the present study introduced a new batch bioreactor model for phenolic pollutant removal that enabled the complete degradation of pollutants by P. aeruginosa STV1713 in both free and immobilized states. The bacterial cells immobilized on different matrices, such as sodium alginate, ACA, and PVA-alginate, were used to treat synthetic wastewater containing phenolic pollutants in order to determine the best matrix for phenolic wastewater treatment. Hybrid immobilized cells demonstrated superior morphological preservation and degradation abilities compared to single immobilized cells. Furthermore, enzyme detection methods revealed that the bacterium employed the ortho cleavage pathway to remove phenol and 2,4-DCP. These findings collectively underscore the indispensable role of our proposed bioreactor and the bacterium P. aeruginosa STV1713 in advancing sustainable solutions for environmental pollution mitigation. Although the study presents significant data to the scientific community, it still requires further research on scaling up the current reactor model for treating industrial wastewater. Also, the potential of the immobilized cells in treating real wastewater samples needs to be investigated. Bridging these knowledge gaps will advance our understanding of phenol degradation and contribute to the development of more effective and sustainable remediation approaches.
Conclusion
In the present study, the bacterium P. aeruginosa STV1713 in both free and immobilized states (sodium alginate, ACA and PVA-alginate) was used for the degradation of phenol and 2,4-DCP in a custom-made biological batch reactor. The degradations were carried out under optimized conditions. When synthetic phenolic wastewater was employed as the substrate for degradation in the bioreactor, a gradual decrease in both COD and BOD was evident, indicating effective pollutant removal. Although phenol and 2,4-DCP (1000 mg/L) were completely eliminated, residual traces of COD and BOD persisted to a certain extent. This retention could be related to the release of soluble metabolites into the synthetic wastewater during the degradation process. While using immobilized cells, the hybrid immobilized cells showed better morphological preservations and degradation capabilities under high phenolic pollutant concentrations compared to single immobilized cells. The PVA immobilized cells could degrade maximum phenol and 2,4-DCP concentrations up to 2500 mg/L and 2000 mg/L, respectively. Furthermore, Enzyme assay and enzyme zymography revealed that the bacterium used the ortho degradation mechanism to remove phenolic pollutants. The present study primarily involved the in vitro removal of phenolic pollutants in synthetic wastewater by the bacterium in a lab-scale bioreactor. Therefore, we need more research and advancements in this field, including the scale-up of the current reactor system and the development of onsite pollutant removal strategies.
Data availability
The data supporting this study's findings are available from the corresponding author, Suchithra Tharamel Vasu, upon reasonable request.
References
Abarian M, Hassanshahian M, Esbah A (2019) Degradation of phenol at high concentrations using immobilization of Pseudomonas putida P53 into sawdust entrapped in sodium-alginate beads. Water Sci Technol 79:1387–1396. https://doi.org/10.2166/wst.2019.134
Afzal M, Iqbal S, Rauf S, Khalid ZM (2007) Characteristics of phenol biodegradation in saline solutions by monocultures of Pseudomonas aeruginosa and Pseudomonas pseudomallei. J Hazard Mater 149:60–66. https://doi.org/10.1016/j.jhazmat.2007.03.046
Ahmad SA, Shamaan NA, Arif NM et al (2012) Enhanced phenol degradation by immobilized Acinetobacter sp. strain AQ5NOL 1. World J Microbiol Biotechnol 28:347–352. https://doi.org/10.1007/s11274-011-0826-z
Ahmad SA, Shamaan NA, Syed MA et al (2017) Meta-cleavage pathway of phenol degradation by Acinetobacter sp. strain AQ5NOL 1. Rend Lincei 28:1–9. https://doi.org/10.1007/s12210-016-0554-2
Asimakoula S, Marinakos O, Tsagogiannis E, Koukkou AI (2023) Phenol degradation by Pseudarthrobacter phenanthrenivorans Sphe3. Microorganisms 11:524. https://doi.org/10.3390/microorganisms11020524
Bala S, Garg D, Thirumalesh BV et al (2022) Recent strategies for bioremediation of emerging pollutants: a review for a green and sustainable environment. Toxics 10:484. https://doi.org/10.3390/toxics10080484
Barik M, Das CP, Kumar Verma A et al (2021) Metabolic profiling of phenol biodegradation by an indigenous Rhodococcus pyridinivorans strain PDB9T N-1 isolated from paper pulp wastewater. Int Biodeterior Biodegrad 158:105168. https://doi.org/10.1016/j.ibiod.2020.105168
Basak B, Jeon BH, Kurade MB et al (2019) Biodegradation of high concentration phenol using sugarcane bagasse immobilized Candida tropicalis PHB5 in a packed-bed column reactor. Ecotoxicol Environ Saf 180:317–325. https://doi.org/10.1016/j.ecoenv.2019.05.020
Chatzisymeon E, Droumpali A, Mantzavinos D, Venieri D (2011) Disinfection of water and wastewater by UV-A and UV-C irradiation: application of real-time PCR method. Photochem Photobiol Sci 10:389–395. https://doi.org/10.1039/c0pp00161a
Chris Felshia YA, Varadharajan K, Baran A, Mandal AB, Ganesan S, Arumugam G (2014) Maximum phenol tolerance and subsequent degradation profile of a Bacillus pumilus strain mcg03an isolate from tannery wastewater contaminated soil. Int J Curr Res 6:9734–9744
Dell’ Anno F, Rastelli E, Sansone C et al (2021) Bacteria, fungi and microalgae for the bioremediation of marine sediments contaminated by petroleum hydrocarbons in the omics era. Microorganisms 9:1695. https://doi.org/10.3390/microorganisms9081695
Guo F, Chai L, Zhang S et al (2020) Computational biotransformation profile of emerging phenolic pollutants by cytochromes P450: phenol-coupling mechanism. Environ Sci Technol 54:2902–2912. https://doi.org/10.1021/acs.est.9b06897
Hosseini SH, Borghei SM (2005) The treatment of phenolic wastewater using a moving bed bio-reactor. Process Biochem 40:1027–1031. https://doi.org/10.1016/j.procbio.2004.05.002
Keweloh H, Heipieper HJ, Rehm HJ (1989) Protection of bacteria against toxicity of phenol by immobilization in calcium alginate. Appl Microbiol Biotechnol 31:383–389. https://doi.org/10.1007/BF00257609
Krastanov A, Alexieva Z, Yemendzhiev H (2013) Microbial degradation of phenol and phenolic derivatives. Eng Life Sci 13:76–87. https://doi.org/10.1002/elsc.201100227
Lee GL, Zakaria NN, Futamata H et al (2022) Metabolic pathway of phenol degradation of a cold-adapted Antarctic bacteria Arthrobacter sp. Catalysts 12:1422. https://doi.org/10.3390/catal12111422
Lin YH, Cheng YS (2020) Phenol degradation kinetics by free and immobilized Pseudomonas putida BCRC 14365 in batch and continuous-flow bioreactors. Processes 8:721. https://doi.org/10.3390/toxics10080484
Lin YH, Gu YJ (2023) Phenol degradation performance in batch and continuous reactors with immobilized cells of Pseudomonas putida. Processes 11:739. https://doi.org/10.3390/pr11030739
Lowry O, Rosebrough N, Farr AL, Randall R (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193:265–275. https://doi.org/10.1016/S0021-9258(19)52451-6
Lu D, Zhang Y, Niu S et al (2012) Study of phenol biodegradation using Bacillus amyloliquefaciens strain WJDB-1 immobilized in alginate–chitosan–alginate (ACA) microcapsules by electrochemical method. Biodegradation 23:209–219. https://doi.org/10.1007/s10532-011-9500-2
Mahiudddin M, Fakhruddin ANM, Abdullah-Al M (2012) Degradation of phenol via meta cleavage pathway by Pseudomonas fluorescens PU1. ISRN Microbiol 2012:741820. https://doi.org/10.5402/2012/741820
Mishra V (2017) Microbial degradation of phenol: a review. J Water Pollut Purif Res 32:12–28. https://doi.org/10.1504/IJEP.2008.016895
Mollaei M, Abdollahpour S, Atashgahi S et al (2010) Enhanced phenol degradation by Pseudomonas sp. SA01: gaining insight into the novel single and hybrid immobilizations. J Hazard Mater 175:284–292. https://doi.org/10.1016/j.jhazmat.2009.10.002
Nikel PI, Pérez-Pantoja D, de Lorenzo V (2013) Why are chlorinated pollutants so difficult to degrade aerobically? Redox stress limits 1,3-dichloprop-1-ene metabolism by Pseudomonas pavonaceae. Philos Trans R Soc B Biol Sci 368:20120377. https://doi.org/10.1098/rstb.2012.0377
Oluwasanu AA (2018) Fate and toxicity of chlorinated phenols of environmental implications: a review. Med Anal Chem Int J. https://doi.org/10.23880/macij-16000126
Panigrahy N, Priyadarshini A, Sahoo MM et al (2022) A comprehensive review on eco-toxicity and biodegradation of phenolics: recent progress and future outlook. Environ Technol Innov 27:102423. https://doi.org/10.1016/j.eti.2022.102423
Patel A, Sartaj K, Arora N et al (2017) Biodegradation of phenol via meta cleavage pathway triggers de novo TAG biosynthesis pathway in oleaginous yeast. J Hazard Mater 340:47–56. https://doi.org/10.1016/j.jhazmat.2017.07.013
Pinheiro PF, Menini LAP, Bernardes PC et al (2018) Semisynthetic phenol derivatives obtained from natural phenols: antimicrobial activity and molecular properties. J Agric Food Chem 66:323–330. https://doi.org/10.1021/acs.jafc.7b04418
Ruan B, Wu P, Chen M et al (2018) Immobilization of Sphingomonas sp. GY2B in polyvinyl alcohol–alginate–kaolin beads for efficient degradation of phenol against unfavorable environmental factors. Ecotoxicol Environ Saf 162:103–111. https://doi.org/10.1016/j.ecoenv.2018.06.058
Samudro G, Mangkoedihardjo S (2010) Review on BOD, COD and BOD/COD ratio: a triangle zone for toxic, biodegradable and stable levels. Int J Acad Res 2:235–239
Sasi R, Suchithra TV (2023) Transcriptome analysis of a novel and highly resistant hydrocarbon-degrading bacteria during the removal of phenol and 2,4-dichlorophenol. Biomass Convers Biorefinery 11:1–21. https://doi.org/10.1007/s13399-023-04509-x
Sengupta A, Jebur M, Kamaz M, Wickramasinghe SR (2022) Removal of emerging contaminants from wastewater streams using membrane bioreactors: a review. Membranes 12:60. https://doi.org/10.3390/membranes12010060
Seung HS, Suk SC, Park K, Yoo YJ (2005) Novel hybrid immobilization of microorganisms and its applications to biological denitrification. Enzyme Microb Technol 37:567–573. https://doi.org/10.1016/j.enzmictec.2005.07.012
Shourian M, Noghabi KA, Zahiri HS et al (2009) Efficient phenol degradation by a newly characterized Pseudomonas sp. SA01 isolated from pharmaceutical wastewaters. Desalination 246:577–594. https://doi.org/10.1016/j.desal.2008.07.015
Subramaniam K, Athirrah T, Mazuki T et al (2019) Isolation and optimisation of phenol degradation by antarctic isolate using one factor at time. Malaysian J Biochem Mol Biol 2019:79–86
Subramaniam K, Shaharuddin NA, Tengku-Mazuki TA et al (2020) Statistical optimisation for enhancement of phenol biodegradation by the Antarctic soil bacterium Arthrobacter sp. strain AQ5–15 using response surface methodology py. J Environ Biol 41:1560–1569. https://doi.org/10.22438/JEB/41/6/MRN-1496
Surendra SV, Mahalingam BL, Velan M (2017) Degradation of monoaromatics by Bacillus pumilus MVSV3. Braz Arch Biol Technol 60:1–18. https://doi.org/10.1590/1678-4324-2017160319
Tripathi M, Garg SK (2013) Co-remediation of pentachlorophenol and Cr6+ by free and immobilized cells of native Bacillus cereus isolate: spectrometric characterization of PCP dechlorination products, bioreactor trial and chromate reductase activity. Process Biochem 48:496–509. https://doi.org/10.1016/j.procbio.2013.02.009
Acknowledgements
We want to acknowledge the Kerala State Council for Science, Technology, and Environment for the financial support throughout this study. Also, we would like to acknowledge Dr. K. Haribabu for his valuable suggestions during the design of the batch bioreactor.
Funding
Kerala State Council for Science Technology and Environment (Sasthra Bhavan, Pattom, Thiruvananthapuram, Kerala, India-695004) (Research Fellowship- Sanction order No.29/FSHP/2016/KSCSTE dated 24.03.2017).
Author information
Authors and Affiliations
Contributions
All authors, RS and STV, contributed to the study's conception and design. RS performed material preparation, data collection and experimental analyses. RS wrote the first draft of the manuscript and all authors commented on previous versions. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sasi, R., Vasu, S.T. Batch-mode degradation of high-strength phenolic pollutants by Pseudomonas aeruginosa strain STV1713 immobilized on single and hybrid matrices. Biodegradation 35, 423–438 (2024). https://doi.org/10.1007/s10532-023-10067-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-023-10067-w