Abstract
Rhodococcus erythropolis U23A is a polychlorinated biphenyl (PCB)-degrading bacterium isolated from the rhizosphere of plants grown on a PCB-contaminated soil. Strain U23A bphA exhibited 99% identity with bphA1 of Rhodococcus globerulus P6. We grew Arabidopsis thaliana in a hydroponic axenic system, collected, and concentrated the plant secondary metabolite-containing root exudates. Strain U23A exhibited a chemotactic response toward these root exudates. In a root colonizing assay, the number of cells of strain U23A associated to the plant roots (5.7 × 105 CFU g−1) was greater than the number remaining in the surrounding sand (4.5 × 104 CFU g−1). Furthermore, the exudates could support the growth of strain U23A. In a resting cell suspension assay, cells grown in a minimal medium containing Arabidopsis root exudates as sole growth substrate were able to metabolize 2,3,4′- and 2,3′,4-trichlorobiphenyl. However, no significant degradation of any of congeners was observed for control cells grown on Luria–Bertani medium. Although strain U23A was unable to grow on any of the flavonoids identified in root exudates, biphenyl-induced cells metabolized flavanone, one of the major root exudate components. In addition, when used as co-substrate with sodium acetate, flavanone was as efficient as biphenyl to induce the biphenyl catabolic pathway of strain U23A. Together, these data provide supporting evidence that some rhodococci can live in soil in close association with plant roots and that root exudates can support their growth and trigger their PCB-degrading ability. This suggests that, like the flagellated Gram-negative bacteria, non-flagellated rhodococci may also play a key role in the degradation of persistent pollutants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Although the use of polychlorinated biphenyls (PCBs) has been banned in many countries since the late 1970s (Vasilyeva and Strijakova 2007; Van Aken et al. 2010), they still persist in the environment due to their chemical and physical properties for which they were appreciated in the first place. PCBs mainly accumulate in soils, sediments, but also in animal and human adipose tissues (Borja et al. 2005), which provided the incentive to find ways of eradicating these compounds from the environment. Because of the relatively high cost associated with land filling or incineration, biological degradation of PCBs has received increasing interest over the years. Many studies have now shown that a wide range of bacteria that can utilize or co-metabolize PCBs through the biphenyl catabolic pathway are promising tools to remediate these compounds (see review by Vasilyeva and Strijakova 2007).
Generally, the bacteria capable of aerobically degrading PCB congeners do so via the 2,3-dioxygenase pathway (Abramowicz 1995). Although analogue enrichment with biphenyl can increase bacterial PCB degradation rates (Fava and Bertin 1999; Luo et al. 2007), the use of biphenyl to enrich PCB-contaminated soils is not a viable idea. Because of the similarity between xenobiotics (e.g., PCBs) and natural occurring compounds in the soil (e.g., plant secondary metabolites—PSMs), it has been suggested that PSMs could be suitable candidates to be exploited in order to stimulate bacterial degradation of PCBs and other persistent organic pollutants such as polycyclic aromatic hydrocarbons (Miya and Firestone 2001; Singer et al. 2003). This is supported by several studies showing that PSMs such as flavonoids and terpenes can support the growth of PCB-degrading bacteria and trigger their PCB-degrading abilities (Donnelly et al. 1994; Hernandez et al. 1997; Leigh et al. 2002, 2006; Narasimhan et al. 2003). For example, PCB degradation was stimulated in soils containing natural sources of terpenes (e.g., orange peels, eucalyptus leaves, pine needles, spearmint leaves, and ivy leaves) (Gilbert and Crowley 1997; Hernandez et al. 1997), or in liquid culture, when biphenyl-degrading bacteria were grown in the presence of flavonoids (Donnelly et al. 1994). Furthermore, PCB depletion was significantly higher in soil sown with an Arabidopsis mutant overproducing root PSMs and inoculated with the PCB-degrading and flavonoid-degrading rhizobacteria Pseudomonas putida PML2, than in soil sown with the wild-type plants (Narasimhan et al. 2003). These experiments provide evidence that plant chemicals can trigger bacterial and notably rhizobacterial PCB degradation in soil.
These observations encourage the development of processes exploiting plants and their associated rhizobacteria to degrade PCBs (Sylvestre and Toussaint 2011). However, the mechanism by which PSMs trigger the PCB-degrading ability of rhizobacteria is still unclear since no study has yet demonstrated that these chemicals can act as non-specific inducers of the bacterial biphenyl degradation enzymes when they are used as growth substrate (Shaw et al. 2006). It is also unclear whether these chemicals act at low concentrations, as signals to induce PCB degradation (Singer et al. 2003), or if they act as substrates in a co-metabolism process as suggested by Hernandez et al (1997). Answers to these questions will help in designing new enrichment strategies based on plant–microbe interactions to increase PCB degradation in the rhizosphere.
On the other hand, although Gram-negative bacteria are traditionally believed to play a major role in pollutant removal in soil and in the rhizosphere, many rhodococcal bacteria capable of degrading PCBs have been described (Chebrou et al. 1999; Iwasaki et al. 2006; Kim et al. 2003; McKay et al. 2003). In addition, rhodococci were recently found to represent the dominant population in the rhizosphere of plants grown in a PCB-contaminated site (Leigh et al. 2006). However, a very limited number of studies examining plant–rhodococcal bacteria interactions and their impact on PCB degradation have been reported (Francis et al. 2010).
In this work, we have isolated a PCB-degrading rhodococcal strain. We provide evidence that this strain is able to colonize plant roots and exhibits a chemotactic response toward root exudates. We have examined the ability of Arabidopsis thaliana PSMs to promote growth and PCB-degrading ability of this bacterium, and we have assessed the ability of flavanone, one of the major root exudate components to induce its biphenyl catabolic pathway. Data provide more insights into how plant flavonoids might interact with rhodoccocal rhizobacteria to trigger their PCB-degrading ability.
Materials and methods
Bacterial strains, culture media, and chemicals
The bacterial strains used in this study were isolate U23A identified based on 16S rRNA gene sequencing as Rhodococcus erythropolis (see below) and Pseudomonas fluorescens F113 (Brazil et al. 1995) (obtained from Prof. David Dowling—Institute of Technology, Carlow, Ireland). The culture media used were Luria–Bertani (LB) broth (Sambrook et al. 1989), basal medium M9 (Sambrook et al. 1989), or minimal mineral medium no. 30 (MM30) (Sylvestre 1980) amended with various sources of carbon depending on the experiment (see below). All strains were maintained on LB agar medium at 28°C for experimental purposes. The flavonoids and other chemical standards used in this study were obtained from Sigma-Aldrich and the PCB congeners were purchased from UltraScientific.
R. erythropolis U23A was isolated by enrichment on biphenyl from the rhizosphere of a PCB-contaminated microcosm set in the laboratory. To prepare the microcosm, a beaker (250 ml) was filled to approximately 10-cm height with a PCB-contaminated soil obtained from Netolice, Czech Republic, and this soil was covered with the 2-cm top portion of a vegetated soil from St-Hippolyte, QC, Canada which contained a mixture of monocotyledon plants and their roots. The microcosm was kept under laboratory conditions for 6 months at 20°C under a fluorescent light with a 16/8-h day/night cycle so that the plant could grow. Then, the rhizosphere (i.e., the remaining soil adhering to plant roots after vigorous shaking) of this mixture of monocotyledon plants was collected. One gram of freshly collected rhizosphere material was then transferred in 10 ml sterile NaCl (0.85% w/v). The suspension was vigorously shaken by vortexing, diluted serially, and inoculated onto solid MM30 agar plates exposed to biphenyl vapor as sole growth substrate. The cultures were grown at 25°C for 48 to 72 h. The colonies able to degrade chlorobiphenyls were identified using the 4-chlorobiphenyl sprayed-plate assay described previously (Sylvestre 1980). From this screening step, seven bacterial strains were retained based on colony morphology (size, color, and colony surface roughness) and were further purified by streaking on MM30 agar plates exposed to biphenyl vapor. Based on their ability to use root exudates as sole growth substrate (see below), three of these isolates U22, U23A, and U24 were selected. Because of their inability to grow on root exudates, the other isolates were not considered for this study and discarded.
Identification of strains U22, U23A, and U24 and amplification of bphA gene
Strains U22, U23A, and U24 were identified based on their 16S rRNA gene sequence, and strain U23A was further characterized on the basis of morphological examination and biochemical tests. Genomic DNA was isolated using the QIAGEN QIAamp DNA Mini kit and the protocol suggested by the manufacturer. The 16S rRNA gene was amplified from total genomic DNA using primers pA (AGAGTTTGATCCTGGCTCAG) and pH (AAGGAGGTGATCCAGCCGCA) (Edwards et al. 1989). Genomic DNA was also probed for the presence of bphA gene using a previously described set of three pairs of degenerated primers that amplify the C-terminal portion of bphA gene (Vézina et al. 2007) and specific primers designed to amplify the complete bphA genes from Burkholderia xenovorans LB400 (Hurtubise et al. 1998), from Pandoraea pnomenusa B-356 (Hurtubise et al. 1996) and from Rhodococcus globerulus P6 (Chebrou et al. 1999). The PCR conditions used to amplify the genes were as recommended by QIAGEN for the HotStart High Fidelity polymerase kit (QIAGEN), with the following modifications of the program: 15 min at 95°C to activate the polymerase, then for 30 cycles, 94°C for 1 min, 56°C for 1 min and 72°C for 2.5 min, and 1 cycle at 72°C for 10 min. The amplicons from the PCR were sequenced at the Génome Québec DNA Sequencing Center (Montreal, Quebec, Canada). BLASTn from NCBI data bank was used to analyze the 16S rRNA gene sequence and BLASTp for the deduced sequence of BphA.
Axenic culture for the production of root exudates
A. thaliana was grown in a hydroponic axenic culture system containing 100 ml of a tenth-strength Hoagland’s solution (Hoagland’s No. 2 Basal Salt Mixture from Sigma-Aldrich; see Fig. 1). The system in which the plants were grown was made out of a pipette tip support and 0.5 ml Eppendorf tubes without lids. Holes were punched at the bottom of the Eppendorf tubes in order to let A. thaliana roots grow through. The tubes were then fitted in a pipette tip support rack, which was sterilized by autoclaving for 1 h at 121°C, and put in sterile transparent plastic Magenta containers. Each Eppendorf tube was filled with a tenth-strength Hoagland’s solution containing 1% agar. Once the medium was solidified, sterilized seeds of A. thaliana were deposited on the top of the solidified medium. The seeds were sterilized by soaking into a 2% hypochloric solution for 5 min and rinsing with deionized sterile water for 5 min. There was one seed per Eppendorf tube, and the whole system comprised approx. 30 tubes (plants). The plants were grown for 18 weeks under a fluorescent light with a 16/8-h day/night cycle. Root exudates were collected under sterile conditions periodically starting at the eighth week after seed germination. Preliminary experiments allowed us to determine that out of all the exudates harvested and tested, the ones collected after 16 weeks of A. thaliana growth were the ones that allowed optimal bacterial growth. Hence, those exudates were used for all of the experiments described thereafter.
Axenic culture system for the growth of A. thaliana and the collection of root exudates. Sterile plastic Magenta containing approx. 30 Eppendorf tubes punched with holes at the bottom that were mounted on a pipette rack and filled with a tenth strength Hoagland’s solution containing 1% agar. Sterilized seeds of A. thaliana were grown for several weeks in 100 ml of a tenth strength Hoagland solution, under a fluorescent lamp with a 16/8-h day/night cycle
Flavonoid detection and identification from root exudates by HPLC, LC–MS, and GC–MS
For root exudate fractionation, 10 ml of root exudates from A. thaliana were collected and concentrated approx. 15 times using a Büchi roto-evaporator prior injection to HPLC. The HPLC apparatus was an Agilent 1100 Series liquid chromatography system comprising a UV lamp detector. The analysis was performed on a reverse-phase Eclipse XBD-C8 column (Agilent 4·6 × 150 mm, 5-μm pore size). The column was eluted with a non-linear gradient starting from 80% A (HPLC grade water with 0.085% ortho-phosphoric acid) and 20% B (acetonitrile) to 50% B at 12 min, 70% B at 17 min, 80% at 21 min, 100% B at 25 min, then back to 80% A at 29 min. Fifty microliters of the root exudates was injected and the column was eluted at a flow rate of 1 ml min−1 and detected at 280 nm.
For further detection of the compounds, root exudates were hydrolyzed for 1 h at 90°C with 1 M HCl to hydrolyze the conjugated plant metabolites. The hydrolysate was then neutralized (pH = 7–8) before extracting the metabolites twice with ethyl acetate. The extract was dried over ammonium sulfate and evaporated to dryness under N2. This hydrolyzed extract was analyzed by liquid chromatography–mass spectrometry (LC–MS) and by gas chromatography–mass spectrometry (GC–MS). For LC–MS analysis, the residues were dissolved in a solution containing 30% methanol and 1% acetic acid in HPLC grade water. Fifty microliters was injected into an Agilent HP111/Quatro Micromass LC–MS. The LC analysis was performed on a reverse-phase Eclipse XBD-C8 column (Agilent 4·6 × 150 mm, 5-μm pore size), and the column was eluted with a gradient consisting of solvent A, water and solvent B, 1% acetic acid in acetonitrile (0–1 min 0% solvent B; 1–5 min 50% B; 5–10 min 50% B; 10–19 min 100% B; 19–24 min 100% B; and 24–29 min 0% B). The mass spectrometry detection was done using a positive mode (ESP+) with a capillary value of 3.50, cone value of 28, and extractor value of 5.
For GC–MS analysis, the dried residues from the ethyl acetate extract of the hydrolyzed exudates were dissolved in N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) (Supelco, Sigma-Aldrich), and the solution was heated for 30 min at 70°C (Massé et al. 1989). The trimethysilyl (TMS) derivatized extract was then injected in a Hewlett Packard HP6890 gas chromatograph equipped with a SPB-5 capillary column (30 m × 0.2 mm i.d.) (Sigma-Aldrich Biotechnology LP) and a HP5973 mass selective detector. The initial oven temperature (100°C) was held for 2 min and programmed at 15°C per min to 200°C and at 10°C per min to 300°C. The injector and the detector were maintained at 250°C and 300°C, respectively. The mass spectrometer was operated in the electron impact ionization mode.
Growth of R. erythropolis U23A and other isolates on root exudates
To test for the ability of isolates to grow on root exudates, the exudates from a 16-week-old A. thaliana hydroponic culture were concentrated 15 times using a Büchi roto-evaporator. The concentrated preparation was filter-sterilized, aliquoted, and frozen at −20°C until used. Cells grown overnight in LB broth were washed twice in 0.85% NaCl and suspended in 0.85% NaCl to an OD600 nm of 0.5. Ten-milliliter screw-capped tubes containing 2 ml of MM30 were supplemented with 1% (v/v) (20 μl) of the 15 times concentrated exudates. Concentrated exudates were used in order to avoid diluting the culture medium, but the final medium contained the equivalent of 15 parts of exudates per 100 parts of culture medium. These media were inoculated with 100 μl of the above cell suspension. The cultures were incubated 28°C at 100 rpm. Cell growth was monitored at OD600 nm.
PCB congener degradation
R. erythropolis U23A was grown overnight at 28°C on MM30 containing 0.1% (w/v) sodium acetate, in LB or in MM30 amended with either 3.4 mM biphenyl or with 1% (v/v) of the 15 times concentrated root exudates described above. Cultures growing on biphenyl were filtered through packed glass wool to remove crystals of the substrate. Cells from overnight cultures were centrifuged, washed with MM30 medium, and suspended in a final volume of 1 ml. The bacterial suspensions were adjusted to OD600 nm = 1 with MM30 and distributed in 2-ml Teflon-lined screw-capped tubes by portions of 200 μl containing 1 μl of a mixture of 18 PCB congeners prepared in ethanol such as to give the final concentration of each congener which is indicated in Table 1. Tubes were incubated overnight at 28°C and shaken at 100 rpm. The cultures were then extracted twice with hexane and the extracts were analyzed by gas chromatography using an electron capture detector to quantify PCB depletion using previously published protocols (Barriault et al. 2004).
Metabolism of flavanone by biphenyl-induced cells of strain U23A
R. erythropolis U23A was grown at 28°C on MM30 containing 3.4 mM biphenyl. Cells from overnight cultures were harvested by centrifugation and washed with M9 medium. The cells were suspended in M9 to an OD600 nm = 5. This cell suspension was proportionally distributed (5 ml) among 50 ml glass tubes covered with Teflon-lined screw caps. Flavanone was added to a final concentration of 200 μM and the cell suspensions were incubated at 28°C. After 30 min, or periodically between 3 and 18 h, the cell suspensions were extracted twice with 5 ml of ethyl acetate, and the solvent phases were combined, dehydrated over ammonium sulfate, and evaporated to dryness under a stream of nitrogen. The residues were dissolved in 250 μl of anhydrous acetone plus 5 μl of a 25-mM solution of n-butylboronate (nBuB). The mixture was incubated at 20°C for 30 min and then evaporated and the residues were dissolved in 50 μl hexane for GC–MS analysis. Alternatively, the extracts were treated with BSTFA as above to generate the TMS derivatives. The GC–MS conditions were identical to those described above.
As a control to facilitate the identification of the metabolites produced from flavanone by U23A cells, we also examined the ability of a reconstituted purified preparation of P. pnomenusa B356 biphenyl dioxygenase (BPDO) to metabolize flavanone. In this case, each of the three purified enzyme components were produced as His-tagged proteins by recombinant Escherichia coli cells and they were purified by affinity chromatography using previously described protocols (L’Abbée et al. 2011). The enzyme assays were performed in a volume of 200 μl in 50 mM morpholinethanesulfonic buffer pH 5.5, at 37°C as described previously (Hurtubise et al. 1996). The metabolites generated after 10 min of incubation were extracted with ethyl acetate and treated with nBuB or BSTFA for GC–MS analysis.
4-Chlorobiphenyl degradation assay
A resting cell assay using 4-chlorobiphenyl as substrate was performed to assess the ability of root exudates to induce the biphenyl catabolic pathway of R. erythropolis U23A. 4-Chlorobiphenyl metabolism was determined by monitoring 4-chlorobenzoate which accumulates as end metabolite of the biphenyl catabolic pathway. Cells of R. erythropolis U23A were grown overnight at 28°C on LB medium or M9 medium amended with biphenyl (3.4 mM) or with 1% (v/v) of the 15 times concentrated root exudates described above. The bacterial cultures were centrifuged at 7,000 rpm for 10 min and washed twice in M9. The suspensions were adjusted to OD600 nm of 1 with M9 and distributed by portions of 200 μl into 1.5 ml Eppendorf tubes. The tubes were vortexed quickly and incubated at 28°C in an Eppendorf Thermomixer 5436 for 5 min. Five microliters of 4-chlorobiphenyl in acetone (final concentration of 1.25 mM) or acetone (negative control) was added and the reaction vials were further incubated for 120 min. The suspensions were then acidified with HCl before extracting the metabolites twice with ethyl acetate. The extracts were dried over ammonium sulfate, evaporated to dryness under N2, and derivatized with BSFTA for GC–MS analysis according to published protocols (Massé et al. 1989).
A similar experiment was set up to examine the ability of flavanone to induce the biphenyl catabolic pathway of strain U23A when it was used as co-substrate. In this case, cells were grown overnight in MM30 containing 30 mM sodium acetate as sole growth substrate or in MM30 containing 30 mM sodium acetate plus variable amounts (1, 0.01, or 0.001 mM) of flavanone or of biphenyl. The latter two concentrations were in the range of those estimated to be present in the culture media containing 15 times concentrated root exudates described above. Cells from each of the overnight grown cultures were then prepared to evaluate their ability to metabolize 4-chlorobiphenyl to 4-chlorobenzoate using the 4-chlorobiphenyl assay described above.
Motility assays
Although R. erythropolis U23A is a Gram-positive bacterium, it can likely be motile by gliding (Jarrell and McBride 2008). Therefore, it was tested for its capacity to exhibit phenotypic traits that are likely to contribute in the root colonization process. Hence, mobility, chemotaxis, cell hydrophobicity, cell adhesion, and root colonization capacities were tested as described below.
Chemotaxis assay
The chemotaxis assay was based on a protocol described by Gordillo et al. (2007). Tryptone 1% (positive control), sterile water (negative control), and a sterile concentrated root exudates were tested for their capacity to induce a chemotaxis response. R. erythropolis U23A was grown overnight at 28°C in Petri dishes on solid LB medium. Cells were then collected and suspended in a 0.5-M phosphate buffered solution (PBS), pH 7.3 containing 0.1% agar and the suspension was adjusted to an OD600 nm of 5. The suspension was poured into 50 mm Petri dishes and allowed to settle for a few minutes before transferring the disk soaked with the substrates to be tested. Sterile filter paper discs were soaked individually with 20 μl of each preparation to be tested, the filters were air-dried in a sterile laminar-flow cabinet, and then gently deposited on the surface of the soft agar medium containing the bacterial suspension. The formation of a turbidity ring around the disc, reflecting the movement of the bacterial suspension towards the chemo-attractant, was observed regularly for 2 h and pictures were taken to document the assay.
Binding assays: microbial adhesion test on hydrocarbons/sand
Microbial adhesion to hydrocarbons (MATH) (Rosenberg et al. 1980) and microbial adhesion to silica sand (MATS) (Déziel et al. 2001) assays were performed according to previously described protocols (Déziel et al. 2001; Rosenberg et al. 1980). R. erythropolis U23A and P. fluorescens F113 (a positive control) were grown on liquid LB medium overnight at 28°C, centrifuged at 7,000 rpm and washed twice in 0.85% NaCl, and suspended in 3 ml of 0.5 M PBS, pH 7.3, and OD600 nm was adjusted to 0.5. For the MATH assay, 500 μl of the bacterial suspension was mixed with 500 μl of mineral oil in 2 ml glass vials, vortexed for 1 min at maximum speed and allowed to settle for 30 min at room temperature. Two hundred microliters of the aqueous phase was transferred into the wells of a 96-well plate and OD600 nm was recorded. For the MATS assay, 500 μl of the bacterial suspension was mixed in 500 mg of washed sand in an Eppendorf tube and mixed on a rotation shaker for 30 min at room temperature. The tubes were then put aside to let the sand settle at the bottom, and the supernatant was transferred in a spectrophotometer tube to read OD600 nm against a blank of PBS. For both assays, OD600 nm was measured prior and after completing the test in order to compare and calculate the absorption percentage on either mineral oil or sand. Comparisons were also made between the two strains for each assay.
Root colonization assay
The root colonization assay was partly adapted from that of Scher et al. (1984) and Villacieros et al. (2005). For this assay, alfalfa (Medicago sativa L. var. Geneva) seeds were used as their root system is bigger and easier to handle than that of A. thaliana. Seeds were surface-sterilized by soaking in a 50% hypochlorite solution (commercial bleach 4%) for 5 min and rinsed with sterile distilled water. Bacterial cultures of R. erythropolis U23A and P. fluorescens F113 were grown at 28°C overnight in 2 ml LB medium, and OD600 nm was adjusted to 1 in sterile NaCl (0.85%). The sterilized seeds were soaked in the bacterial suspension for 30 min on a horizontal agitator, after which they were inoculated in 50 ml test tubes filled with 40 g sterilized Ottawa sand. Prior seed inoculation, the sand was watered with 7 ml of half-strength Hoagland’s solution. Triplicate tubes were seeded for each treatment. Triplicate control tubes seeded with uninoculated seeds were also included in the experiment to confirm the sterility of the plant cultures. The seeds were allowed to germinate and grow for 4 weeks before harvesting. For each treatment, three replicated seeds that had been soaked in the bacterial suspensions were washed in sterile 0.85% NaCl and vortexed for 10 s. Serial tenfold dilutions were made in sterile 0.85% NaCl from which 0.1 ml was inoculated on LB agar plates in triplicate. Colonies were counted following incubation for 24 h at 28°C to determine the total number of colony-forming units per inoculated seed. The extent of root colonization was evaluated using a protocol similar to the one described by Sanchez-Contreras et al. (2002). Four-week-old plants (including the roots) were carefully removed from the sands, and their roots were excised, weighted, transferred in a tube containing 5 ml sterile 0.85% NaCl, and vortexed vigorously. Similarly, the sand from each tube was collected, weighted, and vortexed into 5 ml 0.85% NaCl to release the bacteria remaining in the sand fraction. Both bacterial suspensions (roots and sands) were diluted and plated on LB agar as above to count the CFUs from the roots or sand fractions. The CFUs were expressed per gram of roots or of sand.
Results
Isolation and identification
Based on 16S rRNA sequence analysis, strains U22 and U24 clustered with R. globerulus. Their PCB-degrading patterns, based on their ability to metabolize the mixture of 18 PCB congeners described in Table 1, were very similar to the one obtained for strain U23A described below (data not shown). However, both of these strains exhibited a poor growth when root exudates were used as the sole carbon source. Therefore, these two strains were not further considered in the study. Strain U23A is an aerobic, non-sporulating, Gram-positive bacterium. It is catalase-positive, and based on microscopic observation, it forms rods to extensively branched vegetative chains. On the basis of its morphological and biochemical features, it was identified as a Rhodococcus. The identity was confirmed as R. erythropolis based on 16S rRNA gene sequence. The 16S rRNA gene sequence of R. erythropolis U23A is available under the GenBank accession number HQ412801 and it is 99% identical to that of R. erythropolis strain LG12 and OUCZ211 16S rRNA gene (GenBank: AY785750.1 and EU852376.1). Strain U23A has been deposited at the American Type Culture Collection under the accession number ATCC BAA-2259.
Furthermore, using the primers to amplify the gene encoding for the large subunit of the biphenyl dioxygenase’s oxygenase component (bphA), we amplified a 1.2-kb fragment with the primers that are specific for R. globerulus P6 bphA1. Sequence analysis of the amplified 1.2-kb fragment confirmed the presence of bphA and its deduced amino acid sequence was closely related (99% identity) to BphA1 of R. globerulus strain P6 (Barriault et al. 2002). The two proteins differ by only one amino acid residue, where the alanine at position 446 of P6 BphA1 is replaced by a glycine in U23A BphA. The presence of a single bphA in the genome of strain U23A was supported by the fact that the previously described sets of degenerated pairs of primers designed to amplify a broad range of bphA genes (Vézina et al. 2007) did not amplify any amplicon other than the one corresponding to the sequence described above and the primers specific for B. xenovorans LB400 or P. pnomenusa B-356 bphA did not amplify any gene.
Bacterial growth on root exudates and flavonoid detection by HPLC
R. erythropolis U23A was able to grow in MM30 containing 1% (v/v) of the 15 times concentrated root exudates as sole growth substrate, reaching an OD600 nm of approximately 0.7 within 48 h at 28°C. Under the same conditions, when grown on 3.4 mM biphenyl, strain U23A reached an OD600 nm of 2.5 after 24 h of incubation. The root exudates were fractionated by HPLC according to the protocol described in the “Materials and methods” section. Approximately 40 HPLC chromatographic peaks were obtained from this separation. In order to facilitate identification of the components, the exudates were acid-hydrolyzed before separation by HPLC. On the basis of LC–MS, GC–MS analyses and with comparison to commercially available flavonoids and phenylpropanoids few peaks were identified, including coumarin, flavanone, hydroxybenzoic acid, hydroxytyrosol, naringenin, syringic acid, and vanillic acid (not shown). However, under our experimental conditions, we were unable to identify the other peaks, many of which were very low, indicating they were minor components of the mixture or they were lost during acid hydrolysis.
PCB and 4CB degradation by cells grown on root exudates
Resting cell suspensions of biphenyl-induced R. erythropolis U23A were able to degrade seven congeners of the mix of 18 congeners described in Table 1, ranging from di- to tetra-chlorinated biphenyls (Table 2). More importantly, when cells were grown in a minimal medium containing 1% (v/v) of the 15 times concentrated root exudates as sole growth substrate, the resting cells were able to degrade three chlorobiphenyls more significantly than cells grown on MM30 containing sodium acetate as sole growth substrate or grown on LB medium (Table 2).
A resting cell assay described in the “Materials and methods” section was performed to determine the effect of root exudates on the ability of R. erythropolis U23A to convert 4-chlorobiphenyl to 4-chlorobenzoate. When cells were grown on MM30 containing biphenyl as growth substrate, the resting cell suspension converted approximately half (750 μM) of the 1.25 mM of 4-chlorobiphenyl added to the medium into 4-chlorobenzoate (Fig. 2) which was detected as sole metabolite by GC–MS analysis (not shown). When cells were grown on LB medium, approximately 7 μM 4-chlorobenzoate was produced and no other intermediate metabolites were detected in the medium, indicating that the biphenyl-degrading enzymes are constitutively expressed at a very low level in strain U23A. However, when cells were grown using root exudates as sole growth substrate, using identical conditions, resting cells of R. erythropolis produced 3.5 times more 4-chlorobenzoate than those grown on LB broth. This observation provides further evidence that U23A bph gene cluster responds to the Arabidopsis root exudates (although to a lesser extent than for biphenyl) by increasing the level of the biphenyl catabolic enzymes above the basal level found in cells grown in LB broth.
Amount (micromolar) of 4-chlorobenzoic acid produced when standardized resting cell suspensions of R. erythropolis U23A were incubated with 1,250 μM 4-chlorobiphenyl for 2 h. R. erythropolis was previously grown overnight at 28°C in either LB Broth, MM30 amended with 3.4 mM biphenyl, or a 1% (v/v) of a 15 times concentrated root exudates. Bars represent mean (n = 2) and SD are shown. Different letters indicate significant differences according to Tukey’s test (P ≤ 0.05). The protocol for the standardized 4-chlorobiphenyl assay is described in the “Materials and methods” section
Metabolism of flavonoids by strain U23A
None of the compounds that were identified from root exudates (listed above) were able to sustain the growth of strain U23A when used as sole growth substrate. Since flavanone was among the most abundant chemicals detected in the root exudates (its concentration was estimated between 0.5 and 1 mM based on the peak area of HPLC and GC–MS chromatography), we have examined the ability of biphenyl-induced cells to metabolize this flavonoid. When a resting cell suspension was incubated for 30 min in the presence of 200 μM flavanone under the conditions described in the “Materials and methods” section, two dihydrodiol metabolites were detected in the culture media (Fig. 3a). The mass spectral features of the butylboronate derivatives of both metabolites were very similar and corresponded to the 2-(2,3-dihydro-2,3-dihydroxyphenyl)chromane-4-one and the 2-(3,4-dihydro-3,4-dihydroxyphenyl)chromane-4-one. Both metabolites were characterized by a high molecular ion at m/z 324 and diagnostically important ions at m/z 308 (M+–O), 267 (M+–n-Bu), 240 (M+–n-BuBO), 224 (M+–n-BuBO2), 147 (M+–n-BuBO2–C6H5), 120 (M+–n-BuBO2–C6H5–CH–CH2). The identity of these two metabolites as dihydrodihydroxy compounds was confirmed by the fact that a reconstituted purified preparation of P. pnomenusa B356 biphenyl dioxygenase produced the same two metabolites from flavanone (Fig. 3a). Although GC–MS analysis did not allow us to distinguish between the 2,3- and 3,4-dihydroxylated metabolites, the mass spectral fragmentation pattern (presence of ions at 147 and 120) was consistent with a dihydroxylation reaction occurring on the phenyl ring, at both positions. This ring was also the one that an engineered biphenyl dioxygenase produced by Pseudomonas pseudoalcaligenes KF707 was shown to hydroxylate (Shindo et al. 2003).
a Total ion chromatogram showing the peaks of the two metabolites produced from flavanone after 30 min by a resting cell suspension of R. erythropolis U23A (gray curve) or after 10 min by a purified preparation of P. pnomenusa biphenyl dioxygenase (BPDO) (black curve). The inset shows the mass spectra of one of the two metabolites; the second mass spectrum is not shown but very similar. b Total ion chromatogram showing the single peak of metabolite produced after 18 h by a resting cell suspension of R. erythropolis U23A. The inset shows the mass spectra of the metabolite
When the cell suspensions were incubated from 3 to 18 h under the same conditions, a single metabolite was detected. This metabolite accumulated in the culture medium until the substrate was completely depleted and it was not further degraded by the cells. The TMS-derived metabolite was detected by GC–MS and its mass spectral features corresponded to the 4-oxo-2-chromanecarboxylic acid (Fig. 3b). It was characterized by diagnostically important ions at m/z 264 (M+), 249 (M+–CH3), 219 (M+–3CH3), 205 (M+–CH3–O–CO), 174 (M+–COOTMS), and 131 (M+–COOTMS–O).
Together, the data show the unique biphenyl dioxygenase of the biphenyl catabolic pathway of strain U23A oxidized flavanone on the phenyl ring and the ring is then cleaved to ultimately generate 4-oxo-2-chromanecarboxylic acid. However, since the hydroxylation occurs on carbons 2,3 and 3,4 of the phenyl ring and a single metabolite was detected as the end product of this pathway, more work will be required to determine the exact steps involved in the metabolism of flavanone dihydrodiols. The biphenyl dioxygenase, the 2,3-dihydro-2,3-dihydroxybiphenyl 2,3-dehydrogenase and the 2,3-dihydroxybiphenyl 2,3-dioxygenase of P. alcaligenes K707 were found to metabolize flavanone to the meta-cleavage metabolite (Shindo et al. 2004) through an initial oxygenation on carbons 2 and 3 of the phenyl ring. Therefore, it is likely that 2,3-dihydroxylated metabolite generated by U23A biphenyl dioxygenase would be transformed to the 4-oxo-2-chromanecarboxylic acid using the four enzymatic steps of the biphenyl catabolic pathway. However, it is not clear how the 3,4-dihydroxylated metabolite was converted to this final metabolite. Trace amounts of a metabolite whose TMS derivative exhibited mass spectral features that were consistent with the 2-hydroxy(4-oxo-2-chromane) acetaldehyde or the 4-oxo-2-chromane acetic acid were detected. Diagnostically important ions were observed at m/z 278 (M+), 263 (M+–CH3), 147 (M+–TMS–O–CH–CH) (not shown). This metabolite is likely to be generated from the cleavage of the 3,4-dihydroxylated metabolite. However, it is not clear what are the enzymes involved in the ring cleavage and in the oxidation of the aliphatic chain to generate the 4-oxo-2-chromanecarboxylic acid.
Induction of U23A biphenyl catabolic enzymes by flavanone
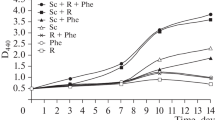
Since flavanone alone was unable to serve as growth substrate for strain U23A, we verified if this chemical could induce the biphenyl catabolic enzymes when the cells were grown co-metabolically on sodium acetate plus flavanone. The resting cell assay described in the “Materials and methods” section was performed to determine the effect of growing cells co-metabolically on sodium acetate plus variable concentrations of flavanone on the ability of R. erythropolis U23A to convert 4-chlorobiphenyl to 4-chlorobenzoate. By growing the cells co-metabolically on sodium acetate plus flavanone and collecting them after overnight growth (late log phase), we were reproducing the conditions that were used to prepare the cells grown on root exudates. Under these conditions, flavanone was metabolized during growth on sodium acetate to generate principally 4-oxo-2-chromanecarboxylic acid (not shown). When cells were grown overnight on MM30 containing sodium acetate alone and tested for their ability to metabolize 4-chlorobiphenyl, only traces of 4-chlorobenzoate were detected in the assay medium after 2 h of incubation. However, cells grown co-metabolically on sodium acetate plus flavanone metabolized 4-chlorobiphenyl to 4-chlorobenzoate and the amounts of 4-chlorobenzoate produced varied depending on the amount of flavanone added to the growth medium (Fig. 4). Cells grown in the presence of 1 mM flavanone produced significantly more 4-chlorobenzoate than those grown on 0.01 mM. However, it is noteworthy that in spite of the fact that flavanone was metabolized and its concentration decreased during growth on sodium acetate, cells grown in medium containing as little as 10 or 1 μM flavanone produced more chlorobenzoate than the control without flavanone. Cells grown on sodium acetate plus biphenyl responded similarly to those grown on sodium acetate plus flavanone, where the catabolic activity of overnight grown cells toward 4-chlorobiphenyl decreased as the level of biphenyl was lowered in the medium (Fig. 4). However, the amount of 4-chlorobenzoate produced by the cells grown in the presence of flavanone was in the same range and even higher than for those grown in the presence of biphenyl.
Amount (micromolar) of 4-chlorobenzoic acid produced when standardized resting cell suspensions of R. erythropolis U23A were incubated with 1,250 μM 4-chlorobiphenyl for 2 h. R. erythropolis was previously grown overnight at 28°C in MM30 amended with 30 mM acetate or with 30 mM acetate plus the indicated concentrations of flavanone or of biphenyl. Bars represent mean (n = 2) and SD are shown. The protocol for the standardized 4-chlorobiphenyl assay is described in the “Materials and methods” section
Together, the data provided strong evidence for the ability of the root exudates to promote PCB degradation, which can be attributed to the capacity of flavanone (and perhaps other unidentified phenylpropanoids) to serve as inducer of U23A biphenyl catabolic pathway. Flavanone was unable to serve as growth substrate, most likely because the chromane-4-one portion was not metabolized. However, the data show that the flavonoids, (especially flavanone found in the exudates) are metabolized by the enzymes of the biphenyl catabolic pathway and can serve as inducers of the biphenyl catabolic pathway. The exudate components supporting cell growth could not be identified but could be anyone of the many peaks, including glycosylated compounds, sugars, and/or amino acids that are likely to be present in exudates.
Motility, binding, and colonization assays
In order to assess the ability of R. erythropolis U23A to colonize plant roots, we have examined some of the traits that are normally associated with bacterial adhesion to surface, formation of biofilm, and colonization of plant roots. In this respect, when 500 μl of a cell suspension of strain U23A adjusted to 0.5 OD600 nm was mixed with 500 mg of sand, 30 ± 2% of cells remained associated to the sand fraction which was much higher than the 8 ± 1% of P. fluorescens F113 cells. On the other hand, when the same cell suspensions of strain U23A were mixed with an equal amount of mineral oil (MATH assay), 52 ± 4% cells remained associated to the oil fraction compared to 98 ± 8% of strain F113 cells that remained in the hydrocarbon fraction. The cell surface of strain U23A was quite hydrophobic in the MATH assay since 50% of the cells remained associated with the hydrocarbon fraction, although this was to a lesser extent than cells from strain F113, which is a well-characterized Gram-negative root colonizer (Brazil et al. 1995).
Notably, R. erythropolis U23A exhibited a chemotactic response to root exudates, which was significantly stronger than that of the positive control using tryptone (Fig. 5). No chemotaxis movement was observed in the presence of water (negative control; results not shown). Finally, as shown on Fig. 6, although R. erythropolis U23A did not colonize alfalfa roots as well as the well-characterized P. fluorescens F113 strain, the number of cells associated to the plant roots fraction (5.7 × 105 CFU g−1) compared to the sand fraction (4.5 × 104 CFU g−1) was high enough for strain U23A to conclude that this strain can either colonize the roots or adhere to them.
Chemotaxis movement of R. erythropolis U23A towards a filter paper soaked with 1% tryptone (positive control; left) or A. thaliana root exudates (concentrated 15 times; right) as indicated by the white arrows. The bacterial suspension was adjusted to OD600 nm = 5 in a phosphate buffered saline solution containing 0.1% agar
Number of colony-forming units (CFU) of R. erythropolis U23A or P. fluorescens F113 remaining in sand or associated to roots of M. sativa (alfalfa) after 4 weeks growth in vitro. The experimental conditions are described in the “Materials and methods” section; CFUs are expressed per unit of seed or per gram of roots or sand. Mean (n = 3) and SE are shown
Discussion
Rhizoremediation is becoming increasingly popular as a means to treat soils contaminated with xenobiotics such as PCBs (Van Aken et al. 2010; Sylvestre et al. 2009; Field and Sierra-Alvarez 2008; Daar et al. 2002). Several reports have provided evidence that plants promote the bacterial PCB degradation in soil (Chekol et al. 2004; Dercova et al. 2003; Leigh et al. 2006; Narasimhan et al. 2003). However, the mechanism by which plants associate with rhizobacteria to remove the contaminants in the soil is poorly understood. Understanding how plants stimulate the bacterial degradation of pollutants in soil will help design novel approaches for soil remediation. Here we have isolated and characterized a rhodococcal rhizobacterium whose PCB-degrading pathway was shown to be triggered by root exudate components.
The presence of bphA encoding for the large subunit of biphenyl dioxygenase’s oxygenase component and its ability to transform 4-chlorobiphenyl into 4-chlorobenzoate are evidences that R. erythropolis U23A metabolizes PCB congeners and flavanone through the biphenyl catabolic pathway. The detection of 2-(dihydro-dihydroxy-phenyl) chromane-4-ones and 4-oxo-2-chromanecarboxylic acid in resting cell assay performed with biphenyl-induced cells provided strong evidence that flavanone was metabolized by the unique biphenyl dioxygenase found in strain U23A and most likely by the other three enzymes of the biphenyl catabolic pathway.
Other R. erythropolis strains have previously been shown to carry the bph gene cluster (Pieper 2005; Yang et al. 2004, 2007). However, sequence analysis of U23A bphA shows that the dioxygenase that initiates the catabolic pathway is closely related to that of R. globerulus P6, which is distinct from those of previously described R. erythropolis PCB-degrading strains. Because of the similarity between the biphenyl dioxygenase of strains P6 and U23A, it was expected that both strains would degrade the same range of PCB congeners. This was confirmed since the pattern of PCB congeners degraded by biphenyl-induced R. erythropolis U23A cells (inability to metabolize 2,2′-dichlorobiphenyl or 2,2′,5,5′-tetrachlorobiphenyl and ability to metabolize 3,3′-dichlorobiphenyl and 2,2′,3,3′-tetrachlorobiphenyl) is identical to the one previously reported by Bedard et al. (1986) for Corynebacterium strain MB1 (later reclassified and renamed as R. globerulus P6) (Asturias et al. 1994).
Many species belonging to the Rhodococcus genus are known to degrade PCBs (Warren et al. 2004; Arai et al. 1998; Seto et al. 1996). Chlorobenzoate degradation by R. erythropolis strain S-7 and Rhodococcus sp. R04 has also been reported previously (Chung et al. 1994; Yang et al. 2004; Yun et al. 2009; Zhang et al. 2009). However, unlike these strains, U23A was unable to metabolize 4-chlorobenzoate produced from 4-chlorobiphenyl and it was unable to metabolize 4-oxo-2-chromanecarboxylic acid derived from flavanone.
Being non-flagellated, R. erythropolis U23A is not expected to be motile, which is an important trait for root colonization (Lugtenberg and Kamilova 2009). However, our data show that the strain was able to colonize (or at least to adhere) the roots of alfalfa, as well as to move towards a disk soaked with a solution containing root exudates in a chemotaxis assay. Motility is traditionally associated with bacteria that possess flagella and/or pili that confer them the ability to swarm (Kearns 2010) or to move by swimming or twitching (O’Toole et al. 2000), thus giving them the potential to colonize plant roots. Rhodococcal are not flagellated and there is no evidence indicating that they possess pili on their surface. However, we cannot exclude the possibility that R. erythropolis U23A is able to move by gliding, as reported for other bacteria that lack flagella (Jarrell and McBride 2008), or by sliding, as reported for a Pseudomonas aeruginosa mutant lacking both pili and flagella (Murray and Kazmierczak 2008).
On the other hand, the binding capacity of R. erythropolis in the MATH test indicated that the hydrophobicity of this strain is in the same range as found for R. erythropolis strain CCM 2595 in a similar assay (Masak et al. 2005). Although the role of cell surface properties on microbial adhesion to surface is not clearly understood, surface hydrophobicity appears as an important factor (Rosenberg 2006). In the case of strain U23A, hydrophobicity might facilitate aggregation on or around plant roots.
An important finding in this work is that cells of strain U23A grown in a medium containing Arabidopsis root exudates as sole growth substrate could metabolize 3,3′-dichlorobiphenyl, 2,3,4′-trichlorobiphenyl, and 2,3′,4-trichlorobiphenyl more efficiently than cells grown on LB medium or on MM30 with sodium acetate as sole growth substrate (see Table 2). This suggests that root exudates contain components not present in LB medium that can trigger (or induce) the expression of bph genes. The inability of cells grown on root exudates to degrade all the congeners that are degraded by biphenyl-induced cells is most likely due to the very low expression of the biphenyl catabolic genes in the PSM-induced cells. The first enzyme of the bph pathway, the three component biphenyl dioxygenase, determines which PCBs are at least partially transformed by the microorganism (Mohammadi et al. 2011). In a previous investigation with a reconstituted purified BPDO preparation, dihydroxylation of the more resistant congeners, such as 2,2′-dichlorobiphenyl, was only detected when the levels of enzyme components were high and optimized, i.e., when the proportions of the ferredoxin and ferredoxin-reductase were much higher than the oxygenase component (Imbeault et al. 2000). Although no explanation has yet been provided for this observation, it would explain why cells grown on root exudates only degraded the low and most labile congeners (4-chlorobiphenyl and 2,3′,4-trichlorobiphenyl).
Our results indicated that flavanone, one of the major components of A. thaliana exudates, is an inducer of the biphenyl catabolic pathway. It is noteworthy to say that flavanone is not only an inducer of the biphenyl catabolic pathway, but is also metabolized by the biphenyl catabolic pathway. However, despite the ability of the biphenyl catabolic enzymes to metabolize flavanone, this compound could not support growth of strain U23A, most likely because of the inability of the cells to metabolize the 4-oxo-2-chromanecarboxylic acid. However, we cannot exclude an inhibitory effect of flavanone on one or more of the steps of the biphenyl assimilatory process since strain U23A was unable to grow in a culture medium containing biphenyl plus flavanone (results not shown). A similar inhibitory effect of phenylterpenoids was made by Gilbert and Crowley (1997) where carvone, a terpenoid present in spearmint (Mentha spicata), was shown to promote the PCB-degrading ability of an Arthrobacter isolate. However, this terpene was toxic to this organism and it was unable to support its growth.
It has long been suspected that PSMs may allow the growth of bacteria in the rhizosphere as well as triggering their biphenyl pathway, resulting in enhanced PCB degradation (Bais et al. 2004; Donnelly et al. 1994; Fletcher and Hegde 1995; Leigh et al. 2002; Shaw et al. 2006). Recently, Leigh et al. (2006) demonstrated that rhodococci isolates were associated with the roots of certain tree species (e.g., pine and willow) in a contaminated site and that some of these isolates had strong PCB degradation potential. As highlighted by the authors, rhodococci seem to be able to use PSMs as growth substrate in soil (Donnelly et al. 1994; Hernandez et al. 1997) and this feature might explain why they are well adapted to degrade PCBs. However, this hypothesis has not yet been demonstrated.
Although the interactions between Actinobacteria and plants and their effect on plant growth and bacterial metabolism have been much less documented than the interaction involving other bacterial groups (Francis et al. 2010), it is clear that many members of the Actinobacteria including members of the non-mobile genus Streptomyces have been associated to plant diseases or been used to control plant diseases. Our observation with strain U23A is corroborated by the recent observation whereby R. erythropolis (MtCC 7905) (Trivedi et al. 2007) was shown to exhibit plant growth promoting activity. In addition to its plant growth promoting ability, strain MtCC 7905 was also shown to be a useful chromate-reducing bacterium in soil. Therefore, our data strongly support the idea that Gram-positive rhodoccoci may play an active role in the rhizosphere microbiota.
This is strengthened by the fact that flavanone induces the biphenyl catabolic enzymes of R. erythropolis U23A and is metabolized by this pathway. In a previous metabolomic analysis of A. thaliana exudates, 125 PSMs were identified (Narasimhan et al. 2003), 76% of which belonged to the phenylpropanoid class of compounds. Therefore, flavanone might not be the sole exudate component to exert an inducing effect on the biphenyl catabolic enzymes. However, by singling out flavanone, our study clearly shows that although each single component of root exudates cannot support growth, individually, some of the exudates components can serve as inducer of this pathway. de Lorenzo and Pérez-Martin (1996) have introduced the notion of fortuitous inducers to explain why several substrate analogs can induce catabolic pathway enzymes. Traditionally, the level of induction obtained from fortuitous inducers is significantly lower than for the natural substrate of the pathway. However, it is quite intriguing that during co-metabolic growth, flavanone was found to be as efficient as biphenyl to induce the biphenyl catabolic pathway enzymes and that flavanone phenyl ring was metabolized completely by this pathway.
The ability of the biphenyl catabolic enzymes to metabolize flavanone and other flavonoids has been reported by several investigators (Shindo et al. 2003, 2004; Seeger et al. 2003; Seo et al. 2011). Until now, these reactions were believed to be fortuitous and explained on the basis of the ability of the pathway enzymes to bind biphenyl analogs productively. However, on the basis of our investigation and others that provided evidence that PSMs act as inducers of the biphenyl catabolic enzymes (Dercova et al. 2003; Donnelly et al. 1994; Gilbert and Crowley 1997; Hernandez et al. 1997; Miya and Firestone 2001; Singer et al. 2003), the question can be raised whether this pathway plays a role in the maintenance or recycling of phenylpropanoids and phenylterpenoids in the rhizosphere.
Altogether, our results highlight the fact that even though R. erythropolis is considered a non-mobile bacterium, it still possesses several traits conferring the capacity to colonize (or grow on) plant roots surface (or in the rhizosphere). Our data do not allow us to distinguish between the increase in bacterial density in the rhizosphere that would be associated to root colonization and growth promotion due to the presence of PSMs that promote growth of bacteria adhering to root surfaces. Further work will be required to clearly show that rhodoccoci can colonize plant roots. However, our data clearly show that strain U23A exhibits a chemotaxic response to root exudates and that flavanone and perhaps other chemicals present in the root exudates can trigger the biphenyl catabolic enzymes. We can thereby propose a model for explaining how plants promote PCB degradation in soil. In the rhizosphere, labile chemicals such as the sugar moiety of the conjugated PSMs might provide a substrate on which to grow, whereas the flavonoids or other phenylpropanoids would then induce the biphenyl pathway of strain U23A. The ability of flavanone to act as strong inducer of the biphenyl pathway raises the question of whether this pathway plays a role in phenylpropanoid metabolism in soil. However, an important contribution of this process is the finding that flavanone triggers the metabolism of PCBs in soil.
References
Abramowicz DA (1995) Aerobic and anaerobic PCB biodegradation in the environment. Environ Health Persp 103:97–99
Arai H, Kosono S, Taguchi K, Maeda M, Song E, Fuji F, Chung SY, Kudo T (1998) Two sets of biphenyl and PCB degradation genes on a linear plasmid in Rhodococcus erythropolis TA421. J Ferment Bioeng 86:595–599
Asturias JA, Moore E, Yakimov MM, Klatte S, Timmis KN (1994) Reclassification of the polychlorinated biphenyl-degraders Acinetobacter sp. strain P6 and Corynebacterium sp. strain MB1 as Rhodococcus globerulus. Syst Appl Microbiol 17:226–231
Bais HP, Park SW, Weir TL, Callaway RM, Vivanco JM (2004) How plants communicate using the underground information superhighway. Trends Plant Sci 9:26–32
Barriault D, Plante MM, Sylvestre M (2002) Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J Bacteriol 184:3794–3800
Barriault D, Lépine F, Mohammadi M, Milot S, Leberre N, Sylvestre M (2004) Revisiting the regiospecificity of Burkholderia xenovorans LB400 biphenyl dioxygenase toward 2,2′-dichlorobiphenyl and 2,3,2′,3′-tetrachlorobiphenyl. J Biol Chem 279:47489–47496
Bedard DL, Unterman R, Bopp LH, Brennan MJ, Haberl ML, Johnson C (1986) Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol 51:761–768
Borja J, Taleon DM, Auresenia J, Gallardo S (2005) Polychlorinated biphenyls and their biodegradation. Proc Biochem 40:1999–2013
Brazil GM, Kenefick L, Callanan M, Haro A, Delorenzo V, Dowling DN, Ogara F (1995) Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated-biphenyls and detection of bph gene-expression in the rhizosphere. Appl Environ Microbiol 61:1946–1952
Chebrou H, Hurtubise Y, Barriault D, Sylvestre M (1999) Heterologous expression and characterization of the purified oxygenase component of Rhodococcus globerulus P6 biphenyl dioxygenase and of chimeras derived from it. J Bacteriol 181:4805–4811
Chekol T, Vough LR, Chaney RL (2004) Phytoremediation of polychlorinated biphenyl-contaminated soils: the rhizosphere effect. Environ Int 30:799–804
Chung SY, Maeda M, Song E, Horikoshi K, Kudo T (1994) A gram-positive polychlorinated biphenyl-degrading bacterium, Rhodococcus erytrhopolis strain TA421, isolated from a termite ecosystem. Biosci Biotechnol Biochem 58:2111–2113
Daar AS, Thorsteinsdottir H, Martin DK, Smith AC, Nast S, Singer PA (2002) Top ten biotechnologies for improving health in developing countries. Nat Genet 32:229–232
de Lorenzo V, Pérez-Martin J (1996) Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol Microbiol 19:1177–1184
Dercova K, Tandlich R, Brezna B (2003) Application of terpenes as possible inducers of biodegradation of PCBs. Fresen Environ Bull 12:286–290
Déziel E, Comeau Y, Villemur R (2001) Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 183:1195–1204
Donnelly PK, Hegde RS, Fletcher JS (1994) Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere 28:981–988
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853
Fava F, Bertin L (1999) Use of exogenous specialised bacteria in the biological detoxification of a dump site-polychlorobiphenyl-contaminated soil in slurry phase conditions. Biotechnol Bioeng 64:240–249
Field JA, Sierra-Alvarez R (2008) Microbial degradation of chlorinated dioxins. Chemosphere 71:1005–1018
Fletcher JS, Hegde RS (1995) Release of phenols by perennial plant-roots and their potential importance in bioremediation. Chemosphere 31:3009–3016
Francis I, Holsters M, Vereecke D (2010) The Gram-positive side of plant–microbe interactions. Environ Microbiol 12:1–12
Gilbert ES, Crowley DE (1997) Plant compounds that induce polychlorinated biphenyl biodegradation by Arthrobacter sp. strain B1B. Appl Environ Microbiol 63:1933–1938
Gordillo F, Chavez FP, Jerez CA (2007) Motility and chemotaxis of Pseudomonas sp B4 towards polychlorobiphenyls and chlorobenzoates. FEMS Microbiol Ecol 60:322–328
Hernandez BS, Koh SC, Chial M, Focht DD (1997) Terpene-utilizing isolates and their relevance to enhanced biotransformation of polychlorinated biphenyls in soil. Biodegradation 8:153–158
Hurtubise Y, Barriault D, Sylvestre M (1996) Characterization of active recombinant his-tagged oxygenase component of Comamonas testosteroni B-356 biphenyl dioxygenase. J Biol Chem 271:8152–8156
Hurtubise Y, Barriault D, Sylvestre M (1998) Involvement of the terminal oxygenase beta subunit in the biphenyl dioxygenase reactivity pattern toward chlorobiphenyls. J Bacteriol 180:5828–5835
Imbeault NY, Powlowski JB, Colbert CL, Bolin JT, Eltis LD (2000) Steady-state kinetic characterization and crystallization of a polychlorinated biphenyl-transforming dioxygenase. J Biol Chem 275:12430–12437
Iwasaki T, Miyauchi K, Masai E, Fukuda M (2006) Multiple-subunit genes of the aromatic-ring-hydroxylating dioxygenase play an active role in biphenyl and polychlorinated biphenyl degradation in Rhodococcus sp strain RHA1. Appl Environ Microbiol 72:5396–5402
Jarrell KF, McBride MJ (2008) The surprisingly diverse ways that prokaryotes move. Nature Rev Microbiol 6:466–476
Kearns DB (2010) A field guide to bacterial swarming motility. Nature Rev Microbiol 8:634–644
Kim BH, Oh ET, So JS, Ahn Y, Koh SC (2003) Plant terpene-induced expression of multiple aromatic ring hydroxylation oxygenase genes in Rhodococcus sp strain T104. J Microbiol 41:349–352
L’Abbée JB, Tu Y, Barriault D, Sylvestre M (2011) Insight into the metabolism of 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) by biphenyl dioxygenases. Arch Biochem Biophys 516:35–44
Leigh MB, Fletcher JS, Fu XO, Schmitz FJ (2002) Root turnover: an important source of microbial substrates in rhizosphere remediation of recalcitrant contaminants. Environ Sci Technol 36:1579–1583
Leigh MB, Prouzova P, Mackova M, Macek T, Nagle DP, Fletcher JS (2006) Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol 72:2331–2342
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting Rhizobacteria. Ann Rev Microbiol 63:541–556
Luo WS, D’Angelo EM, Coyne MS (2007) Plant secondary metabolites, biphenyl, and hydroxypropyl-beta-cyclodextrin effects on aerobic polychlorinated biphenyl removal and microbial community structure in soils. Soil Biol Biochem 39:735–743
Masak J, Cejkova A, Jirku V, Kotrba D, Hron P, Siglova M (2005) Colonization of surfaces by phenolic compounds utilizing microorganisms. Environ Int 31:197–200
Massé R, Messier F, Ayotte C, Lévesque M-F, Sylvestre M (1989) A Comprehensive gas chromatographic/Mass spectrometric analysis of 4-chlorobiphenyl bacterial degradation products. Biomed Environ Mass 18:27–47
McKay DB, Prucha M, Reineke W, Timmis KN, Pieper DH (2003) Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6. J Bacteriol 185:2944–2951
Miya RK, Firestone MK (2001) Enhanced phenanthrene biodegradation in soil by slender oat root exudates and root debris. J Environ Qual 30:1911–1918
Mohammadi M, Viger JF, Kumar P, Barriault D, Bolin JT, Sylvestre M (2011) Retuning Rieske-type oxygenases to expand substrate range. J Biol Chem 286:27612–27621
Murray TS, Kazmierczak BI (2008) Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190:2700–2708
Narasimhan K, Basheer C, Bajic VB, Swarup S (2003) Enhancement of plant–microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132:146–153
O’Toole G, Kaplan HB, Kolter R (2000) Biofilm formation as microbial development. Ann Rev Microbiol 54:49–79
Pieper DH (2005) Aerobic degradation of polychlorinated biphenyls. Appl Microbiol Biot 67:170–191
Rosenberg M (2006) Microbial adhesion to hydrocarbons: twenty-five years of doing MATH. FEMS Microbiol Lett 262:129–134
Rosenberg M, Gutnick D, Rosenberg E (1980) Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9:29–33
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sanchez-Contreras M, Martin M, Villacieros M, O’Gara F, Bonilla I, Rivilla R (2002) Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J Bacteriol 184:1587–1596
Scher FM, Ziegle JS, Kloepper JW (1984) A method for assessing the root-colonizing capacity of bacteria on maize. Can J Microbiol 30:151–157
Seeger M, Gonzalez M, Camara B, Munoz L, Ponce E, Mejias L, Mascayano C, Vasquez Y, Sepulveda-Boza S (2003) Biotransformation of natural and synthetic isoflavonoids by two recombinant microbial enzymes. Appl Environ Microbiol 69:5045–5050
Seo J, Kang SI, Kim M, Han J, Hur HG (2011) Flavonoids biotransformation by bacterial non-heme dioxygenases, biphenyl and naphthalene dioxygenase. Appl Microbiol Biot 91:219–228
Seto M, Ida M, Okita N, Hatta T, Masai E, Fukuda M (1996) Catabolic potential of multiple PCB transformation systems in Rhodococcus sp strain RHA1. Biotechnol Lett 18:1305–1308
Shaw LJ, Morris P, Hooker JE (2006) Perception and modification of plant flavonoids signals by rhizosphere microorganisms. Environ Microbiol 8:1867–1880
Shindo K, Kagiyama Y, Nakamura R, Hara A, Ikenaga H, Furukawa K, Misawa N (2003) Enzymatic synthesis of novel antioxidant flavonoids by Escherichia coli cells expressing modified metabolic genes involved in biphenyl catabolism. J Mol Catal B-Enzym 23:9–16
Shindo K, Osawa A, Nakamura R, Kagiyama Y, Sakuda S, Shizuri Y, Furukawa K, Misawa N (2004) Conversion from arenes having a benzene ring to those having a picolinic acid by simple growing cell reactions using Escherichia coli that expressed the six bacterial genes involved in biphenyl catabolism. J Am Chem Soc 126:15042–15043
Singer AC, Crowley DE, Thompson IP (2003) Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol 21:123–130
Sylvestre M (1980) Isolation method for bacterial isolates capable of growth on p-chlorobiphenyl. Appl Environ Microbiol 39:1223–1224
Sylvestre M, Toussaint JP (2011) Engineering microbial enzymes and plants to promote PCB degradation in soil: current state of knowledge. In: Koukkou AI (ed) Microbial bioremediation of non-metals current research. Caister Academic, Norfolk, pp 177–196
Sylvestre M, Macek T, Mackova M (2009) Transgenic plants to improve rhizoremediation of polychlorinated biphenyls (PCBs). Curr Opin Biotechnol 20:242–247
Trivedi P, Pandey A, Sa TM (2007) Chromate reducing and plant growth promoting activies of psychrotrophic Rhodococcus erythropolis MtCC 7905. J Basic Microb 47:513–517
Van Aken B, Correa PA, Schnoor JL (2010) Phytoremediation of polychlorinated biphenyls: new trends and promises. Environ Sci Technol 44:2767–2776
Vasilyeva GK, Strijakova ER (2007) Bioremediation of soils and sediments contaminated by polychlorinated biphenyls. Microbiology 76:639–653
Vézina J, Barriault D, Sylvestre M (2007) Family shuffling of soil DNA to change the regiospecificity of Burkholderia xenovorans LB400 biphenyl dioxygenase. J Bacteriol 189:779–788
Villacieros M, Whelan C, Mackova M, Molgaard J, Sanchez-Contreras M, Lloret J, de Carcer DA, Oruezabal RI, Bolanos L, Macek T, Karlson U, Dowling DN, Martin M, Rivilla R (2005) Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Appl Environ Microbiol 71:2687–2694
Warren R, Hsiao WWL, Kudo H, Myhre M, Dosanjh M, Petrescu A, Kobayashi H, Shimizu S, Miyauchi K, Masai E, Yang G, Stott JM, Schein JE, Shin H, Khattra J, Smailus D, Butterfield YS, Siddiqui A, Holt R, Marra MA, Jones SJM, Mohn WW, Brinkman FSL, Fukuda M, Davies J, Eltis LD (2004) Functional characterization of a catabolic plasmid from polychlorinated-biphenyl-degrading Rhodococcus sp strain RHA1. J Bacteriol 186:7783–7795
Yang XQ, Sun Y, Qian SJ (2004) Biodegradation of seven polychlorinated biphenyls by a newly isolated aerobic bacterium (Rhodococcus sp R04). J Ind Microbiol Biotechnol 31:415–420
Yang X, Liu X, Song L, Xie F, Zhang G, Qian S (2007) Characterization and functional analysis of a novel gene cluster involved in biphenyl degradation in Rhodococcus sp strain R04. J Appl Microbiol 103:2214–2224
Yun Q, Lin Z, Xin T (2009) Cometabolism and immobilized degradation of monochlorobenzoate by Rhodococcus erythropolis. Afr J Microbiol Res 3:482–486
Zhang GQ, Yang XQ, Xie FH, Chao YP, Qian SJ (2009) Cometabolic degradation of mono-chloro benzoic acids by Rhodococcus sp R04 grown on organic carbon sources. World J Microbiol Biotechnol 25:1169–1174
Acknowledgments
This work was supported by grant STPSC 356996-07 from the Natural Sciences and Engineering Research Council of Canada. We thank Prof. David Dowling, Institute of Technology, Carlow, Ireland, who generously provided Pseudomonas fluorescens F113. We thank Prof. Martina Mackova, Prague Institute of Chemical Technology, Prague, Czech Republic, for providing the PCB-contaminated soil used in this investigation. We also thank Inspec-Sol Inc., Montreal, Quebec, Canada for providing an in-kind contribution to this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Toussaint, JP., Pham, T.T.M., Barriault, D. et al. Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl Microbiol Biotechnol 95, 1589–1603 (2012). https://doi.org/10.1007/s00253-011-3824-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3824-z