Abstract
Five methylotrophic strains (UCDFST 71-1024T, UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1 and UCDFST 74-1030) from the Phaff Yeast Culture Collection (University of California Davis, USA) that were originally designated as Pichia pastoris were found to represent a novel Komagataella species. Strains of Komagataella mondaviorum sp. nov. UCDFST 71-1024T(type strain) = CBS 15017, UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1, and UCDFST 74-1030 were isolated in USA, respectively, from cottonwood tree Populus deltoides in 1971 (Davis, CA), slime flux of Quercus sp. in 1954 (CA), exudate of black oak Q. kelloggii in 1954 (Central Sierra Nevada. CA), dry frass from Salix sp. in 1968 (Soleduck Road, Olympic National Park, WA) and from flux of hackberry tree Celtis sp. in 1974 (CA). The new species was differentiated from Komagataella kurtzmanii, Komagataella pastoris, Komagataella phaffii, Komagataella populi, Komagataella pseudopastoris and Komagataella ulmi by divergence in gene sequences for D1/D2 LSU rRNA, ITS1-5.8S-ITS2, RNA polymerase subunit I and translation elongation factor-1α. Komagataella mondaviorum sp. nov. is registered in MycoBank under MB 821789.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The methanol assimilating yeasts of the genus Komagataella Y. Yamada et al., better known under the name Pichia pastoris (Guilliermond) Phaff, have a long and complex taxonomic history. P. pastoris has been also classified in five different genera: Petasospora, Saccharomyces, Zygosaccharomyces, Zymopichia and Zygowillia (Kurtzman 2011). The genus Komagataella was proposed based on the partial 18S and 26S rRNAs sequences (Yamada et al. 1995). However, the genus has been generally accepted only after discovery of Komagataella phaffii and recognition of European species Komagataella pseudopastoris (Kurtzman 2005, 2011), initially described as Pichia pseudopastoris (Dlauchy et al. 2003). Recently, three more Komagataella species have been proposed based on single strains isolated in North America: Komagataella populi, Komagataella ulmi and Komagataella kurtzmanii (Kurtzman 2012; Naumov et al. 2013, 2016). During multigene sequence analysis of methanol assimilating strains maintained at the Phaff Yeast Culture Collection as Komagataella (Pichia) pastoris, a new Komagataella species was found. In the present study, we describe the species as Komagataella mandaviorum sp. nov.
Materials and methods
Yeast strains, cultivation and phenotypic characterization
The strains and their origins are listed in Table 1. Phenotypic characterization of strains UCDFST 71-1024T, UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1 and UCDFST 74-1030, representing a novel species of the genus Komagataella, was carried out according to Yarrow (1998) and Kurtzman et al. (2011). Yeast cells were grown at 25 °C on YPD complete medium (20 g glucose, 20 g peptone, 10 g yeast extract and 20 g agar in 1 l of distilled water). Sporulation and zygote formation were induced at 22 and 25 °C on three different acetate agar media used in genetics and taxonomy of yeasts: (1) 10 g CH3COONa, 5 g KCl and 20 g agar in 1 l of distilled water; (2) 8.2 g CH3COONa, 1.8 g KCl, 2.5 g yeast extract, 1 g glucose and 15 g agar in 1 l of distilled water (McClary et al. 1959); (3) 5 g CH3COONa, 10 g KCl, 10 g glucose and 20 g agar in 1 l of distilled water (Chen et al. 2012). Also, ME medium was used (50 g malt extract and 20 g agar in 1 l of distilled water).
DNA extraction, sequencing and phylogenetic analysis
Genomic DNA was extracted from yeast cells using the Genomic DNA Purification Kit (Fermentas, Lithuania). The genes for the D1/D2 region of the large (26S) ribosomal rRNA subunit, translation elongation factor-1α (EF-1α), RNA polymerase II (subunit RPB1), and ITS1-5.8S-ITS2 were amplified and sequenced using standard oligonucleotide primers (Kurtzman 2009; Kurtzman and Robnett 1998, 2003). Amplification reactions were performed in a volume of 30 μl containing 100 ng of genomic DNA template, Taq polymerase (0.05 U, Syntol, Moscow), and the primers (50 pmol each). A Bio-Rad (USA) thermal cycler was programmed for 30 cycles of 45 s at 94 °C, 30 s at 52 °C and 2 min at 72 °C. Amplification products were separated by electrophoresis in 1% agarose gels and detected by staining with ethidium bromide. For sequencing, the amplified products were purified using the GeneClean Purification Kit (Bio101, USA) according to the manufacturer’s instructions. Direct sequencing of both strands was performed using an Applied Biosystems 3730 automated DNA sequencer according to the manufacturer’s instructions. The sequences obtained were compared with those in the GenBank database using BLAST (http://www.ncbi.nlm.nih.gov). An alignment was done visually with the program BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). A neighbour-joining tree using the Kimura 2-parameter correction was generated with the MEGA 6 software package (Tamura et al. 2013). A total of 1000 bootstrap replicates were used for analysis using 2482 aligned nucleotide positions. Alignment gaps were treated pairwise. Type cultures of Ogataea glucozyma NRRL YB-2185 and Pichia membranifaciens NRRL Y-2026 were used as outgroup species. The nucleotide sequences determined in this study have been deposited in GenBank (Table 1). The reference sequences used in the phylogenetic analysis were retrieved from GenBank under the accession numbers indicated in Table 1.
Results and discussion
The D1/D2 sequences obtained for North American strains UCDFST 71-1024T, UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1 and UCDFST 74-1030 were compared with the corresponding sequences from the GenBank database. The D1/D2 sequences of strains UCDFST 54-11.16, UCDFST 54-11.141 and UCDFST 68-967.1 were identical and differed from the corresponding sequences of UCDFST 71-1024T and UCDFST 74-1030 by one nucleotide substitution. Strains UCDFST 71-1024T and UCDFST 74-1030 had D1/D2 sequences, which differed from the corresponding sequences of the type strains of K. pseudopastoris NRRL Y-27603T and K. populi NRRL YB-455T by one and two nucleotides, respectively (Table 2). On the other hand, UCDFST 71-1024T and UCDFST 74-1030 shared identical D1/D2 sequences with K. pseudopastoris strains NRRL Y-27602 and NRRL Y-27604. The D1/D2 sequences of strains UCDFST 54-11.16, UCDFST 54-11.141 and UCDFST 68-967.1 differed by two nucleotide substitutions from the corresponding sequences of the type strains of K. populi NRRL YB-455T and K. pseudopastoris NRRL Y-27603T, and by one substitution from K. pseudopastoris NRRL Y-27602 and NRRL Y-27604 (Table 2).
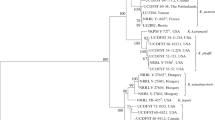
To elucidate the taxonomic status of UCDFST 71-1024T, UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1 and UCDFST 74-1030 we conducted comparative analyses of D1/D2, ITS1-5.8S-ITS2, EF-1α and RPB1 nucleotide sequences. The mitochondrial SSU rRNA gene was not used in multigene sequence analysis, since this molecular marker does not distinguish three out of the six known Komagataella species: K. phaffii, K. ulmi and K. kurtzmanii (Kurtzman 2012; Naumov et al. 2013). Based on the multigene sequence comparisons of the thirteen Komagataella strains listed in Table 1, a phylogenetic tree was generated (Fig. 1). Strains UCDFST 71-1024T, UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1 and UCDFST 74-1030 formed a clearly separated clade from the closest relatives K. pseudopastoris and K. populi (Fig. 1). The five strains shared identical RPB1 sequences and differed by not more than one substitution in D1/D2, by one substitution and two indels in ITS, and by two substitutions in EF-1α, suggesting they are conspecific. It should be noted that K. pseudopastoris is characterized by intraspecific sequence divergences. NRRL Y-27602 differed from the type strain NRRL Y-27603T and NRRL Y-27604 by 5 and 11 nucleotide substitutions, respectively, in EF-1α and RPB1, and by two substitutions and four indels in ITS1-5.8S-ITS2. On the other hand, the number of nucleotide substitutions in ITS1-5.8S-ITS2, EF-1α and RPB1 sequences among K. populi, K. pseudopastoris and the strains under examination was markedly higher and can be interpreted as interspecific sequence divergences (Table 2). Note that only D1/D2 and ITS are usually used for description of new yeast species. Phylogenetic tree based on the analysis of concatenated D1/D2 and ITS sequences is depicted in Fig. S1 (available in the online Supplementary Material). The five Komagataella strains studied is clearly differentiated by divergence in nucleotide sequences for ITS1-5.8S-ITS2. The results indicate that strains UCDFST 71-1024T, UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1 and UCDFST 74-1030 represent a distinct species. The novel species differs from its closest relatives, K. populi and K. pseudopastoris, by 10–14 nucleotide substitutions in the EF-1α gene, by 27–32 nucleotide substitutions in the RPB1 gene, and by more than 7 nucleotide substitutions and 5 indels in the ITS–5.8S region (Table 2).
Neighbour-joining tree showing phylogenetic relationship between seven species of the genus Komagataella based on combined sequences for D1/D2 and ITS1-5.8S-ITS2 rDNA, translation elongation factor-1α, and RNA polymerase II (subunit RPB1). The analysis is based on 2482 aligned positions. Bootstrap percentages > 70% for 1000 replicates are shown. Pichia membranifaciens and Ogataea glucozyma were used as the outgroup. Bar, 20 estimated base substitutions per 1000 nucleotide positions. T—type culture
Based on the multigene sequence analysis, we propose to describe a novel species: Komagataella mondaviorum sp. nov. (type strain UCDFST 71-1024T, other investigated strains UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1 and UCDFST 74-1030).
Using induced complementary auxotrophic mutants and selective growth of prototrophic hybrids on minimal medium, we demonstrated earlier that K. kurtzmanii, K. pastoris, K. phaffii, K. populi, K. pseudopastoris and K. ulmi possess a common mating type system allowing them to be crossed (Naumov 2015). Due to postzygotic isolation, the resulting interspecies hybrids are sterile, having non-viable ascospores (Naumov et al. 2016). Biogeographical factor may play a role in genetic isolation of Komagataella species. K. pastoris and K. pseudopastoris are known from Europe, whereas K. kurtzmanii, K. mondaviorum sp. nov, K. phaffii, K. populi, and K. ulmi are isolated in North America (Dlauchy et al. 2003; Kurtzman 2011; present study).
Multigene phylogenetic analysis conducted by us separated the seven Komagataella species into two main groups (Fig. 1). The first combines K. pastoris, K. kurtzmanii, K. phaffii and K. ulmi. The second includes K. mondaviorum, K. populi and K. pseudopastoris. There are numerous strains of Komagataella isolated from various tree exudates (Ulmus fulva, U. caprinifolia, Populis fremontii, P. trichocarpa, Salix sp., Quercus agrifolia, Q. emorye, Q. kelloggii and Q. suber) in North America (Miller et al. 1962; Phaff et al. 1972; Ganter et al. 1986) and Europe (Dlauchy et al. 2003). Large-scale screening of methanol-assimilating ascomycetous yeasts using a methanol-enrichment technique failed to isolate Komagataella strains in Thailand and Brazil (Limtong et al. 2013; Santos et al. 2015). Extremely rare isolates of Komagataella yeasts were documented from Japan (Phaff et al. 1972; Kodama 1974) and from Argentina (Spencer et al. 1995, 1996).
The seven species of the genus Komagataella are phenotypically very similar (Table 3). Some physiological differences are strain-variable. Taking into account that K. kurtzmanii, K. populi and K. ulmi are represented by single strains, it seems difficult to separate all seven Komagataella species from one another solely by conventional physiological tests. Consequently, multigene sequence comparisons should be used for reliable species identification. Of the four molecular markers used in this study, the ITS1-5.8S-ITS2, EF-1α and RPB1 gave congruent resolutions for species differentiation. It should be underlined that the key taxonomic molecular marker (D1/D2 LSU rRNA) fails to separate all Komagataella species (Table 2). K. mondaviorum sp. nov., K. populi and K. pseudopastoris are indistinguishable by D1/D2 sequences. Recent study of several thousand strains maintained in the CBS-KNAW collection showed that 9.5% of yeast species could not be distinguished by LSU (Vu et al. 2016).
The ITS region, currently used as one of the universal DNA barcode marker for species and genera discrimination in fungi (Schoch et al. 2012; Vu et al. 2016), is characterized by a significant interspecies divergence and a low level of intraspecies polymorphism. Indeed, all seven Komagataella species can be delineated by the ITS sequences, which differ by 7–35 substitutions and numerous indels. Note that sequence divergence of the ITS1-5.8S-ITS2 between K. pseudopastoris strains does not exceed two nucleotide substitutions (Table 2), and there was no intraspecific variation for the ITS sequences in K. pastoris and K. phaffii (Kurtzman 2009). Thus, multigene sequence divergence separating Komagataella mondaviorum sp. nov. from the other members of the genus is similar to those of the other six Komagataella species.
Description of Komagataella mondaviorum G. I. Naumov, E. S. Naumova and K. L. Boundy-Mills sp. nov.
Komagataella mondaviorum (mon.da.vio’rum. N. L. gen. masc. plur. n. mondaviorum is named in honor of the late Robert and Margrit Mondavi, honoring their tremendous impact on the CA wine industry and their generous and forward-thinking support of facilities and programs at the University of California Davis).
Growth on 5% malt extract agar
After 3 days on 5% malt extract (ME) agar at 25 °C, cells divide by multilateral budding and are spherical (2–6 μm) to ovoid (2–5 × 4–7 μm), occur singly and in pairs (Fig. 2a). Colony growth is white, butyrous and with a smooth semi-glistening surface.
Dalmau plate culture on morphology agar
After 7 days on morphology agar at 25 °C, growth under the coverglass formed neither hyphae nor pseudohyphae.
Formation of ascospores
Ascosporulation occurs on different acetate agar media after 6 days at 22 °C. The best sporulation is observed on the medium containing 5 g CH3COONa, 10 g KCl, 10 g glucose and 20 g agar in 1 l of distilled water. Asci may be unconjugated or show conjugation between a cell and its bud or between independent cells. One to four hat-shaped ascospores are formed in each ascus and they are soon liberated (Fig. 2b). In view of conjugation between cells and their buds, the species appears to be homothallic.
Fermentation and growth reactions
Glucose is fermented. Galactose, maltose, sucrose, trehalose, lactose and raffinose are not fermented. Carbon compounds: glucose, l-rhamnose, trehalose, glycerol, d-glucitol, d-mannitol (variable), dl-lactate, succinate, methanol and ethanol are assimilated; no growth occurs on galactose, l-sorbose, d-glucosamine, d-ribose, d-xylose, l-arabinose, d-arabinose, sucrose, maltose, methyl α-d-glucoside, cellobiose, salicin, melibiose, lactose, raffinose, melezitose, inulin, soluble starch, erythritol, ribitol, xylitol, l-arabinitol, galactitol, myo-inositol, d-glucono-1,5-lactone, 2-keto-d-gluconate, 5-keto-d-gluconate, d-gluconate, d-glucoronate, d-galacturonate, citrate and N-acetyl-d-glucosamine. Nitrogen compounds: ethylamine, l-lysine and cadaverine are assimilated; no growth occurs on potassium nitrate, sodium nitrite, d-glucosamine and imidazole. Growth on vitamin-free medium is negative. Growth with 0.1% cycloheximide is positive. Growth is absent with 1% acetic acid, on YM agar with 10% NaCl and on 50% w/w glucose/yeast extract (0.5%) agar. Growth at 35°°C is negative. Physiological data are from strains UCDFST 71-1024T, UCDFST 54-11.16, UCDFST 54-11.141, UCDFST 68-967.1 and UCDFST 74-1030.
The type strain is UCDFST 71-1024T isolated in 1971 by H.J. Phaff from exudate of cottonwood (Populus deltoides) in Davis, CA, USA. The strain was originally designated as P. pastoris. It is preserved as a lyophilized preparation in the Phaff Yeast Culture Collection, University of California, Davis, CA, USA. Ex-type culture has been deposited in the CBS yeast collection of the Westerdijk Fungal Biodiversity Institute (Utrecht, the Netherlands) under the designation CBS 15017T, in the All-Russian Collection of Industrial Microorganisms (Moscow, Russia) under the designation VKPM Y-4330T and in the USDA-ARS Culture Collection, National Center for Agricultural Utilization Research (Peoria, Illinois, USA) under the designation NRRL Y-63969T. The Mycobank number is MB 821789.
This study demonstrates the importance of preserving microbes in professionally managed microbe culture collections to enable future discoveries (Boundy-Mills et al. 2016). Because many yeast strains and associated data were preserved in the Phaff Yeast Culture Collection by foresighted researchers, the yeasts continue to be instrumental for discoveries and innovation, such as recent discoveries regarding oleaginous yeast species (Garay et al. 2016), glycolipid-secreting yeasts (Garay et al. 2017) and novel species of industrially important methylotrophic yeasts Komagataella and Ogataea (Kurtzman 2009; Naumov 2015; Naumov et al. 2013, 2016, 2017; Yamada et al. 1995).
References
Boundy-Mills KL, Glantschnig E, Roberts IN, Yurkov A, Casarégola S, Daniel H-M, Turchetti B (2016) Yeast culture collections in the twenty-first century: new opportunities and challenges. Yeast 33:243–260
Chen MT, Lin S, Shandil I, Andrews D, Stadheim TA, Choi BK (2012) Generation of diploid Pichia pastoris strains by mating and their application for recombinant protein production. Microb Cell Fact 11:91
Dlauchy D, Tornai-Lehoczki J, Fülöp L, Péter G (2003) Pichia (Komagataella) pseudopastoris sp. nov., a new yeast species from Hungary. Antonie Van Leeuwenhoek 83:327–332
Ganter PF, Starmer WT, Lachance M-A, Phaff HJ (1986) Yeast communities from host plants and associated Drosophila in southern Arizona: new isolations and analysis of the relative importance of hosts and vectors on community composition. Oecologia 70:386–392
Garay LA, Sitepu IR, Cajka T, Chandra I, Shi S, Lin T, German JB, Fiehn O, Boundy-Mills K (2016) Eighteen new oleaginous yeast species. J Ind Microbiol Biotechn 43:887–900
Garay LA, Sitepu IR, Cajka T, Fiehn O, Cathcart E, Fry RW, Kanti A, Joko Nugroho A, Faulina SA, Stephanandra S, German JB, Boundy-Mills KL (2017) Discovery of synthesis and secretion of polyol esters of fatty acids by four basidiomycetous yeast species in the order Sporidiobolales. J Ind Microbiol Biotechn 44:923–936
Kodama K (1974) Ascosporogenous yeasts isolated from tree exudates in Japan. Ann Microbiol Enzimol 24:215–231
Kurtzman CP (2005) Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella. Int J Sys Evol Microbiol 55:973–976
Kurtzman CP (2009) Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J Ind Microbiol Biotechnol 36:1435–1438
Kurtzman CP (2011) Komagataella Y. Yamada, Matsuda, Maeda, Mikata (1995). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 491–495
Kurtzman CP (2012) Komagataella populi sp. nov. and Komagataella ulmi sp. nov., two new methanol assimilating yeasts from exudates of deciduous trees. Antonie Van Leeuwenhoek 101:859–868
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Kurtzman CP, Robnett CJ (2003) Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res 3:417–432
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 87–110
Limtong S, Kaewwichian R, Groenewald M (2013) Ogataea kanchanaburiensis sp. nov. and Ogataea wangdongensis sp. nov., two novel methylotrophic yeast species from phylloplane in Thailand. Antonie Van Leeuwenhoek 103:551–558
McClary DD, Nulty WL, Miller GR (1959) Effect of potassium versus sodium in the sporulation of Saccharomyces. J Bacteriol 78:362–368
Miller MW, Phaff HJ, Snyder HE (1962) On the occurrence of various species of yeast in nature. Mycopath Mycol Appla 16:1–18
Naumov GI (2015) The yeast Komagataella: a genetic genus in accordance with interspecies hybridization. Microbiology (Moscow) 84:538–543
Naumov GI, Naumova ES, Tjurin OV, Kozlov DG (2013) Komagataella kurtzmanii sp. nov., a new sibling species of Komagataella (Pichia) pastoris in accordance with multigene sequence analysis. Antonie Van Leeuwenhoek 104:339–347
Naumov GI, Kondratieva VI, Meshcheryakova EV, Naumova ES (2016) Taxonomic genetics of methylotrophic yeast genus Komagataella: new biological species K. kurtzmanii. Russ J Genet 52:378–382
Naumov GI, Naumova ES, Lee Ch-Fu (2017) Ogataea haglerorum sp. nov., a new member of the Ogataea (Hansenula) polymorpha species complex. Int J Sys Evol Microbiol 67:2465–2469
Phaff HJ, Miller MW, Yoneyama M, Soneda M (1972) A comparative study of the yeast florae associated with trees on the Japanese islands and on the west coast of North America. In: Terui G (ed) Fermentation Technology Today. Society of Fermentation Technology, Osaka, pp 759–774
Santos AR, Faria ES, Lachance MA, Rosa CA (2015) Ogataea mangiferae sp. nov., a methylotrophic yeast isolated from mango leaves. Int J Syst Evol Microbiol 65:1855–1859
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci (USA) 109:6241–6246
Spencer DM, Spencer JFT, Fengler E, de Figueroa LI (1995) Yeasts associated with algarrobo trees (Prosopis spp.) in northwest Argentina: a preliminary report. J Industrial Microbiol 14:472–474
Spencer DM, Spencer JFT, de Figueroa LI, Garro O, Fengler E (1996) Yeasts associated with pods and exudates of algarrobo trees (Prosopis spp.) and species of columnar cacti in northwest Argentina. App Microbiol Biotechnol 44:736–739
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Vu D, Groenewald M, Szöke S, Cardinali G, Eberhardt U, Stielow B, de Vries M, Verkleij GJ, Crous PW, Boekhout T, Robert V (2016) DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud Mycol 85:91–105
Yamada Y, Matsuda M, Maeda K, Mikata K (1995) The phylogenetic relationships of methanol-assimilating yeasts based on the partial sequences of 18S and 26S ribosomal RNAs: the proposal of Komagataella gen. nov. (Saccharomycetaceae). Biosci Biotechnol Biochem 59:439–444
Yarrow D (1998) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 77–100
Acknowledgements
The authors thank Yuriy A. Rybakov, Sergey E. Cheperegin, Erin Cathcart and Russell Fry for technical assistance. This material is based upon work supported by the Russian Foundation for Basic Research under Grant Number 18-54-52002 MHT_a and the National Science Foundation under Grant Number 1349395, which funded, respectively, nuclear genes (EF-1α and RPB1) and ribosomal sequence analyses of yeasts from the Phaff Yeast Culture Collection. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Naumov, G.I., Naumova, E.S. & Boundy-Mills, K.L. Description of Komagataella mondaviorum sp. nov., a new sibling species of Komagataella (Pichia) pastoris. Antonie van Leeuwenhoek 111, 1197–1207 (2018). https://doi.org/10.1007/s10482-018-1028-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1028-6