Abstract
Polyol esters of fatty acids (PEFA) are amphiphilic glycolipids produced by yeast that could play a role as natural, environmentally friendly biosurfactants. We recently reported discovery of a new PEFA-secreting yeast species, Rhodotorula babjevae, a basidiomycetous yeast to display this behavior, in addition to a few other Rhodotorula yeasts reported on the 1960s. Additional yeast species within the taxonomic order Sporidiobolales were screened for secreted glycolipid production. PEFA production equal or above 1 g L−1 were detected in 19 out of 65 strains of yeast screened, belonging to 6 out of 30 yeast species tested. Four of these species were not previously known to secrete glycolipids. These results significantly increase the number of yeast species known to secrete PEFA, holding promise for expanding knowledge of PEFA synthesis and secretion mechanisms, as well as setting the groundwork towards commercialization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Surfactants, or surface active agents, are amphiphilic organic molecules used as detergents, emulsifiers, wetting agents, dispersants, foaming agents, antifoam agents, and humectants. The annual global production of surfactants was 13 million metric tons in 2008 [3]. Between 40 and 65% of surfactants are derived from petroleum [8, 34]. The remaining feedstocks are primarily plant oils such as palm or coconut oil. Due to environmental and food security concerns, renewable and more environmentally friendly alternatives are being sought. Naturally produced microbial biosurfactants are appealing alternatives because they are produced renewably and sustainably [22], have low ecotoxicity [30], are highly biodegradable [27, 30], and are active under a broad range of conditions [17, 25]. Microbial biosurfactants display a unique combination of market attractiveness and high demand growth, and market prices range $10–$30 per kg, depending on the type, the market and the application [1]. Microbial biosurfactants are used in numerous applications including industrial and household cleaning products, cosmetics, lubricants, adhesives, agrochemicals, mining, petroleum extraction and cleanup, and in pulp and paper industries.
Yeast glycolipids (GL, see Fig. 1) are a group of microbial biosurfactants comprising sophorolipids (SL), mannosylerythritol lipid (MEL), cellobiose lipids (CL) and polyol lipids (PL) [25]. PL comprise two subgroups: (1) liamocins (LM), which consist of a single partially acetylated polyol head group with three or four 3,5-dihydroxydecanoic tails polyesterified through the 5-hydroxy group and are currently the best understood (Fig. 1e) [28, 29], and (2) polyol esters of fatty acids (PEFA) produced by yeasts taxonomically close to the Rhodotorula glutinis/graminis clade [7, 46]. PEFA are amphiphilic molecules composed of an acetylated (R)-3-hydroxy fatty acid esterified through the carboxyl end to a 5 or 6 carbon polyol, typically d-mannitol or d-arabitol, with varying degrees of acetylations (Fig. 1d) [7]. They differ from LM in that the Δ5 carbon is not hydroxylated and thus, only have a single acetylated (R)-3-hydroxylated acyl chain of varying numbers of carbons, usually C16:0 and C18:0. LM polyol head group is also non acetylated, while PEFA’s polyol head group can present different degrees of acetylation. Commercial production of SL, MEL and CL requires provision of both a hydrophobic carbon source such as vegetable oil, and a hydrophilic carbon source such as glucose. In contrast, PL could be synthesized in commercially relevant amounts without a hydrophobic carbon source. A few ascomycete and basidiomycete yeast species have been reported to produce PL (see Table 1). Interestingly, all the basidiomycetes reported to produce PEFA are taxonomically close to the Rhodotorula glutinis/graminis clade [19]. Rhodotorula babjevae UCDFST 04-877 secreted five times more PEFA than its phylogenetic neighbors [7], raising the question on whether this feature could be present in related species as well.
Recommended strategies for improving biosurfactant production include identifying new hyper-producing microorganisms, genetically modifying production strains, optimizing media and growth conditions, using wastes or other low-cost raw materials, improving downstream processing, and capturing multiple co-products [9]. Addressing the first strategy to improve SL production, Kurtzman [21] analyzed 26 strains from the USDA-ARS Culture Collection (http://nrrl.ncaur.usda.gov) belonging to 18 yeast species within the Starmerella clade. This clade contains Starmerella species, several related Candida species, and numerous species that currently lack valid species names. They confirmed that clade member Candida apicola produces SL, which had been previously known [13, 44, 47]. They found three additional SL-secreting yeast species within the Starmerella clade [20, 21]. Targeting this taxonomic clade was therefore a successful strategy. It is important to point out that SL production was strain-specific. For example, of two strains of Candida apicola that were analyzed, one produced very high levels of SL, and one produced no detectable SL.
Emulating the successful strategy used by Kurtzman et al., related yeasts in the order Sporidiobolales from the Phaff Yeast Culture Collection at the University of California Davis (http://phaffcollection.ucdavis.edu) were screened for ability to synthesize and secrete PEFA when cultivated on glucose as the carbon source, without a hydrophobic substrate, under nitrogen limiting conditions. This approach simultaneously addressed several of the recommended strategies to improve production of biosurfactants: (1) Discovery of new production species could lead to development of superior industrial organisms. (2) The cost of raw materials could be lowered because sugars are roughly a third the price per kg compared to oils. (3) Because no residual vegetable oil is present to mix with product, one or more purification steps could be eliminated, reducing downstream processing costs. (4) Yeasts that are able to produce multiple co-products in addition to GL could improve the economics of the process.
Materials and methods
Chemicals and yeast strains
All chemicals were of analytical grade, except glucose (catalog number S25295B, Fisher Science Education PA, USA) which was technical grade.
Sixty-five yeast strains listed in Online Resource 1 were revived from cryopreserved stocks in the Phaff Yeast Culture Collection, University of California Davis (http://phaffcollection.ucdavis.edu) by streaking onto potato dextrose agar and incubating at room temperature up to 7 days. It must be noted that the species names of many yeast strains used in this study, and recent publications [11, 37–41] have been updated due to recent taxonomic revisions [53]. The strain ID number is unchanged. For example, yeast strain UCDFST 04-877 was formerly called Rhodosporidium babjevae [11, 37–41], and is now called Rhodotorula babjevae. The former species name is listed as a synonym in Online Resource 1.
Molecular identification
Strain identities were confirmed using ITS and partial 26S ribosomal sequencing by colony PCR using primers ITS1, ITS4 [54], NL1 and NL4 as previously described [12, 14]. Strains Rhodotorula babjevae UCDFST 67-478, UCDFST 06-542, UCDFST 04-830, UCDFST 67-102, Rhodotorula mucilaginosa UCDFST 10-221, UCDFST 67-64, and UCDFST 05-218 did not amplify with the cell lysis method previously described. Therefore, DNA was extracted and purified using the DNA Wizard Kit® (Cat. No. Promega, NY, USA) from 5 mL liquid PDB cultures inoculated with a loopful of fresh cells (≈0.02 g) streaked from a potato dextrose agar and incubated in 16 cm × 150 mm disposable borosilicate glass culture tubes (cat. number 14-961-31 Fisher Scientific, CA, USA) for 20 h. DNA extracts were successfully amplified and the corresponding PCR products were sequenced with the primers described above at The College of Biological Sciences UCDNA Sequencing Facility (University of California, Davis, CA, USA). The rest of the amplified sequences from the strains not requiring DNA extraction with the kit were sequenced with the primers described above at Beckman Coulter Genomics (Danver, MA, USA). In both cases, single pass Sanger sequencing techniques [16] were used, and identification was performed with NCBI-BLAST [2] as previously described [12, 14].
Yeast growth conditions
A loopful of fresh cells (≈0.2 g) from a freshly grown plate was suspended in 5.0 mL of sterile deionized water. To prepare a seed culture, 500 µL of this inoculum were inoculated into 50 mL bioreaction tubes (part number 229475, Celltreat Scientific Products, Shirley, MA, USA) containing 9.5 mL of Medium A, a medium with high C:N ratio (68:1) known to induce lipid accumulation in oleaginous yeasts [11, 45]. Modified Medium A contains 50 g L−1 glucose, 0.1 g L−1 calcium chloride, 0.5 g L−1 ammonium chloride, 1.5 g L−1 yeast extract, 7.0 g L−1 KH2PO4, 5 g L−1 Na2HPO4·2H2O, 1.5 g L−1 MgSO4·7H2O, 0.08 g L−1 FeCl3·6H2O, 10.0 mg L−1 Zn SO4·7H2O, 0.1 mg L−1 MnSO4·H2O, 0.1 mg L−1 CuSO4, and 0.1 mg L−1 Co(NO3)2. The cultures were then grown for 24 h at 24 °C and 200 rpm in a rotary shaker incubator (Series 25, New Brunswick Scientific Co., Edison, NJ, USA). Five mL of this seed culture were then inoculated into 500 mL baffled Erlenmeyer flasks containing 95 mL Medium A containing 50 g L−1 glucose, and stoppered with foam stoppers (Cat.# L800-D, Identi-Plugs®, Jaece Industrie, Inc., NY, USA). The cultures were incubated for 7 days at 24 °C and 200 rpm.
Cell pellets were harvested by centrifugation at 3220×g for 10 min and washed with sterile deionized water two times by centrifugation. The cell pellets were stored overnight at −80 °C and freeze-dried at −46 °C, 0.133 mbar (Freez-one® 4.5 L Freeze Dry System Model 7750020, Labconco®, Kansas City, MO, USA). The dry cells were weighed to estimate cell mass production (CMP) in terms of grams of recovered dry cells per liter of culture. PEFA were recovered by one of the following methods:
When PEFA droplets were observed in the culture, 10 mL of culture were each transferred to three tared 15 mL conical tubes and centrifuged at 3220×g for 10 min at room temperature. PEFA that sedimented to the bottom of the bottles were aspirated using 9 inch Pasteur pipettes and transferred to tared 15 mL conical tubes. Because some PEFA mix with cells during pipetting, deionized sterile water was added to the mixture, which was then vortexed and centrifuged to further separate the PEFA layer from the cells. The cells were carefully resuspended in water and pipetted out from the PEFA. The PEFA were freeze dried overnight to remove moisture and weighed to estimate grams of recovered PEFA per liter culture.
If PEFA droplets were not observed in the culture, then 10 mL of whole culture were extracted twice with 40 mL of ethyl acetate in duplicate and the solvent was removed overnight using a vacuum speed concentrator (miVac Duo®, Genevac Inc., Stone Ridge, NY, USA) programmed for non-freezing solvents (e.g. ethyl acetate) at minimum working pressure (≤6 mm Hg) and 25 °C. The dry weight of the PEFA residue was recorded.
Product characterization
PEFA products were analyzed as previously described [7]. Native PEFA molecules were analyzed using reversed-phase liquid chromatography-electrospray ionization in positive mode with a quadrupole/time-of-flight mass spectrometer (RPLC-ESI(+)-QTOFMS) in MS and MS/MS modes. Aliquots containing 10 µg mL−1 of crude PEFA in methanol were run through a system consisting of an Agilent 1290 Infinity LC system (Agilent Technologies) with a pump (G4220A), a column oven (G1316C), an autosampler (G4226A), and an Agilent 6550 iFunnel QTOFMS. Diluted samples were separated on an Acquity UPLC CSH C18 column (100 × 2.1 mm; 1.7 µm) coupled to an Acquity UPLC CSH C18 VanGuard pre-column (5 × 2.1 mm; 1.7 µm) (Waters). The column was maintained at 65 °C at a flow-rate of 0.6 mL min−1. The mobile phases consisted of (A) acetonitrile/water (60:40, v/v) with ammonium formate (10 mM) and formic acid (0.1%) and (B) isopropanol/acetonitrile (90:10, v/v) with ammonium formate (10 mM) and formic acid (0.1%). The separation was conducted under the following gradient: 0 min 15% (B); 0–2 min 30% (B); 2–2.5 min 48% (B); 2.5–11 min 82% (B); 11–11.5 min 99% (B); 11.5–12 min 99% (B); 12–12.1 min 15% (B), 12.1–15 min 15% (B). A sample volume of 1 µL was used for the injection. Sample temperature was maintained at 4 °C. The QTOF instrument was operated in ESI(+) with the following parameters: MS1 mass range: m/z 50–1700; MS/MS mass range: m/z 50–1700; collision energy: 20 eV; capillary voltage: 3 kV; nozzle voltage: 1 kV; gas temperature: 200 °C; drying gas (nitrogen): 14 L min−1; nebulizer gas (nitrogen): 35 psi; sheath gas temperature: 350 °C; sheath gas flow (nitrogen): 11 L min−1; acquisition rate MS1: 10 spectra s−1; acquisition rate MS/MS: 13 spectra s−1; total cycle time: 0.508 s; number of precursor ions per cycle: 4; mass range for selection of precursor ions: m/z 500–1200. The instrument was tuned using an Agilent tune mix (mass resolving power ~20,000 FWHM). A reference solution (m/z 121.0509, m/z 922.0098) was used to correct small mass drifts during the acquisition. MassHunter Qualitative (B.05.00) and Quantitative (B.05.01) Analysis (Agilent) software programs were used for the data processing. For the identification of the structural fragments and structural assembly the software Mass Frontier 7.0 (Thermo Scientific, Palo Alto, CA, USA) was used.
Results
Screening of yeasts for PEFA synthesis and secretion
Sixty-five yeast strains were used in this study, including PEFA-secreting basidiomycetes Rhodotorula babjevae UCDFST 04-877 [7]. Table 2 lists the 19 strains belonging to 6 species that produced more than 1 g L−1 net PEFA out of the 65 candidate yeasts. The LC–MS data revealed that overlap of triacylglycerols occurred in both strains whose PEFA was separated by physical sedimentation and water washing as well as those extracted with ethyl acetate. Therefore, a “purity percentage” is also included in Table 2, to indicate what percent of the crude extract is PEFA. Some strains carried over a conspicuously large amount of triacylglycerols, and little to no PEFA. Examples include 15 strains from the species R. ruineniae, R. sphaerocarpa, R. aff. lusitaniae, S. salmonicolor, R. dairenensis, R. glutinis, S. pararoseus, S. johnsonii, C. minutum, and O. externus which displayed purity coefficients of less than 80%, ranging from less than 1–71.6%.
The PEFA were either detected visually when the PEFA phase separated from the culture with simple gravity separation such as centrifugation, or when extracted with ethyl acetate. The PEFA that settled in the bottom of the tubes had a density higher than water, approximately 1.08 g L−1 (data not shown). A similar behavior was observed previously for LM [24] and SL [52]. Fifteen strains produced at least 3 g L−1 net PEFA. Rhodotorula aff. paludigena UCDFST 81-84 produced 12.4 g L−1 PEFA and Rhodotorula paludigena UCDFST 81-492 produced 11.7 g L−1 when grown in 50 g L−1 glucose, ranking as the top two PEFA producers identified in the present study. All nineteen PEFA producing strains identified in the present study except Rhodotorula kratochvilovae UCDFST 05-632, and Rhodotorula diobovata UCDFST 08-225, produced at least 1 g L−1 net PEFA that appeared as visible droplets in the culture. For the latter two strains, PEFA was successfully extracted with ethyl acetate.
Identification of PEFA products
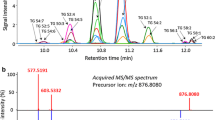
Each PEFA-secreting yeast produced a mixture of nine to thirteen structurally similar PEFA, differing in the type of sugar alcohol attached (either d-mannitol or d-arabitol), the degree of acetylation in the sugar alcohol, and the chain length of the 3-hydroxy fatty acid. The hydroxy group on position 3 of the fatty acid is the only one present throughout the fatty acid chain and is acetylated in all PEFA congeners. A total of 19 different PEFA molecular congeners with different molecular weights and retention times were detected (Table 3). An example of a chromatogram showing the PEFA profile for R. paludigena UCDFST 81-492 and the MS/MS spectra of PEFA 12 (acetylated C16:0 3-hydroxy fatty acid esterified to d-arabitol with 4 acetylations) with fragment annotations are provided in (Fig. 2a, b). PEFA were detected and PEFA profiles were determined for 20 strains that produced less than 1 g L−1 PEFA, with varying purity percentages. The PEFA profiles of the yeast species fall into 3 clusters of PEFA species (Fig. 3). Details of relative abundance of each PEFA species for the 39 strains that secreted detectable amounts are presented in Online Resource 2. The most abundant and prevalent PEFA species among all the yeast PEFA profiles is PEFA 12 (C31H52O12), present in at least 5% relative abundance across all strains that secreted trace or higher amounts of PEFA, and up to 12% relative abundance in 34 out of the 39 strains. Four more PEFA, namely PEFA 8 (C29H50O11), PEFA 9 (C32H54O13), PEFA 14 (C34H58O13), and PEFA 17 (C33H56O12) follow PEFA 12 in terms of abundance and prevalence. They are all present in at least 5% relative abundance in 30 strains and 12% relative abundance in 14 strains. PEFA 8, 12 and 17 contain d-arabitol in the structure, and PEFA 9 and 14 have d-mannitol. PEFA 12 and 17 are fully acetylated, while PEFA 8, 9 and 14 have one available hydroxy group. These characteristics are consistent with more hydrophobic physical properties.
a Overlay of RPLC-ESI(+)-MS extracted ion chromatograms of extracellular lipids isolated from Rhodotorula paludigena UCDFST 81-492. For masses and peak annotations see Table 3; remaining unannotated lipids in this figure were either not detected or at low signal intensity. b MS/MS spectrum acquired at a collision energy of 20 eV and predicted fragment structures corresponding to c acetylated 3-hydroxypalmitic acid condensed with completely acetylated d-arabitol (C31H52O12; peak 12 in Fig. 2a)
Unsupervised hierarchical clustering and heat map of yeast strains and PEFA species. The row z-score represents the deviation from the mean by standard deviation unit; red color indicates high percentage of PEFA; blue color indicates low percentage of PEFA. Yeast strain ID numbers are from the Phaff Yeast Culture Collection, University of California, Davis (color figure online)
The 19 PEFA identified in this study can be grouped into three categories in terms of their free hydroxy groups: (a) a more polar group with at least two free hydroxy groups. Six molecules belong to this group: PEFA 3 (C28H50O11), PEFA 4 (C27H48O10), PEFA 7 (C30H52O12), PEFA 10 (C30H54O11), PEFA 13 (C32H56O12) and PEFA 18 (C34H60O12). The strain producing the most abundant polar PEFA was Rhodotorula babjevae UCDFST 68-916.1, with 26.6% relative abundance; (b) a less polar group with only one free hydroxy group. Seven out of the 19 molecules are grouped here, namely PEFA 1 (C27H46O11), PEFA 2 (C30H50O13), PEFA 8 (C29H50O11), PEFA 9 (C32H54O13), PEFA 14 (C34H58O13), PEFA 15 (C31H54O11) and PEFA 19 (C36H62O13). The strain producing the most abundant fraction of these molecules was Occultifur externus UCDFST 68-934.2, with 66.9% relative abundance; (c) a non-polar group with all the hydroxy groups being acetylated. Six molecules classify in this group: PEFA 5 (C29H48O12), PEFA 6 (C32H52O14), PEFA 11 (C34H56O14), PEFA 12 (C31H52O12), PEFA 16 (C36H60O14) and PEFA 17 (C33H56O12). The strain displaying the highest abundance is Rhodotorula dairenensis UCDFST 68-257 with 88.4% relative abundance. Seven out of the 19 molecules contain d-arabitol, while the rest contain d-mannitol. No other sugar alcohols were detected. Fatty acid chain length ranged from 12 to 20 carbons, in even number increments. The most abundant chain length identified was hexadecanoate (C16:0), and the second most abundant was octadecanoate (C18:0). The highest abundance of medium chain length (C12:0 and C14:0) molecules was seen in Rhodotorula kratochvilovae UCDFST 05-632, while the highest abundance of C20:0 was observed for Cystobasidium minutum UCDFST 68-280. In average, seven molecular species accounted for 78.5% of PEFA abundance across the whole panel, namely PEFA 12 (C29H50O11), PEFA 8 (C32H54O13), PEFA 17 (C34H56O14), PEFA 9 (C31H52O12), PEFA 14 (C34H58O13), PEFA 15 (C31H54O11) and PEFA 11 (C33H56O12) with decreasing relative abundance percentages: 21.4, 12.6, 11.5, 10.5, 9.7, 7.7 and 5.1%, respectively. No unsaturated fatty acyl chains were present in the 19 PEFA identified. This is a striking difference from the intracellular oils reported for these strains, where the bulk of the acyl chains in the intracellular triacylglycerols are unsaturated [11, 41], in some cases with up to 3 double bonds in the acyl chains. The nineteen structures are described in Table 3.
Unsupervised hierarchical clustering and heat map of strains and PEFA species revealed three types of patterns (see Fig. 3). The first denotes an association between Rhodotorula paludigena/Rhodotorula aff. paludigena species and a PEFA profile where PEFA 8 is highly predominant, followed by PEFA 12. These PEFA contain d-arabitol. The second pattern involves Rhodotorula babjevae strains and a PEFA profile dominated by PEFA 12, which is fully acetylated and contains d-arabitol as well. The third pattern includes Rhodotorula kratochvilovae and Rhodotorula diobovata, the other two newly identified secreting yeasts, and a high percentage of PEFA 11, 16 and 17. The first two PEFA contain d-mannitol. There are two exceptions to these tendencies, namely R. babjevae UCDFST 68-916.1 and R. paludigena UCDFST 09-163, which display different patterns compared to the other strains from the same species. These results show that PEFA secretion is primarily species specific, but both qualitative and quantitative strain differences exist as well. The applications for the different PEFA compositions may differ: PEFA from R. babjevae should be more hydrophobic than those coming from R. paludigena due to fewer free hydroxy groups. More research is needed to identify optimal applications for each PEFA profile.
Discussion
Before this study, only seven species of yeasts were known to naturally synthesize and secrete PL (see Table 1). This study increased the number of known species by adding species Rhodotorula diobovata, Rhodotorula kratochvilovae, Rhodotorula paludigena, Rhodotorula aff. paludigena. The taxonomic term “aff.” or affinis means it is a new species; the closest valid species is R. paludigena. The UCDFST (Phaff Yeast Culture Collection) strain used in this study is the only known representative of this novel species. These yeast species belong to a taxonomic clade that was recently revised [53], which resulted in genus name changes, as described above.
These four yeasts produce at least 1 g L−1 with PEFA purities above 80%. Additional yeasts producing trace amounts of PEFA are also reported. The two highest PEFA producers in the present study, namely R. aff. paludigena UCDFST 81-84 and R. paludigena UCDFST 81-492, achieved glucose conversions of 0.25 and 0.23 g of PEFA per gram of glucose respectively. Cajka et al. obtained values equivalent to a glucose conversion of 0.086 g of PEFA per gram of glucose from R. babjevae UCDFST 04-877 and Tulloch et al. in the 1960s documented values equivalent to a glucose conversion of 0.05 g of PEFA per gram of glucose from R. azoricum (previously called R. glutinis or R. graminis) CBS 4648 [7, 46]. The R. paludigena and R. aff. paludigena strains thus converted 2.9 times and 2.7 times more glucose into PEFA compared to Cajka et al. and 5 times and 4.6 times more compared to Tulloch et al. [7, 46]. All these new PEFA-secreting yeasts present significant opportunity to expand our knowledge of synthesis and secretion of structurally complex products, which may be beneficial for development of tools for synthesis and secretion of a broad variety of oleochemicals beyond PEFA and other GL.
The function of PEFA as well as other yeast GL in natural ecosystems has been the subject of speculation. For example, the GL may aid in modifying leaf surface permeability, possibly facilitating uptake of hydrophobic carbon sources such as the long chain waxes present in the cuticle on the surface of leaves [6, 35]. Other theories include secretion of yeast GL as an external form of carbon storage [51]. Others have suggested the use of GL as anti-microbial and anti-fungal compounds [36, 42]. Indeed, LM were shown to display selective antibacterial activity against six Streptococcus species [4], even with different polyol head groups [28]. Many of the PEFA-secreting yeasts identified in this study were isolated from plant surfaces and insects, consistent with the first theory. PEFA production is more apparent in the Rhodotorula glutinis/graminis clade, comprising R. glutinis, R. graminis, R. babjevae and R. diobovata species, as well as in the R. paludigena clade, which appear in the top branches in Fig. 4. PEFA production was negligible or undetected in other phylogenetic branches, suggesting an evolutionary pattern. Furthermore, the highest three producers (R. aff. paludigena UCDFST 81-84, R. paludigena UCDFST 81-492 and R. babjevae UCDFST 04-877) in the present work were originally isolated from plant surfaces (Table 2). Six more PEFA producing strains (R. paludigena UCDFST 09-163, R. babjevae UCDFST 67-458, R. graminis 05-503, R. paludigena 82-646.2, R. babjevae UCDFST 68-916.1, R. graminis UCDFST 05-613, see Table 2) originated from plant surfaces/plant surface interactions as well. All these strains agglomerate in the R. glutinis/graminis/babjevae and the R. paludigena clades, which, as mentioned before, are located in the top branches of the phylogenetic tree depicted in Fig. 4. Only two PEFA secreters identified in this study had source habitats unrelated to plant surfaces/plant surface interactions.
Phylogenetic tree describing the different yeast species involved in the study. The tree was made using version 7.6 of the CLC Sequence Viewer® (QIAGEN Aarhus, Denmark), using the following parameters: Gap open cost value equal to 10, a gap extension cost equal to one, Neighbor Joining construction method, Jukes-Cantor nucleotide and bootstrap analysis based on 1000 replicates. The yeast located at the top branches of the tree, were active PEFA secreters, while those strains on the bottom branches of the tree had little to no PEFA secretion. The tree was built based on differences and similarities in the yeast ITS rDNA. The number in brackets corresponds to the Genbank accession number for the ITS rDNA sequence. Samples with less than 1% pure PEFA are expressed as <1 g L−1. T stands for type strain. NT stands for neotype strain
The production of PEFA by these yeasts differs significantly from that of previously known GL secreting yeasts such as SL-secreting Starmerella bombicola [51, 52] in that significant levels of PEFA are produced when cultivated on glucose as the carbon source, without a hydrophobic co-substrate. Similar results were reported previously for LM, where yields of up to 6.0 g L−1 were obtained from 50 g L−1 sucrose by Aureobasidium pullulans strain CU 39 (NRRL 58551) [24], and up to 35 g L−1 from 120 g L−1 glucose by Aureobasidium sp. strain A-21M (although the latter value came from collecting 30 mL cultures to add up a total volume of about 1 L) [18]. The consequences of this property are quite significant for commercial production of PEFA for two reasons. First, the cost of glucose and other carbohydrates is significantly lower per kilogram than fatty acids and other hydrophobic co-substrates. Second, because residual lipids do not mingle with the GL product, purifying the product would involve simpler, less costly downstream processing. Commercial production of SL requires an ethyl acetate extraction to separate the SL and other hydrophobic materials from the spent media, then a hexane extraction to remove residual input vegetable oils or fatty acids from SL [3, 51, 52]. Elimination of one or both of these steps would reduce both capital expenses for constructing facilities appropriate to handle flammable solvents, and operating expenses. Phase separation of PEFA produced by the basidiomycete yeasts described in the study suggests that a production technology with fewer or no solvent extraction steps is feasible.
These newly identified PEFA-secreting yeast species all are classified in phylum Basidiomycota. Basidiomycetes are significantly different from ascomycetes morphologically, genetically, and physiologically. The phylum names derive from differences in spore formation. Basidiomycetes have much higher GC content, often over 60%, which can lead to difficulties with PCR and DNA sequencing. Basidiomycetes have a very different cell wall structure [33], which affects methods used to harvest intracellular products as well as genetic transformation techniques. As a result, fewer genome sequences and genetic tools are available for basidiomycetous yeasts [26]. However, some genomes have been sequenced, and genetic tools are being developed for some species with biotechnology value such as oleaginous yeast Rhodotorula toruloides [55], which, like the PEFA-secreting yeasts reported here, is in the taxonomic order Sporidiobolales.
Basidiomycetes do have some advantages. In general, basidiomycetes, including those in the order Sporidiobolales, are able to assimilate a broader range of carbon sources, particularly pentoses that are utilized by few of the GL-secreting ascomycete species [37]. Some basidiomycetous yeasts can also grow without supplemented vitamins, reducing culture medium costs.
The basidiomycetous yeasts identified in this study simultaneously synthesize high levels of intracellular triacylglycerols, accumulating 40–65% oil by dry weight [11, 41], indicating efficient citrate flux from mitochondria to generate a rich pool of acetyl CoA, that will maintain the cytosolic fatty acid synthase actively producing fatty acids. This might explain why the yeast uses fatty acids as building blocks for PEFA synthesis [10]. In addition there are three main questions that remain unsolved and required further work towards elucidating the biosynthetic pathway of PEFA. First, further research is needed to understand how the hydroxylation on position Δ3 of the fatty acyl moiety occurs. In other yeast GL systems, like SL production by species Starmerella bombicola, hydroxylation on the ω or ω–1 position occurs through a cytochrome P-450 enzyme that presumably is bound to the outer leaflet of the endoplasmic reticulum [48, 49]. Second, there might be a reductive step in the biosynthetic pathway to convert glucose to d-mannitol. Stodola et al. reported the existence of an NADP dependent mannitol dehydrogenase displaying high activity in cell free extracts from extracellular PEFA producing strains of Rhodotorula glutinis [44]. More research is needed to identify the particular location of the enzyme, and how glucose is reduced to mannitol as part of the biosynthetic pathway [44]. In the case of LM, it was shown that Aureobasidium pullulans strain NRRL 50380 was able to directly incorporate (d-arabitol, d-xylitol, d-ribitol and l-threitol) into the LM structures, whereas dosing of sugars like sucrose, d-fructose, d-mannose and d-arabinose resulted in LM having mannitol head groups [28]. Therefore A. pullulans is able to convert the sugars to mannitol, and incorporate certain polyols into the LM backbone. It is not yet known whether the basidiomycetous yeasts involved in the present study are capable of such behavior, though structural similarities between LM and PEFA suggest this may be possible. Finally, the genomes of those yeasts secreting either SL, MEL or CL have been sequenced, and genes responsible for synthesis of GL have been identified [31], as has the transporter responsible for secretion in the case of SL [43]. The SL transporter protein has been described as an ABC transmembrane transporter [50]. Further analysis is required to determine whether these newly identified PEFA-secreting yeasts possess similar synthesis and/or secretion gene clusters.
This study validates the strategy of screening taxonomic relatives to identify additional yeast species with targeted properties. For example, Kurtzman measured SL production by 26 strains belonging to 18 yeast species in the taxonomic clade that includes Starmerella bombicola [21]. SL production was not detected in most strains tested. Because Candida is a polyphyletic genus, with hundreds of species spread across numerous taxonomic clades within Ascomycota, a strong knowledge of yeast taxonomy enabled selection of appropriate species within the targeted clade. Access to a large and diverse culture collection (the USDA-ARS Culture Collection, http://nrrl.naur.usda.gov/cgi-bin/usda/) also enabled that study.
There were many parallels in the current study. A large and diverse culture collection was tapped to acquire a diversity of native yeast strains. Roughly half of the known yeast species in the targeted clade within the order Sporidiobolales [19] were available from the Phaff Yeast Culture Collection (http://phaffcollection.udcavis.edu). Taxonomic expertise enabled selection of appropriate yeast species for this study, as the genus Rhodotorula is polyphyletic. Novel species currently lacking valid species names were discovered to produce PEFA in the current study. To indicate new species, the taxonomic term “affinis” (abbreviated “aff.”) is placed between the genus and the species names of the closest valid species. A novel species found to synthesize and secrete PEFA in this study is Rhodotorula aff. paludigena. The Phaff collection strain is the only known strain of this species. Further work is needed to fully describe these species and assign a valid species name.
Variation of properties among strains of a given yeast species is often observed, such as enzymatic activity [12], inhibitor tolerance [40] and lipid content [11, 41]. When Kurtzman measured SL production by two strains of Candida apicola, one produced the highest levels seen in their study, and the other did not produce detectable levels. Similarly, we found different PEFA secretion levels when comparing multiple strains of Rhodotorula babjevae. Strain UCDFST 04-877 secreted 8.7 g L−1, strain UCDFST 05-775 secreted 6.3 g L−1 and strain UCDFST 68-916.1 secreted 1.1 g L−1 crude PEFA extract, all with purity above 97% suggesting different phenotypes within the same species.
The first two digits of the Phaff collection ID number indicate the year the strain was deposited in the collection. Yeasts listed in Table 2 were acquired over the last five decades. These observations dramatically demonstrate the importance of preserving a broad variety of microbes in professionally managed collections to enable future discoveries [5], of clearly designating the yeast collection and strain ID number in publications, and of utilizing the same strain in subsequent studies to ensure consistent results.
Discovery of these PEFA-secreting yeasts may aid in improving production of renewable, sustainable, environmentally friendly surfactants for use in household and industrial cleaning products, as well as many other applications. Further studies including optimization of production, technoeconomic analysis and examination of surfactant activities will help determine whether these yeasts have the potential to alter global surfactant production and use.
References
Transparency Market Research (2014) Microbial biosurfactants market: global industry analysis, size, share, growth, trends and forecast 2014– 2020. Transparency Market Research, Albany, pp 1–74
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ashby RD, McAloon AJ, Solaiman DK, Yee WC, Reed M (2013) A process model for approximating the production costs of the fermentative synthesis of sophorolipids. J Surfactants Deterg 16:683–691
Bischoff KM, Leathers TD, Price NP, Manitchotpisit P (2015) Liamocin oil from Aureobasidium pullulans has antibacterial activity with specificity for species of Streptococcus. J Antibiot 68:642–645
Boundy-Mills KL, Glantschnig E, Roberts IN, Yurkov A, Casaregola S, Daniel HM, Groenewald M, Turchetti B (2016) Yeast culture collections in the twenty-first century: new opportunities and challenges. Yeast 33:243–260
Bunster L, Fokkema NJ, Schippers B (1989) Effect of surface-active Pseudomonas spp. on leaf wettability. Appl Environ Microbiol 55:1340–1345
Cajka T, Garay LA, Sitepu IR, Boundy-Mills KL, Fiehn O (2016) Multiplatform mass spectrometry-based approach identifies extracellular glycolipids of the yeast Rhodotorula babjevae UCDFST 04-877. J Nat Prod 79:2580–2589
Deleu M, Paquot M (2004) From renewable vegetables resources to microorganisms: new trends in surfactants. C R Chim 7:641–646
Dhanarajan G, Sen R (2014) Cost analysis of biosurfactant production from a scientist’s perspective. Biosurfactants 159:153
Garay L, Boundy-Mills K, German J (2014) Accumulation of high value lipids in single cell microorganisms: a mechanistic approach and future perspectives. J Agric Food Chem 62:2709–2727
Garay LA, Sitepu IR, Cajka T, Chandra I, Shi S, Lin T, German JB, Fiehn O, Boundy-Mills KL (2016) Eighteen new oleaginous yeast species. J Ind Microbiol Biotechnol 43:887–900
Golomb BL, Morales V, Jung A, Yau B, Boundy-Mills KL, Marco ML (2013) Effects of pectinolytic yeast on the microbial composition and spoilage of olive fermentations. Food Microbiol 33:97–106
Gorin P, Spencer J, Tulloch A (1961) Hydroxy fatty acid glycosides of sophorose from Torulopsis magnoliae. Can J Chem 39:846–855
Hamby KA, Hernández A, Boundy-Mills K, Zalom FG (2012) Associations of yeasts with spotted-wing Drosophila (Drosophila suzukii; Diptera: Drosophilidae) in cherries and raspberries. Appl Environ Microbiol 78:4869–4873
Johnson V, Singh M, Saini VS, Adhikari DK, Sista V, Yadav NK (1992) Bioemulsifier production by oleaginous yeast Rhodotorula glutinis IIP-30. Biotechnol Lett 14:487–490
Khan AS, Wilcox AS, Polymeropoulos MH, Hopkinsz IA (1992) Single pass sequencingand physical and genetic mapping of human brain cDNAs. Nat Genet 2:180–185
Khan MSA, Singh B, Cameotra SS (2014) Biological applications of biosurfactants and strategies to potentiate commercial production. Biosurfactants 159:269
Kurosawa T, Sakai K, Nakahara T, Oshima Y, Tabuch T (1994) Extracellular accumulation of the polyol lipids, 3, 5-dihydroxydecanoyl and 5-hydroxy-2-decenoyl esters of arabitol and mannitol, by Aureobasidium sp. Biosci Biotechnol Biochem 58:2057–2060
Kurtzman C, Fell J, Boekhout T (2011) The yeasts: a taxonomic study. Elsevier, Oxford
Kurtzman CP (2012) Candida kuoi sp. nov., an anamorphic species of the Starmerella yeast clade that synthesizes sophorolipids. Intl J Syst Evol Microbiol 62:2307–2311
Kurtzman CP, Price NP, Ray KJ, Kuo T-M (2010) Production of sophorolipid biosurfactants by multiple species of the Starmerella (Candida) bombicola yeast clade. FEMS Microbiol Lett 311:140–146
Makkar RS, Cameotra SS, Banat IM (2011) Advances in utilization of renewable substrates for biosurfactant production. AMB Express 1:1–19
Manitchotpisit P, Leathers TD, Peterson SW, Kurtzman CP, Li X-L, Eveleigh DE, Lotrakul P, Prasongsuk S, Dunlap CA, Vermillion KE, Punnapayak H (2009) Multilocus phylogenetic analyses, pullulan production and xylanase activity of tropical isolates of Aurebasidium pullulans. Mycol Res 113:1107–1120
Manitchotpisit P, Price NP, Leathers TD, Punnapayak H (2011) Heavy oils produced by Aureobasidium pullulans. Biotechnol Lett 33:1151–1157
Marchant R, Banat IM (2012) Microbial biosurfactants: challenges and opportunities for future exploitation. Trends Biotechnol 30:558–565
McCluskey K, Wiest A, Boundy-Mills K (2014) Genome data drives change at culture collections. In: Nowrousian M, Esser K (eds) Fungal genomics. Springer, Berlin, pp 81–96
Mulligan CN (2005) Environmental applications for biosurfactants. Environ Pollut 133:183–198
Price NP, Bischoff KM, Leathers TD, Cossé AA, Manitchotpisit P (2016) Polyols, not sugars, determine the structural diversity of anti-streptococcal liamocins produced by Aureobasidium pullulans strain NRRL 50380. J Antibiot. doi:10.1038/ja.2016.92
Price NP, Manitchotpisit P, Vermillion KE, Bowman MJ, Leathers TD (2013) Structural characterization of novel extracellular liamocins (mannitol oils) produced by Aureobasidium pullulans strain NRRL 50380. Carbohydr Res 370:24–32
Renkin M (2003) Environmental profile of sophorolipid and rhamnolipid biosurfactants. Riv Ital Sostanze Gr 80:249–252
Roelants SL, De Maeseneire SL, Ciesielska K, Van Bogaert IN, Soetaert W (2014) Biosurfactant gene clusters in eukaryotes: regulation and biotechnological potential. Appl Microbiol Biotechnol 98:3449–3461
Ruinen J, Deinema MH (1964) Composition and properties of the extracellular lipids of yeast species from the phyllosphere. Antonie Van Leeuwenhoek 30:377–384
Ruiz-Herrera J, Ortiz-Castellanos L (2010) Analysis of the phylogenetic relationships and evolution of the cell walls from yeasts and fungi. FEMS Yeast Res 10:225–243
Rust D, Wildes S (2008) Surfactants: a market opportunity study update. OmniTech International Ltd, Midland
Schreiber L, Krimm U, Knoll D, Sayed M, Auling G, Kroppenstedt RM (2005) Plant–microbe interactions: identification of epiphytic bacteria and their ability to alter leaf surface permeability. New Phytol 166:589–594
Shah V, Badia D, Ratsep P (2007) Sophorolipids having enhanced antibacterial activity. Antimicrob Agents Chemother 51:397–400
Sitepu I, Garay L, Sestric R, Levin D, Block DE, German J, Boundy-Mills K (2014) Oleaginous yeasts for biodiesel: current and future trends in biology and production. Biotechnol Adv 32:1336–1360
Sitepu I, Ignatia L, Franz A, Wong D, Faulina S, Tsui M, Kanti A, Boundy-Mills K (2012) An improved high-throughput Nile red fluorescence assay for estimating intracellular lipids in a variety of yeast species. J Microbiol Methods 91:321–328
Sitepu I, Jin M, Fernandez J, Sousa L, Balan V, Boundy-Mills K (2014) Identification of oleaginous yeast strains able to accumulate high intracellular lipids when cultivated in alkaline pretreated corn stover. Appl Microbiol Biotechnol 98:7645–7657
Sitepu I, Selby T, Zhu S, Lin T, Boundy-Mills K (2014) Carbon source utilization and inhibitor tolerance of 45 oleaginous yeast species. J Ind Microbiol Biotechnol 41:1061–1070
Sitepu IR, Sestric R, Ignatia L, Levin D, Bruce German J, Gillies LA, Almada LA, Boundy-Mills KL (2013) Manipulation of culture conditions alters lipid content and fatty acid profiles of a wide variety of known and new oleaginous yeasts species. Bioresour Technol 144:360–369
Sleiman JN, Kohlhoff SA, Roblin PM, Wallner S, Gross R, Hammerschlag MR, Zenilman ME, Bluth MH (2009) Sophorolipids as antibacterial agents. Ann Clin Lab Sci 39:60–63
Soetaert W, Van Bogaert I (2010) Sophorolipid transporter protein. United States Patent Application #US 20120311741 A1
Stodola FH, Deinema MH, Spencer J (1967) Extracellular lipids of yeasts. Bacteriol Rev 31:194
Suutari M, Priha P, Laakso S (1993) Temperature shifts in regulation of lipids accumulated by Lipomyces starkeyi. J Am Oil Chem Soc 70:891–894
Tulloch A, Spencer J (1964) Extracellular glycolipids of Rhodotorula species: the isolation and synthesis of 3-d-hydroxypalmitic and 3-d-hydroxystearic acids. Can J Chem 42:830–835
Tulloch A, Spencer J, Deinema M (1968) A new hydroxy fatty acid sophoroside from Candida bogoriensis. Can J Chem 46:345–348
Van Bogaert IN, Demey M, Develter D, Soetaert W, Vandamme EJ (2009) Importance of the cytochrome P450 monooxygenase CYP52 family for the sophorolipid-producing yeast Candida bombicola. FEMS Yeast Res 9:87–94
Van Bogaert IN, Develter D, Soetaert W, Vandamme EJ (2007) Cloning and characterization of the NADPH cytochrome P450 reductase gene (CPR) from Candida bombicola. FEMS Yeast Res 7:922–928
Van Bogaert IN, Holvoet K, Roelants SL, Li B, Lin YC, Van de Peer Y, Soetaert W (2013) The biosynthetic gene cluster for sophorolipids: a biotechnological interesting biosurfactant produced by Starmerella bombicola. Mol Microbiol 88:501–509
Van Bogaert IN, Saerens K, De Muynck C, Develter D, Soetaert W, Vandamme EJ (2007) Microbial production and application of sophorolipids. Appl Microbiol Biotechnol 76:23–34
Van Bogaert IN, Zhang J, Soetaert W (2011) Microbial synthesis of sophorolipids. Process Biochem 46:821–833
Wang Q-M, Yurkov A, Göker M, Lumbsch HT, Leavitt S, Groenewald M, Theelen B, Liu X-Z, Boekhout T, Bai F-Y (2015) Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Stud Mycol 81:149–189
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc 18:315–322
Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, Zhou YJ, Jin G, Ye M, Zou H (2012) A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun 3:1112
Acknowledgements
The authors are grateful to Enrique Fernandez, Silviana Tjahyono, and Ting Lin for technical assistance. This research was funded by Grant Number U01TW008160 from the NIH Fogarty International Center, the NIH Office of Dietary Supplements, the National Science Foundation and the Department of Energy. This project was supported by the USDA Agricultural Food Research Initiative of the National Food and Agriculture, USDA, Grant Number 35621-04750. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Fogarty International Center or the National Institutes of Health, the Office of Dietary Supplements, the National Science Foundation, the Department of Energy, or the Department of Agriculture. Funding by NIH HL113452 and NIH DK097154 (to OF) is greatly appreciated. NIH instrument funding by NIH S10-RR031630 (to OF) is acknowledged. This work was supported by the Science Translation and Innovation Research (STAIR) Grant Program of the University of California Davis, and by the Consejo Nacional de Ciencia y Tecnología (CONACYT) Grant Number 291795. Yeasts Rhodosporidiobolus poonsookiae UCDFST 10-441, Sporidiobolus ruineniae UCDFST 10-1058, UCDFST 10-1109, UCDFST 12-776, Rhodotorula mucilaginosa UCDFST 13-478, Sporobolomyces bannaensis UCDFST 10-421, 10-451, 10-453 and 11-470 used in this study were isolated and identified as part of a collaborative project with Indonesian Institute of Sciences (LIPI); Research, Development and Innovation Agency, the Ministry of Environment and Forestry; and the Government of the Republic of Indonesia. All authors have agreed to submit this manuscript to the “Journal of Industrial Microbiology and Biotechnology”. The authors are grateful to the anonymous reviewers, whose suggestions greatly improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Luis A. Garay, Irnayuli R. Sitepu, and Tomas Cajka have contributed equally to the realization of the manuscript, and are co-first authors.
Agustinus Joko Nugroho: Deceased.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Garay, L.A., Sitepu, I.R., Cajka, T. et al. Discovery of synthesis and secretion of polyol esters of fatty acids by four basidiomycetous yeast species in the order Sporidiobolales. J Ind Microbiol Biotechnol 44, 923–936 (2017). https://doi.org/10.1007/s10295-017-1919-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-017-1919-y