Abstract
Two biphenyl-degrading bacterial strains, SS1 and SS2, were isolated from polychlorinated biphenyl (PCB)-contaminated soil. They were identified as Rhodococcus ruber and Rhodococcus pyridinivorans based on the 16S rRNA gene sequence, as well as morphological, physiological and biochemical characteristics. SS1 and SS2 exhibited tolerance to 2000 and 3000 mg/L of biphenyl. And they could degrade 83.2 and 71.5% of 1300 mg/L biphenyl within 84 h, respectively. In the case of low-chlorinated PCB congeners, benzoate and 3-chlorobenzoate, the degradation activities of SS1 and SS2 were also significant. In addition, these two strains exhibited chemotactic response toward TCA-cycle intermediates, benzoate, biphenyl and 2-chlorobenzoate. This study indicated that, like the flagellated bacteria, non-flagellated Rhodococcus spp. might actively seek substrates through the process of chemotaxis once the substrates are depleted in their surroundings. Together, these data provide supporting evidence that SS1 and SS2 might be good candidates for restoring biphenyl/PCB-polluted environments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Polychlorinated biphenyls (PCBs) have been used extensively for a variety of industrial purposes, such as the manufacture of flame retardants, oil condensers, dielectrics, plasticizers, heat exchangers and hydraulic fluids (Abraham et al. 2002; Pieper 2005; Pieper and Seeger 2008; Vasilyeva and Strijakova 2007). Although the use of PCBs is now banned (Aken et al. 2010), they are still recalcitrant in the environment due to low bioavailability and stable structure. PCBs are toxic and cause serious effects on the immune, endocrine, nervous and reproductive systems in animals, often leading to cancer (Aoki 2001; Faroon et al. 2000). These deleterious effects have motivated the search for ways to remove these organic pollutants from contaminated environments.
Biological degradation of PCBs is an attractive clean-up strategy because it is environmentally friendly and cost-effective (Ohtsubo et al. 2004). Many studies have shown that biphenyl-degrading bacteria can utilize or co-metabolize various PCB congeners through the biphenyl catabolic pathway (Kohler et al. 1988; Bartels et al. 1999; Bopp 1986; Taguchi et al. 2007). In fact, a complete biphenyl catabolic pathway includes two parts: biphenyl upper pathway (transformation of biphenyl/PCBs into benzoate/chlorobenzoates (CBAs) and aliphatic acids) and biphenyl lower pathway (further mineralization of benzoate/CBAs and aliphatic acid) (Aken et al. 2010). Some biphenyl-degrading bacteria do not contain a complete biphenyl catabolic pathway (Pieper and Seeger 2008), which might lead to accumulation of dead-end intermediates and cause potential damage during biphenyl/PCBs biodegradation. The most easily accumulated dead-end intermediates are benzoate and CBAs (Adebusoye et al. 2008a; Furukawa et al. 1979). It has been proved that the intermediates (benzoate and CBAs) formed during the process of biphenyl/PCB biodegradation might inhibit the growth of microorganisms (Eklund 1985). And the inhibition effects of CBAs on PCBs degradation have been also reported (Adebusoye et al. 2008a; Stratford et al. 1996). Thus, it is necessary to screen excellent strains which have the ability to degrade biphenyl, PCBs and their intermediates.
Nevertheless, biodegradation efficiency is not only associated with the degrading capability of bacteria, but also depends on the bioavailability of pollutants (Yang et al. 1995), which is influenced by microbial mobility in addition to the soil medium and the nature of the pollutants. The biodegradation efficiency of biphenyl can be limited by its strong hydrophobicity and low biodegradability. In the past few years, many studies have shown that most motile bacteria can sense and access pollutants through the process of chemotaxis (Pandey and Jain 2002; Parales and Harwood 2002). The chemotactic motility of bacteria could increase pollutant bioavailability, which in turn was found to have a key role in bioremediation (Krell et al. 2013; Marx and Aitken 1999, 2000; Pandey and Jain 2002; Paul et al. 2006; Parales et al. 2015). The extent to which biphenyl, PCB congeners and their intermediates act as chemoattractants for biphenyl-degrading bacteria plays an important role in improving the degradation efficiency of PCBs. There have been reports of biphenyl-degrading bacteria showing chemotactic response to benzoate, biphenyl (Gordillo et al. 2007; Wu et al. 2003), PCBs (Gordillo et al. 2007; Tremaroli et al. 2010), 2-chlorobenzoate (2-CBA), 3-chlorobenzoate (3-CBA) and 4-chlorobenzoate (4-CBA) (Harwood 1989; Harwood et al. 1990). Notably, most of the reported biphenyl-degrading bacteria with chemotactic capability are Pseudomonas spp., which are motile and have flagellums. Interestingly, although Rhodococcus spp. are considered non-motile bacteria, some studies have shown that they exhibit chemotaxis towards Arabidopsis thaliana root exudates (Toussaint et al. 2012), biphenyl (Wu et al. 2003), and natural crude oil (Tanase et al. 2012). In fact, Rhodococcus spp., which can utilize biphenyl, are the dominant microorganisms found in a PCB-contaminated site (Leigh et al. 2006). It is unclear how these flagella-independent mechanisms contribute to overcome the restricted bioavailability of pollutants.
In this study, two biphenyl-degrading strains SS1 and SS2 were isolated from soil samples in electronic waste recycling area, Taizhou (28.5605°N, 121.3852°E, PCB concentration of 3.60 mg/kg), Zhejiang Province, China. The degradation ability of PCBs, biphenyl, benzoate and CBAs of SS1 and SS2 were investigated, and the chemotaxis of SS1 and SS2 toward TCA-cycle intermediates, biphenyl, benzoate, PCBs and CBAs were preliminarily studied.
Materials and methods
Chemicals and culture media
Biphenyl and benzoic acid were obtained from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). PCB congeners were purchased from Accustandard Co., Ltd (USA), while 2-CBA, 3-CBA, and 4-CBA were purchased from Aladdin Chemical Co., Ltd (Shanghai, China). TCA-cycle intermediates were purchased from Beijing Solarbio Science & Technology Co., Ltd (Beijing, China). Other chemical standards used in this study were obtained from Sangon Biotech Co., Ltd (Shanghai, China).
The culture media used in this study were Luria–Bertani (LB) medium (NaCl 10.0, tryptone 10.0, and yeast extract 5.0 g/L, pH 7.2), mineral salts medium (MSM, KH2PO4 1.0, K2HPO4·3H2O 3.0, MgSO4 0.15, FeSO4 0.01, CaCl2 0.005, NaCl 1.0, (NH4)2SO4 0.5 g/L), and trace elements solution 0.1% (v/v) at pH 7.2. The trace elements solution contained the following (g/L): Na2MO4·H2O 6.7, ZnSO4·5H2O 28.0, CuSO4·5H2O 2.0, H3BO4 4.0, MnSO4·5H2O 4.0, and CoSO4·7H2O 4.7 at pH 7.2, drop assay medium (KH2PO4 1, K2HPO4·3H2O 3, FeSO4·7H2O 0.02, (NH4)2SO4 0.5, NaCl 2 g/L, containing 0.5% agar), and M9 minimal salts medium (Na2PO4·12H2O 15.13, KH2PO4 3.0, NaCl 0.5, NH4Cl 1.0, MgSO4·7H2O 0.246, CaCl2 0.01, g/L). All media were sterilized by autoclaving at 121 °C for 20 min.
Bacterial strains
Rhodococcus ruber SS1 and Rhodococcus pyridinivorans SS2 were isolated in our laboratory by enrichment on biphenyl from PCB-contaminated soil samples as described earlier by Hu et al. (2015). Strains SS1 and SS2 were identified based on their 16S rRNA gene sequence and further characterized on the basis of morphological examination and biochemical tests. The 16S rRNA genes of SS1 and SS2 were amplified by PCR with a pair of forward and reverse primers: 27f (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492r (5′-TAC CTT GTT ACG ACT T-3′) (Heuer et al. 1997). The 16S rRNA gene sequence and related sequences acquired from GenBank were aligned by BIOEDIT version 7.0 software. A phylogenetic tree was built by the neighbor-joining method as implemented in MEGA version 4.0 software.
Biphenyl degradation experiments
SS1 and SS2 were pre-cultured for 24 h in LB medium containing 5 mg/L biphenyl at 30 °C and 180 rpm on a shaker. Then cells were collected by centrifugation at 6000 rpm for 5 min at room temperature, washed twice with phosphate buffers (0.05 M, pH 7.2) and suspended in MSM. The cell suspensions were adjusted to an OD600 of 1.0 for the following test.
For biphenyl tolerance concentration experiment, biphenyls (dissolved in acetone) were added to sterile flasks and the acetone evaporated, then 20 mL cell suspensions were added to the flasks. The initial concentrations of biphenyl were 50, 100, 500, 1000, 2000 and 3000 mg/L. Biphenyl is poorly soluble in water and therefore the indicated concentrations in the test are only nominal. Residual biphenyl was collected with an equal volume of ethyl acetate after incubation for 84 h at 30 °C and 180 rpm. The organic phases were diluted appropriately, dried with anhydrous sodium sulphate and measured using gas chromatography-mass spectrometry (GC–MS) as described by Hu et al. (2015).
To investigate the biphenyl degradation curve of SS1 and SS2, biphenyls (dissolved in acetone) were added to glass tubes and the acetone evaporated, then 5 mL cell suspensions were inoculated into the glass tubes at a final biphenyl concentration of 1300 mg/L. The glass tubes were cultured at 30 °C and 180 rpm. Residual biphenyl was extracted with an equal volume of ethyl acetate at 12 h intervals within 108 h. The growth of SS1 and SS2 were monitored by measuring the OD600 at the time of extraction. Concentrations of biphenyl were determined as previously described.
PCB and CBAs degradation experiments
Before 5 mL cell suspensions (OD600 1.0) of SS1 and SS2 were separately inoculated into sterile glass tubes (control groups without SS1 and SS2), PCB congeners (dissolved in hexane) were added to these tubes. The initial concentration of 2-CB, 3-CB, 2,4′-DCB, 3,3′-DCB, 2,4,4′-TrCB, 2,4′,5-TrCB, 2,2′,3,3′-TeCB, 2,2′,4,5′-TeCB, 2,3′,4′,5-TeCB, 2,2′,4,4′,5,5′-HCB were 5.57, 6.41, 7.18, 7.93, 7.43, 9.36, 16.73, 4.53, 2.44, 5.51 mg/L, respectively. This treatment method was also adopted to CBAs degradation experiment. Residual PCBs and CBAs were extracted after incubation for 72 h at 30 °C and 180 rpm and analyzed by gas chromatography (GC) as described by Hu et al. (2015).
The degradation efficiency was calculated by the following equation: degradation efficiency (%) = (1 − residual concentration/initial concentration) × 100.
Chemotaxis assays
The chemotactic behaviour was studied by modified drop assays as described by Pandey et al. (2002). Cells were grown in LB medium at the logarithmic phase, washed twice, resuspended in drop assay medium (OD600 0.6) and poured into petri dishes. A small amount of crystal attractant was placed in the center of the petri dish to create a concentration gradient. For the biphenyl assay, cells ware grown in M9 minimal salts medium with 1 mM biphenyl. Chemotactic rings were assessed for TCA-cycle intermediates (succinate, malate, α-ketoglutarate, citrate, fumarate, oxaloacetate, aconitate), benzoate and biphenyl after incubating 24 h at 30 °C.
All experiments were performed in triplicate and the averages were calculated. SPSS version 16.0 software was used for data analysis. One-way analysis of variance was used for statistical comparisons. The significance level was set to P < 0.05.
Results
Identification of R. ruber SS1 and R. pyridinivorans SS2
Two biphenyl-degrading strains, SS1 and SS2, were isolated from the soil sample. Both colonies were observed to be round and convex with a smooth, opaque surface on LB agar plates. Colonies were orange red and flesh pink for SS1 and SS2, respectively. Observation by scanning electron microscopy revealed that both cells were short rods or cocci (Fig. 1). Both strains were aerobic gram-positive bacteria with no spore formed. Based on the 16S rRNA sequence analysis, morphological features, physiological and biochemical characteristics (Tables S1 and S2), strains SS1 and SS2 were identified as R. ruber (Accession Number: KY082044) and R. pyridinivorans (accession number: KY082045), respectively. The 16S rRNA gene sequence of SS1 and SS2 were 99% similar to R. ruber DSM 43338T (Accession Number: X80625) and 99% similar to R. pyridinivorans PDB9T (accession number: AF173005), respectively (Fig. 2).
Metabolism of biphenyl
Tolerance concentration of biphenyl
To study the biphenyl tolerance ability of SS1 and SS2, a series of biphenyl concentrations from 100 to 3000 mg/L were adopted. The results are showed in Fig. 3. With an increase in biphenyl concentration from 100 to 1000 mg/L, almost all biphenyls were removed by SS1 within 84 h. At an initial concentration of 2000, 1840 mg/L biphenyl was degraded. Nevertheless, when the biphenyl concentration reached 3000 mg/L, only 269 mg/L biphenyl was removed (Fig. 3a). Over 99% of the biphenyl was degraded by SS2 when the biphenyl concentration increased from 100 to 3000 mg/L (Fig. 3b). These results indicated that SS1 could tolerate 2000 mg/L biphenyl, and SS2 could tolerate more than 3000 mg/L biphenyl. The biphenyl tolerance of SS2 appears to be better than that of SS1.
Degradation characteristics of biphenyl
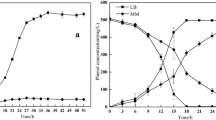
Figure 4 shows the degradation of biphenyl and cell growth at 1300 mg/L biphenyl. SS2 exhibited higher biphenyl degradation ability and cell growth than SS1 did. The degradation efficiency of SS2 reached 48.5% within 12 h, but only 14.5% for SS1. Cell growth for both strains reached a plateau between 48 and 84 h. The general cell growth trend was consistent with the biphenyl degradation rate. The biphenyl degradation efficiencies were 83.2 and 71.8% for SS1 and SS2 after 84 h, respectively.
Degradation of PCBs, benzoate and CBAs
The degradation of different chlorinated PCB congeners by SS1 and SS2 were investigated. Both strains exhibited significant degradation of mono-, di-, and tri-PCBs, while the degradation percentages of tetra- and hex-PCBs were insignificant (Table 1). Table 2 shows the results of CBAs and benzoate degradation by SS1 and SS2. Benzoate was almost completely degraded by both strains after 72 h of incubation. The degradation efficiencies of 3-CBA by SS1 and SS2 were 99.4 and 50.1%, respectively. However, the degradation of 2-CBA and 4-CBA was not remarkable.
Chemotactic responses of SS1 and SS2 towards TCA-cycle intermediates, biphenyl, benzoate, CBAs and PCBs
Strains SS1 and SS2 were tested for chemotaxis toward TCA-cycle intermediates, biphenyl, benzoate, CBAs and PCBs by drop assay. For these qualitative assays, the solid attractant in the middle of the drop plate created a concentration gradient. The accumulation of bacteria around the crystals is measured to assess chemotaxis. Drop assays showed that strain SS1 and SS2 shown chemotaxis toward seven tested TCA-cycle intermediates, biphenyl, benzoate and 2-CBA (Table 3). Cells of SS1 and SS2 grew in LB medium did not respond to biphenyl after incubation for 24 h. However, the biphenyl-grown cells formed a denser ring of turbidity in the middle of the plate around the biphenyl (Fig. 5). These results indicated an induced chemotaxis toward biphenyl. Although SS1 and SS2 can degrade 3-CBA, 2-CB, and 3-CB, they did not show chemotaxis towards these compounds.
Discussion
It has been reported that many species belonging to the Rhodococcus genus have the ability to degrade PCBs, such as Rhodococcus jostii RHA1 (Atago et al. 2016), Rhodococcus sp. M5 (Labbe et al. 1997), R. erythropolis TA421 (Chung et al. 1994), R. globerulus P6 (McKay et al. 2003), Rhodococcus sp. R04 (Yang et al. 2004), Rhodococcus rhodochrous K37 (Taguchi et al. 2004), Rhodococcus sp. TA431 and Rhodococcus sp. HA99 (Taguchi er al. 2007). In this study, Rhodococcus SS1 and SS2 isolated from PCB-contaminated soil also had the capability of depleting biphenyl and PCBs.
Previous studies had shown that Mycobacterium sp. PYR-1 was able to degrade over 98% of 80 mg/L biphenyl within 72 h (Moody et al. 2002), Dyella ginsengisoli LA-4 could degrade 95% of 100 mg/L biphenyl within 36 h (Li et al. 2009), Achromobacter sp. BP3 could completely degrade 50 mg/L biphenyl within 28 h (Hong et al. 2009), and HC3 could grow on 1000 mg/L biphenyl (Hu et al. 2015). In this study, both strains could grow on 3000 mg/L biphenyl and degrade biphenyl more rapidly, transforming almost 100% of 1000 mg/L biphenyl within 84 h (Fig. 3). SS1 and SS2 exhibited higher biphenyl tolerance and degradation abilities than those of the previously reported strains.
In the case of low-PCB congeners, the degradation activities of SS1 and SS2 were significant, as was previously reported for other aerobic degradation bacteria (Hu et al. 2015; Sakai et al. 2005), while the degradation efficiency of tetra- and hex-PCBs were insignificant. Previous studies shown that the number and the position of chlorine substitution greatly affect the biodegradation of PCBs. Degradation efficiency decreased as chlorine substitution increased, and PCB congeners containing more than four chlorines were not easy to be degraded. Furthermore, PCB congeners with double ortho substitutions are also poorly degraded (Furukawa et al. 1979; Bedard and Haberl 1990). Based on the existing results, strains SS1 and SS2 were relatively sensitive to 2- and 3-chlorophenyl groups. And SS1 was insensitive to 2,4-chlorophenyl. This may be influenced by the substrate preference of enzyme. Burkholderia xenovorans LB400 have been proved to exhibit exceptional abilities to attack a wide range of PCB congeners, including hexachlorobiphenyls (Bopp 1986). In this research, the substrate range of SS1 and SS2 are narrow compared to LB400, but SS1 and SS2 have an excellent capacity to degrade biphenyl and low chlorinated biphenyl. Thus, it is significant to investigate the biodegradation genes of SS1 and SS2 for further research.
Furthermore, compounds with two or more halogens per molecule are generally more recalcitrant, they are usually biodegradable via co-metabolic transformation processes (Kang 2014; Zhao et al. 2010). It has been reported that strains Ralstonia sp. SA-5 and Pseudomonas sp. SA-6 were able to degrade tetrachlorobiphenyls in the presence of biphenyl supplementation as the growth substrate (Adebusoye et al. 2008b). It is necessary to investigate whether SS1 and SS2 can transform PCBs that contain more than three chlorines in the presence of biphenyl as co-substrate for follow-up research.
During biodegradation of biphenyl/PCBs, benzoate and different CBAs intermediates are generated (Potrawfke et al. 1998). It has been shown that some of these compounds are toxic to the biodegrading bacteria and inhibit the growth of microorganisms (Camara et al. 2004; Parnell et al. 2006). Therefore, it was necessary to investigate whether SS1 and SS2 were able to degrade the benzoate and different CBAs intermediates. R. erythropolis strain S-7 and Rhodococcus sp. R04 have been reported to utilize chlorobenzoate (Yun et al. 2009; Zhang et al. 2009). Similar to these strains, both SS1 and SS2 have the ability to metabolize benzoate, 3-CBA and 4-CBA (Table 2), suggesting that SS1 and SS2 might have the ability to mineralize biphenyl and PCBs. Thus, SS1 and SS2 may play a more important role in the degradation of biphenyl and PCBs.
Although microbial bioremediation can be environmentally friendly and cost-effective for restoring PCB-contaminated environments, the limited bioavailability of the pollutants could restrict the degradation rate. The current research has shown that bacterial chemotaxis could increase pollutant bioavailability, which in turn would have improve the potential for bioremediation (Law and Aitken 2003; Marx and Aitken 2000; Paul et al. 2006). Being non-flagellated, SS1 and SS2 are not expected to be motile. However, our data indicate that SS1 and SS2 showed chemotaxis toward TCA-cycle intermediates, biphenyl, benzoate and 2-CBA. The chemotaxis of Rhodococcus spp. toward chemical compounds has been previously reported by other researchers (Toussaint et al. 2012; Wu et al. 2003). These flagella-independent bacteria may overcome the restricted bioavailability of pollutants by gliding, and they are possibly motile under certain conditions, as has been reported for other bacteria that lack flagella (Jarrell and McBride 2008). Thus, it is necessary to quantitatively analyze the chemotaxis of SS1 and SS2 in semi-solid medium, other heterogenous medium and in soil in future research. For bioremediation application, it is important to evaluate whether bacteria actually migrate through the heterogenous soil medium towards a gradient of a particular chemoattractant. Therefore, further research should be conducted to investigate whether strains SS1 and SS2 could actually move in soil in a way that can increase the biodegradation efficiency. And much work is needed to uncover the potential integration of chemotaxis with degradation and to identify chemoreceptors and biodegradation genes.
In conclusion, R. ruber SS1 and R. pyridinivorans SS2, which have high biphenyl tolerances and degradation efficiency, were isolated from a PCB-contaminated soil sample. Strains SS1 and SS2 could tolerate up to 2000 and 3000 mg/L biphenyl, respectively, and they were able to degrade benzoate, 3-CBA, 4-CBA and PCBs with three or fewer chlorine atoms. SS1 and SS2 shown chemotaxis toward TCA-cycle intermediates, biphenyl, benzoate and 2-CBA. The results suggest that both bacteria might be good candidates for restoring biphenyl/PCBs-contaminated environments.
References
Abraham WR, Nogales B, Golyshin PN, Pieper DH, Timmis KN (2002) Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr Opin Microbiol 5:246–253
Adebusoye SA, Picardal FW, Ilori MO, Amund OO (2008a) Influence of chlorobenzoic acids on the growth and degradation potentials of PCB-degrading microorganisms. World J Microb Biotechnol 24:1203–1208
Adebusoye SA, Ilori MO, Picardal FW, Amund OO (2008b) Cometabolic degradation of polychlorinated biphenyls (PCBs) by axenic cultures of Ralstonia sp. strain SA-5 and Pseudomonas sp. strain SA-6 obtained from Nigerian contaminated soils. World J Microb Biotechnol 24:61–68
Aken BV, Correa PA, Schnoor JL (2010) Phytoremediation of polychlorinated biphenyls: new trends and promises. Environ Sci Technol 44:2767–2776
Aoki Y (2001) Polychlorinated biphenyls, polychloronated dibenzo-p-dioxins, and polychlorinated dibenzofurans as endocrine disrupters: what we have learned from Yusho disease. Environ Res 86:2–11
Atago Y, Shimodaira J, Araki N, Bin Othman N, Zakaria Z, Fukuda M, Futami J, Hara H (2016) Identification of novel extracellular protein for PCB/biphenyl metabolism in Rhodococcus jostii RHA1. Biosci Biotechnol Biochem 80:1012–1019
Bartels F, Backhaus S, Moore E, Timmis KN, Hofer B (1999) Occurrence and expression of glutathiane-S-transferase-encoding bphK genes in Burkholderia sp. strain LB400 and other biphenyl-utilizing bacteria. Microbiol-SGM 145:2821–2834
Bedard DL, Haberl ML (1990) Influence of chlorine substitution pattern on the degradation of polychlorinated biphenyls by eight bacterial strains. Microb Ecol 20:87–102
Bopp LH (1986) Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J Ind Microbiol Biotechnol 47:247–254
Camara B, Herrera C, Gonzalez M, Couve E, Hofer B, Seeger M (2004) From PCBs to highly toxic metabolites by the biphenyl pathway. Environ Microbiol 6:842–850
Chung SY, Maeda M, Song E, Horikoshij K, Kudo T (1994) A gram-positive polychlorinated biphenyl-degrading bacterium, Rhodococcus erythropolis strain TA421, isolated from a termite ecosystem. Biosci Biotechnol Biochem 58:2111–2113
Eklund T (1985) Inhibition of microbial growth at different pH levels by benzoic and propionic acids and esters of p-hydroxybenzoic acid. Int J Food Microbiol 2:159–167
Faroon O, Jones D, Rosa CD (2000) Effects of polychlorinated biphenyls on the nervous system. Toxicol Ind Health 16:305–333
Furukawa K, Tomizuka N, Kamibayashi A (1979) Effect of chlorine substitution on the bacterial metabolism of various polychlorinated biphenyls. Appl Environ Microbiol 38:301–310
Gordillo F, Chavez FP, Jerez CA (2007) Motility and chemotaxis of Pseudomonas sp. B4 towards polychlorobiphenyls and chlorobenzoates. FEMS Microbiol Ecol 60:322–328
Harwood CS (1989) A methyl-accepting protein is involved in benzoate taxis in Pseudomonas putida. J Bacteriol 171:4603–4608
Harwood CS, Parales RE, Dispensa M (1990) Chemotaxis of Pseudomonas putida toward chlorinated benzoates. Appl Environ Microbiol 56:1501–1503
Heuer H, Krsek M, Baker P, Smalla K, Wellington E (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol 63:3233–3241
Hong Q, Dong XJ, He LJ, Jiang X, Li SP (2009) Isolation of a biphenyl-degrading bacterium, Achromobacter sp. BP3, and cloning of the bph gene cluster. Int Biodeterior Biodegrad 63:365–370
Hu JX, Qian MR, Zhang Q, Cui JL, Yu CN, Su XM, Shen CF, Hashmi MZ, Shi JY (2015) Sphingobium fuliginis HC3: a novel and robust isolated biphenyl- and polychlorinated biphenyls-degrading bacterium without dead-end intermediates accumulation. PLoS ONE 10:e122740. doi:10.1371/journal.pone.0122740
Jarrell KF, McBride MJ (2008) The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476
Kang JW (2014) Removing environmental organic pollutants with bioremediation and phytoremediation. Biotechnol Lett 36:1129–1139
Kohler HP, Kohler-Staub D, Focht DD (1988) Co-metabolism of polychlorinated biphenyls: enhanced transformation of Aroclor 1254 by growing bacterial cells. Appl Environ Microbiol 54:1940–1945
Krell T, Lacal J, Reyes-Darias JA, Jimenez-Sanchez C, Sungthong R, Ortega-Calvo JJ (2013) Bioavailability of pollutants and chemotaxis. Curr Opin Microbiol 24:451–456
Labbe D, Garnon J, Lau P (1997) Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J Bacteriol 179:2772–2776
Law A, Aitken MD (2003) Bacterial chemotaxis to naphthalene desorbing from a nonaqueous liquid. Appl Environ Microbiol 69:5968–5973
Leigh MB, Prouzova P, Mackova M, Macek T, Nagle DP, Fletcher JS (2006) Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol 72:2331–2342
Li A, Qu YY, Zhou JT, Gou M (2009) Isolation and characteristics of a novel biphenyl-degrading bacterial strain, Dyella ginsengisoli LA-4. J Environ Sci (China) 21:211–217
Marx RB, Aitken MD (1999) Quantification of chemotaxis to naphthalene by Pseudomonas putida G7. Appl Environ Microbiol 65:2847–2852
Marx RB, Aitken MD (2000) Bacterial chemotaxis enhances naphthalene degradation in a heterogeneous aqueous system. Environ Sci Technol 34:3379–3383
McKay DB, Prucha M, Reineke W, Timmis KN, Pieper DH (2003) Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6. J Bacteriol 185:2944–2951
Moody JD, Doerge DR, Freeman JP (2002) Degradation of biphenyl by Mycobacterium sp. strain PYR-1. Appl Microbiol Biotechnol 58:364–369
Ohtsubo Y, Kudo T, Tsuda M, Nagata Y (2004) Strategies for bioremediation of polychlorinated biphenyls. Appl Microbiol Biotechnol 65:250–258
Pandey G, Jain RK (2002) Bacterial chemotaxis toward environmental pollutants: role in bioremediation. Appl Environ Microbiol 68:5789–5795
Pandey G, Chauhan A, Samanta SK, Jain RK (2002) Chemotaxis of a Ralstonia sp. SJ98 toward co-metabolizable nitroaromatic compounds. Biochem Biophys Res Commun 299:404–409
Parales RE, Harwood CS (2002) Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr Opin Microbiol 5:266–273
Parales RE, Luu RA, Hughes JG, Ditty JL (2015) Bacterial chemotaxis to xenobiotic chemicals and naturally-occurring analogs. Curr Opin Biotechnol 33:318–326
Parnell JJ, Park J, Denef V, Tsoi T, Hashsham S, Quensen J, Tiedje JM (2006) Coping with polychlorinated biphenyl (PCB) toxicity: physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl Environ Microbiol 72:6607–6614
Paul D, Singh R, Jain RK (2006) Chemotaxis of Ralstonia sp. SJ98 towards p-nitrophenol in soil. Environ Microbiol 8:1797–1804
Pieper DH (2005) Aerobic degradation of polychlorinated biphenyls. Appl Microbiol Biotechnol 67:170–191
Pieper DH, Seeger M (2008) Bacterial metabolism of polychlorinated biphenyls. J Mol Microbiol Biotechnol 15:121–138
Potrawfke T, Lohnert TH, Timmis KN, Wittich RM (1998) Mineralization of low-chlorinated biphenyls by Burkholderia sp. strain LB400 and by a two membered consortium upon directed interspecies transfer of chlorocatechol pathway genes. Appl Microbiol Biotechnol 50:440–446
Sakai M, Ezaki S, Suzuki N, Kurane R (2005) Isolation and characterization of a novel polychlorinated biphenyl-degrading bacterium, Paenibacillus sp. KBC101. Appl Microbiol Biotechnol 68:111–116
Stratford J, Wright MA, Reineke W, Mokross H, Havel J, Knowles CJ, Robinson GK (1996) Influence of chlorobenzoates on the utilization of chlorobiphenyls and chlorobenzoates mixtures by chlorobiphenyl/chlorobenzoate-mineralising hybrid bacterial strains. Arch Microbiol 165:213–218
Taguchi K, Motoyama M, Kudo T (2004) Multiplicity of 2,3-dihydroxybiphenyl dioxygenase genes in the Gram-positive polychlorinated biphenyl degrading bacterium Rhodococcus rhodochrous K37. Biosci Biotechnol Biochem 68:787–795
Taguchi K, Motoyama M, Iida T, Kudo T (2007) Polychlorinated biphenyl/biphenyl degrading gene clusters in Rhodococcus sp. K37, HA99, and TA431 are different from well-known bph gene clusters of Rhodococci. Biosci Biotechnol Biochem 71:1136–1144
Tanase A, Vassu T, Csutak O, Pelinescu D, Robertina I, Stoica I (2012) Phylogenetic analysis of oil polluted soil microbial strains. Rom Biotechnol Lett 17:7093–7103
Toussaint JP, Pham TTM, Barriault D, Sylvestre M (2012) Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl Microbiol Biot 95:1589–1603
Tremaroli V, Suzzi CV, Fedi S, Ceri H, Zannoni D, Turner RJ (2010) Tolerance of Pseudomonas pseudoalcaligenes KF707 to metals, polychlorobiphenyls and chlorobenzoates: effects on chemotaxis-, biofilm- and planktonic-grown cells. FEMS Microbiol Ecol 74:291–301
Vasilyeva GK, Strijakova ER (2007) Bioremediation of soils and sediments contaminated by polychlorinated biphenyls. Microbiology 76:639–653
Wu G, Feng Y, Boyd A (2003) Characterization of bacteria capable of degrading soil-sorbed biphenyl. Bull Environ Contam Toxicol 71:768–775
Yang XQ, Erickson LE, Fan LT (1995) A study of the dissolution rate-limited bioremediation of soils contaminated by residual hydrocarbons. J Hazard Mater 41:299–313
Yang X, Sun Y, Qian S (2004) Biodegradation of seven polychlorinated biphenyls by a newly isolated aerobic bacterium (Rhodococcus sp. R04). J Ind Microbiol Biotechnol 31:415–420
Yun Q, Lin Z, Xin T (2009) Cometabolism and immobilized degradation of monochlorobenzoate by Rhodococcus erythropolis. Afr J Microbiol Res 3:482–486
Zhang GQ, Yang XQ, Xie FH, Chao YP, Qian SJ (2009) Cometabolic degradation of mono-chloro benzoic acids by Rhodococcus sp. R04 grown on organic carbon sources. World J Microb Biotechnol 25:1169–1174
Zhao HP, Schmidt KR, Tiehm A (2010) Inhibition of aerobic metabolic cis-1,2-di-chloroethene biodegradation by other chloroethenes. Water Res 44:2276–2282
Acknowledgements
We gratefully acknowledge the financial support provided by the Fundamental Research Funds for the Central Universities (2016QNA6009) and the Provincial Public Technology and Applied Research Projects by Science and Technology Department of Zhejiang Province (2014C33020, 2017C33020).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, H., Hu, J., Xu, K. et al. Biodegradation and chemotaxis of polychlorinated biphenyls, biphenyls, and their metabolites by Rhodococcus spp.. Biodegradation 29, 1–10 (2018). https://doi.org/10.1007/s10532-017-9809-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-017-9809-6