Abstract
In 2013 and 2014, an extensive survey of bacterial wilt in Myanmar was performed, and 70 strains of Ralstonia solanacearum (Rs) were collected from wilting plants of tomato, potato, chili and eggplant. Myanmar Rs strains were characterized by traditional and molecular methods. Polymerase chain reaction (PCR) test using Rs-specific primer set amplified one specific band (281-bp) from template DNA of all strains. Pathogenicity tests on the four solanaceous plants differentiated the strains into six pathogenic groups. Biovar determination tests showed that biovar 3 strains predominated (63%) among all Rs strains. Biovar 4 strains (7%) were obtained from both tomato and chili strains, whereas biovar 2 (30%) strains were isolated only from potato. Multiplex-PCR analysis indicated that tomato, eggplant and chili strains belonged to phylotype I, whereas potato strains comprised phylotype I and phylotype II. Strains in phylotype I, which was suggested to have originated from Asia, were the most prevalent in all surveyed areas. Phylogenetic analysis based on the endoglucanase (egl) gene sequences revealed that Myanmar strains partitioned into two major clusters that corresponded to phylotype I and II. Strains in phylotype I were further divided into seven subclusters, each corresponding to a distinct sequevar (15, 17, 46, 47, 48, unknown 1 or unknown 2). All strains in phylotype II belonged to sequevar 1. This is the first comprehensive report of the presence of diverse Rs strains in Myanmar.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial wilt caused by Ralstonia solanacearum (Yabuuchi et al. 1995) (Rs) is one of the most devastating bacterial diseases and distributed throughout the world including tropical, subtropical and warm temperate regions (Buddenhagen and Kelman 1964; Elphinstone 2005; Hayward 1991; Kelman 1953). Rs attacks 200 species representing 50 families of plants including Solanaceae, Zingibearaceae and Musaceae (Hayward 1991, 1994). Because of its broad host and geographical range and limited control measures, Rs causes severe crop losses (Elphinstone 2005).
Rs exists as a heterogeneous group of related strains and is classified into subgroups based on pathogenicity, host range and geographical area to predict the behavior of identified Rs strains (Fegan and Prior 2005; Prior and Fegan 2005). Traditionally, Rs has been divided into five races [related to the ability to wilt members of the family Solanaceae (race 1), banana (race 2), potato and tomato in temperate conditions (race 3), ginger (race 4), and mulberry (race 5)] (Buddenhagen et al. 1962; He et al. 1983; Pegg and Moffett 1971), and six biovars (Hayward 1964, 1994; He et al. 1983) based on utilization of different disaccharides and hexose alcohols. Strains belonging to biovars 1 and 2 have a worldwide distribution, and biovars 3, 4 and 5 strains are dominantly present in Asian countries (Ji et al. 2007). Each race does not directly correlate with each biovar, except race 3 is equivalent to biovar 2 (Hayward 1991).
Recent DNA-based approaches have been developed to enhance the understanding of the genetic diversity of Rs. A new hierarchical classification scheme (phylotype) was developed based on the DNA sequences of intergenic transcribed spacer (ITS) region of ribosomal RNA gene (Fegan and Prior 2005). Using this new scheme, strains that broadly originate from the same geographic location are subdivided into four phylotypes namely phylotype I (originating from Asia), II (from the Americas), III (from Africa) and IV (mainly from Indonesia) (Fegan and Prior 2005). Each phylotype further consists of small subgroups (sequevar), which are defined as strains with less than 1% nucleotide variation in the endoglucanase (egl) gene locus (Prior and Fegan 2005; Poussier et al. 2000; Wicker et al. 2012). Based on the comparative analysis of the conserved region of egl, worldwide Rs strains have been divided into more than 50 distinct sequevars. (Wicker et al. 2012; Horita et al. 2014).
In Asian countries such as Japan, China, India, Korea, Indonesia and Malaysia, Rs strains have been analyzed using the classification proposed by Fegan and Prior (2005). In Myanmar, bacterial wilt disease affected about 92.5% of the growers in surveyed villages of Kalaw township, Shan state during the 2008 post-monsoon (Zaw 2010). Bacterial wilt disease can cause complete loss of potato yields in Myanmar (Lwin 2006). But because various solanaceous crops are cultivated under diverse climatic conditions in Myanmar, the distribution and identity of the Rs strains in a particular locale needs to be estimated. Little information is still available to study the ecological view and molecular characterization of Rs species in Myanmar. The objectives of this study were, therefore, to identify the race, biovar, phylotype and sequevar levels of the Myanmar Rs strains from various solanaceous plants in different growing areas inside Myanmar and assess the genetic diversity of the strains.
Materials and methods
Collection of diseased plants
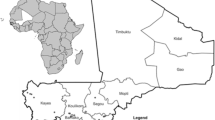
Collection of diseased plants was conducted at Heho, Kalaw in Shan state, Pyin Oo Lwin, Nyaung-U in Mandalay region, Tatkon, Naypyidaw (new capital) in Naypyidaw region (which is newly established and separated from Mandalay region in 2011) and Monywa in Sagaing region (Fig. 1a, b) during 2013 and 2014. The climatological data of each region was indicated in Table 1.
Isolation of R. solanacearum
Wilted tomato, potato, chili, eggplant stems and cuttings of potato tubers were dipped in 70% (v/v) ethanol for 30 s, surface sterilized with 5% (w/w) sodium hypochlorite for 3 min, and rinsed in sterilized distilled water for three times. Thereafter, infected stems or tubers were dipped in sterilized distilled water to obtain turbid bacterial oozes. One loopful of bacterial suspension was streaked on 2,3,5-triphenyltetrazolium chloride (TTC) plate containing 1 g casamino acids, 10 g peptone, 5 g glucose and 50 mg TTC, 15 g agar per liter (Kelman 1954) and incubated at 30 °C for 2 days. Whitish fluidal colonies were serially streaked onto TTC plates to obtain purified bacterial cultures with homogenous colony morphology. Bacterial strains used in this study are listed in Table 2. To identify and comparatively analyze the Myanmar Rs strains with foreign Rs strain, we used one reference strain PD441 (potato isolate in Sweden, belonging to race 3 biovar 2 and phylotype II sequevar 1) (Cellier and Prior 2010; Horita and Tsuchiya 2001).
Pathogenicity test
Tomato (cv. Sekaiichi), potato (cv. Nishiyutaka), chili (cv. Himetougarashi) and eggplant (cv. Hitennaganasu) were used for pathogenicity tests. Two-week-old seedlings of tomato, eggplant and chili were transplanted into a 9-cm-diameter plastic pot containing 3:1 horticultural soil–vermiculite mixture and grown in a greenhouse at 27–30 °C. Potato bud pieces from healthy potato tubers were planted and grown as well. Plants at the fourth to fifth true-leaf stage were inoculated using a stem inoculation technique (Janse 1991) and a sterilized tooth pick dipped in an inoculum suspension [ca. 108 colony-forming unit (cfu)/mL sterilized distilled water]. Three plants of each host were inoculated with each strain. Sterilized distilled water was used as a negative control treatment. Disease severity of wilting was recorded at 2-day intervals (He et al. 1983; Horita and Tsuchiya 2001) after inoculation on the following scale: 0 = healthy, 1 = ≤25% of total plant leaves wilted, 2 = 26–50% of leaves wilted, 3 = 51–75% of leaves wilted, 4 = >76% of leaves wilted, 5 = plant dead by modifying the scoring system from 1 to 5 as described by Winstead and Kelman (1952).
Biovar determination
The biovar of Myanmar strains was determined according to the physiological test of Hayward (1964, 1991). The biovar test is based on the ability to oxidize disaccharides (cellobiose, lactose and maltose) and hexose alcohols (mannitol, dulcitol and sorbitol). Utilization of trehalose, myo-inositol and d-ribose was further checked for determination of biovar 2 or N2 of the strains (Hayward 1991). The standard biovar test medium (basal medium) (Hayward 1964) was prepared by adding 1 g of NH4H2PO4, 0.2 g of KCl, 0.2 g of MgSO4·7H2O, 1 g of peptone and 80 mg of bromothymol blue into 1 L of distilled water. Then the pH was adjusted to 7.4 with NaOH. Each 10% (w/v) carbohydrate was separately prepared and sterilized. The medium was autoclaved after adding 3 g of agar. After cooling, each carbohydrate solution was mixed with basal medium to obtain a final concentration of 1% (w/v) of the carbohydrate. Five milliliters of the mixed medium for each carbohydrate (test medium) was then dispensed into 15 mL sterilized test tubes. A bacterial suspension was prepared in sterilized distilled water at ca.109 cfu/mL (He et al. 1983). Each tube was inoculated with 3 μL sample of the bacterial suspension. The tubes were incubated for 3 weeks at 28 °C. The reactions of Rs strains were then observed weekly. Growth and acid production were observed to determine carbohydrate use (Hayward 1991).
DNA extraction
Rs strains were cultured on TTC medium for 3 days at 30 °C, and total genomic DNA was extracted using the Viogene DNA/RNA Extraction Kit (Viogene Bio Tek, Taipei, Taiwan) according to the manufacturer’s instructions. DNA solution was stored at −20 °C until further use.
PCR-based identification and phylotype-specific-multiplex PCR amplification
Specific primers sets for phylotype identification (Fegan and Prior 2005) were used in combination with Rs-specific primer set 759/760 (Opina et al. 1997) to determine and confirm the phylotypes of Myanmar strains. Each 25 μL reaction mixture contained 1× reaction buffer (supplied by the manufacturer), 200 μM each dNTP, 3 μM each primer (Nmult21:1F, Nmult21:2F, Nmult22: InF, Nmult23:AF, Nmult22:RR, 759 and 760), 0.7 units of Blend Taq DNA polymerase (Toyobo, Osaka, Japan), and 1 μL of extracted DNA.
PCR was performed in an automated thermocycler (model T 100 Thermal Cycler, Bio-Rad Laboratories, Hercules, CA, USA) with an initial denaturation at 95 °C for 5 min; followed by 30 cycles of denaturation at 94 °C for 15 s, annealing at 60 °C for 30 s, and extension at 72 °C for 1 min; with a final extension at 72 °C for 10 min. The amplified DNA was separated by 2% (w/v) agarose gel electrophoresis in 1× TAE buffer solution. Gels were stained with ethidium bromide at 0.5 μg/mL, and DNA was examined and photographed under UV light. Multiplex PCR amplifies the 281-bp universal Rs-specific band and phylotype-specific products such as a 144-bp amplicon from phylotype I strains, a 372-bp amplicon from phylotype II strains, a 91-bp amplicon from phylotype III stains, and a 213-bp amplicon from phylotype IV strains, respectively (Fegan and Prior 2005).
DNA sequencing of endoglucanase (egl) gene
The partial nucleotide sequences of egl gene were determined. PCR amplification of a 750-bp region of the egl gene was performed using the primer set Endo-F and Endo-R (Fegan and Prior 2005). The reaction mixture (total 50 μL volume) contained 1× PCR buffer (supplied), 1.5 mM MgCl2, 200 μM each dNTP, 4 μM each primer, 2 μL of a turbid bacterial suspension as template (about 5 ng/μL), and 1 U of AmpliTaq Gold DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA). PCR was performed in an automated thermocycler (model T-100 Thermal Cycler, Bio-Rad) with an initial denaturation at 96 °C for 9 min; followed by 30 cycles of denaturation at 95 °C, 1 min, annealing at 55 °C for 40 s, and extension at 72 °C for 2 min; with a final extension at 72 °C for 10 min.
The amplified DNA was separated by 2% (w/v) agarose gel electrophoresis in 1× TAE buffer and recovered from the gel using the QIA quick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s instructions. PCR products were sent to Fasmac Co. (Kanagawa, Japan) for sequencing. The egl DNA sequences of both strands were determined using the aforementioned PCR primers.
The sequences were aligned using Sequence Scanner v1.0 (Thermo Fisher Scientific) software. The genetic distances between the sequences were calculated using the Kimura 2-parameter method (Kimura 1980). A phylogenetic tree was constructed from the genetic distance data, using the neighbor-joining method in the Clustal W Program of MEGA 5.2 (Tamura et al. 2011). The strength of tree branches was tested using 1000 bootstrap trials. Sequences of Myanmar strains were deposited into the GenBank database. Reference strains of phylotype I [PSS219 (FJ561167), M2 (FJ561067), Pe11 (FJ561084), GMI1000 (KT629830), MAD17 (GU295040), CFBP2968 (EF371806), MAFF 211266 (AF295250), TB28 (FJ561127), UW363 (DQ657621), Pe465 (FJ561090) and PSS81 (FJ561066)], phylotype II [CMR87 (EF439727), CFBP2958 (AF295266), CFBP2972 (AF295264), CMR121 (EF439725), CMR39 (EF439726), CFBP2047 (AF295262), ICMP7963 (AF295263), UW162 (AF295256), MOLK2 (EF371841), NCPPB3987 (AF295261), UW477 (AF295260), CIP309 (EF647735), CFBP3858 (AF295259), IPO1609 (EF371814) and PD441 (GU295044)], phylotype III [JT525 (AF295272), CFBP734 (AF295274), NCPPB332 (AF295276), CMR66 (EF439729) and CFBP3059 (AF295270)] and phylotype IV [MAFF 301558 (AY465002), R. syzygii R001 (DQ011544), ACH732 (GQ907150), BDB R230 (AF295280), UQRS 635 (KC757121) and PSI7 (EF371804)] were used for the analysis.
Results
Pathogenicity test
The results of the pathogenicity tests for the Myanmar strains are shown in Table 2. Myanmar strains were divided into six pathogenic groups (A–F) according to differences in their pathogenicity to the four solanaceous crops. Group A was pathogenic to tomato and potato but not to eggplant and chili. Group B was pathogenic to tomato and eggplant, but not to potato and chili. Group C was pathogenic to tomato, potato and chili, but not to eggplant. Group D was pathogenic to tomato, potato and eggplant, but not to chili. Group E was pathogenic to tomato, eggplant and chili, but not to potato, whereas group F was pathogenic to all host plants (Table 2). Among tested Myanmar strains, 41 strains (59%) belonged to group F together with the reference strain (PD441). Then, group D (19 strains) and group C (5 strains) were 27 and 7%, respectively. All Myanmar strains were pathogenic to tomato. Most Myanmar strains were pathogenic to potato, but three tomato strains (YT8-1, YT8-2, NTA6) were not pathogenic to the tested plants. Two tomato strains (YT3-1, YZT1-1), two potato strains (HP2-1, HP7-1) and three chili strains (YPE5-1, YPE5-2, YPE9-2) were not pathogenic to eggplant, whereas the other strains were pathogenic. Among all strains, 33% were not pathogenic to chili (Table 2).
Biovar determination
Myanmar strains belonged to either biovar 2 (30%), 3 (63%) or 4 (7%). Biovars 3 and 4 were isolated from many solanaceous plants (tomato, potato, eggplant and chili). On the contrary, biovar 2 strains (reference strain PD441 is also biovar 2) were isolated only from potato (Table 2). Tomato and chili strains belonged to biovars 3 and 4. All eggplant strains were biovar 3. Myanmar biovar 3 strains were found in all tested regions of Myanmar.
Characterization of Rs strains and PCR-based identification
All Myanmar strains produced a universal band of 281-bp with the Rs species specific primers set 759/760 (Opina et al. 1997). In multiplex PCR, Myanmar strains produced either a 144-bp (phylotype I) or a 372-bp amplicon (phylotype II) (Fegan and Prior 2005; Prior and Fegan 2005). Reference strain PD441 also produced both Rs species-specific and phylotype II-specific bands as reported previously (Cellier and Prior 2010). The DNA band was not amplified from the negative control. Among the strains, 83% of the total strains that were isolated from tomato, potato, eggplant and chili belonged to phylotype I, whereas the rest of the strains, all from potato, belonged to phylotype II. The geographical distribution of the biovars and phylotypes of the Rs strains in Myanmar is described in Fig. 1.
Phylogenic analysis based on egl sequences
Partial egl gene sequences were determined from 70 Myanmar strains. A dendrogram was constructed by comparing the nucleotide substitutions of the strains including reference strains named Rs, R. syzygii and blood disease bacterium of banana (BDB) that cover the known diversity within the Rs species. Strains were divided into four main clusters (Fig. 2). Each cluster corresponded to a specific phylotype (I, II, III or IV).
Phylogenetic tree based on neighbor-joining method of distance data from partial endoglucanase gene sequences from Ralstonia solanacearum strains in Myanmar with reference strains, R. syzygii and the blood disease bacterium of banana (BDB) strains. Myanmar strains from different solanaceous crops are indicated with markers. Values at the branches are percentage bootstrap support for 1000 resamplings. The scale bar shows one nucleotide substitution per 100 nucleotides
Myanmar Rs strains were classified into two main clusters, which corresponded to phylotype I and phylotype II (Fig. 2). Strains in phylotype I were further divided into seven subclusters, each corresponded to a separate sequevar (15, 17, 46, 47, 48, unknown 1 or unknown 2). Egl sequences of 11 tomato strains and four chili strains from Nyaung-U and Naypyidaw were identical to the reference strain (MAFF 211266) representing sequevar 15. One potato strain from Kalaw, two eggplant strains and two chili strains from Naypyidaw belonged to sequevar 17 with reference strain Pe11. Three tomato strains from Naypyidaw comprised sequevar 46 with reference strain MAD17. Three tomato strains and five chili strains from Naypyidaw and Pyin Oo Lwin and two potato strains from Heho and Kalaw belonged to sequevar 47 together with reference strain GMI8254. One tomato strain (MT29) from Monywa and one potato strain from Tatkon were included in sequevar 48 with the reference strain M2. Two other subclusters, which do not belong to any previously designated sequevars (unknown 1, unknown 2), were confirmed among the phylotype I strains (Table 3; Fig. 2).
On the contrary, all Myanmar strains in phylotype II cluster were homogenous with the reference strains (CFBP3858, IPO1609 and PD441) representing sequevar 1. These were isolated only from potato-growing areas Heho and Tatkon.
Discussion
In this study, we collected Myanmar Rs strains and assessed their pathogenicity, biovar, phylogeny and genetic diversity. We also surveyed the regions where solanaceous crops are grown in Myanmar such as Sagaing region, Mandalay region, Naypyidaw region and Shan state, a major potato-growing area that is severely affected by bacterial wilt (Myint 2001). We could not collect tobacco Rs strains in this survey, since tobacco cultivation is limited. Thus, a wider survey should include tobacco, ginger and other hosts.
From various solanaceous crops, 70 Rs strains with typical colony characters were isolated on TTC medium. Our pathogenicity tests provided preliminary information concerning differences in pathogenic characters among the Myanmar strains. Each strain was pathogenic to each host plant. Almost all the strains were pathogenic to tomato and potato; three strains were not pathogenic to potato. A pathogenicity test of eggplant and chili did differentiate the strains, which could be divided into six pathogenic groups. In this experiment, we used one cultivar per host plant with three replications. Further large-scale tests will also be required for a quantitative assessment of resistance/susceptibility of Myanmar Rs strains using a wide range of different varieties for the same host.
Usually, potato biovar 2 strain causesa highly destructive brown rot and is equivalent to race 3 (Hayward 1991; Janse 1996). Compared with other strains of Rs, race 3 biovar 2 strain is more adapted to cooler temperatures in temperate climates and at higher elevations and latitudes in the tropics (Huang et al. 2011; Smith et al. 1995). Similarly, Myanmar biovar 2 strains were isolated at higher latitudes at Heho (hilly area) and Tatkon and were guessed to correspond to race 3. Although the biovar 2 and N2 strains are assumed to have adapted to cool and tropical climates, respectively (Horita et al. 2005; Marin and El-Nashaar 1993), we did not obtain any biovar N2 strains in this study.
The multiplex PCR method (Fegan and Prior 2005), based upon sequence information from the ITS region, revealed that Myanmar strains clustered into phylotypes I and II. Phylotypes III and IV were not found in the surveyed areas. Phylotype I strains are widespread and affect a very wide range of economically important crops across Asian countries (Elphinstone 2005) and were suggested to originate from Asia (Fegan and Prior 2005). Our results also indicated that phylotype I strains were mostly distributed throughout the studied areas and belonged to biovar 3 and 4, whereas phylotype II strains were only found as biovar 2 from potato.
Phylogenetic analysis of sequence data is now the most reliable and powerful tool to analyze genetic relationship among Rs strains around the world (Castillo and Greenberg 2007; Jeong et al. 2007; Lewis Ivey et al. 2007; Liu et al. 2009; Mahbou Somo Toukam et al. 2009; Nouri et al. 2009; Poussier et al. 2000; Prior and Fegan 2005; Villa et al. 2005; Horita et al. 2010). A phylotype/sequevar classification system of Rs species can differentiate the strains more precisely and exactly than traditional conventional methods (race and biovar system) (Tsuchiya 2014).
Obviously, phylotype I strains, which included seven different sequevars (15, 17, 46, 47, 48, unknown 1 and unknown 2), predominated in all studied areas. Our results also revealed that multiple sequevars were present even from the same host in the same area. On the contrary, in an egl analysis, phylotype II strains belonging to sequevar 1, which were found to be genetically homogenous in regions Heho and Tatkon (Heho in Shan state is one of the main potato-growing regions) (MAS 1990).
Recent DNA-based analyses (e.g., RFLP, rep-PCR, multi locus sequences) elucidated that the worldwide race 3 biovar 2 strains originated from South America and have been distributed locally and internationally on Rs-infected potato tubers and geranium (Elphinstone 2005; Hayward 1991; Hayward et al. 1998; Janse 1996; Ji et al. 2007; Mahbou Somo Toukam et al. 2009; Nouri et al. 2009; Sagar et al. 2013; Xu et al. 2009). Moreover, egl sequence analysis showed that strains belonging to phylotype II sequevar 1 are equivalent to race 3 biovar 2 (Wicker et al. 2012). Phylotype II sequevar 1 strains were distributed in India (Sagar et al. 2013) and China (Xu et al. 2009), neighbors of Myanmar. Our results suggested that phylotype II sequevar 1 (biovar 2) strains are quite similar to the reference phylotype II sequevar 1 (race 3 biovar 2) strain (PD441) by biochemical, pathological and DNA-based analyses and might have been carried into Myanmar via seed potatoes or infected plant materials. Our findings also suggested that plant quarantine and seed potato management systems should be set up to prevent the dissemination of phylotype II potato strains to all new potato-growing areas of Myanmar as well as other foreign potato-growing areas (Elphinstone 2005; Hayward 1991; Hayward et al. 1998; Nouri et al. 2009; Horita et al. 2010). The development of a quick, exact method to diagnose bacterial wilt pathogen from seed potatoes is urgently needed. Utilization of biochemical, pathological and molecular characters to identify Rs (race 3 biovar 2 and/or phylotype II sequevar 1) will support an approach to manage bacterial wilt disease in Myanmar. For a better understanding of the distribution of other phylotypes, races or biovars in Myanmar, further research on other hosts is necessary.
References
Buddenhagen I, Kelman A (1964) Biological and physiological aspects of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 2:203–230
Buddenhagen I, Sequeria L, Kelman A (1962) Designation of races of Pseudomonas solanacearum. Phytopathology 52:726 (Abstract)
Castillo JA, Greenberg JT (2007) Evolutionary dynamics of Ralstonia solanacearum. Appl Environ Microbiol 73:1225–1238
Cellier G, Prior P (2010) Deciphering phenotypic diversity of Ralstonia solanacearum strains pathogenic to potato. Phytopathology 100:1250–1261
Elphinstone JG (2005) The current bacterial wilt situation: a global overview. In: Allen C, Prior P, Hayward AC (eds) Bacterial wilt: the disease and the Ralstonia solanacearum species complex. APS Press, St. Paul, pp 9–28
Fegan M, Prior P (2005) How complex is the "Ralstonia solanacearum species complex". In: Allen C, Prior P, Hayward AC (eds) Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, St. Paul, pp 449–461
Hayward AC (1964) Characteristics of Pseudomonas solanacearum. J Appl Bacteriol 27:265–277
Hayward AC (1991) Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 29:65–87
Hayward AC (1994) The hosts of Pseudomonas solanacearum. In: Hayward AC, Hartman GL (eds) Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, pp 9–24
Hayward AC, Elphinstone JG, Caffier D, Janse JD, Stefani E, French ER, Wright AJ (1998) Round table on bacterial wilt (brown rot) of potato. In: Prior PH, Allen C, Elphinstone JG (eds) Bacterial wilt disease: molecular and ecological aspects. Springer, Heidelberg, pp 420–430
He LY, Sequiera L, Kelman A (1983) Characteristics of strains of Pseudomonas solanacearum from China. Plant Dis 67:1357–1361
Horita M, Tsuchiya K (2001) Genetic diversity of Japanese strains of Ralstonia solanacearum. Phytopathology 91:399–407
Horita M, Tsuchiya K, Ooshiro A (2005) Characteristics of Ralstonia solanacearum Biovar N2 Strains in Asia. J Phytopathol 153:209–213
Horita M, Suga Y, Ooshiro A, Tsuchiya K (2010) Analysis of genetic and biological characters of Japanese potato strains of Ralstonia solanacearum. J Gen Plant Pathol 76:196–207
Horita M, Tsuchiya K, Suga Y, Yano K, Waki T, Kurose D, Furuya N (2014) Current classification of Ralstonia solanacearum and genetic diversity of the strains in Japan. J Gen Plant Pathol 80:455–465
Huang Q, Xuerui Y, Jaw FW (2011) Improved biovar test for Ralstonia solanacearum. J Microbiol Methods 88:271–274
Janse JD (1991) Infra- and intraspecific classification of Pseudomonas solanacearum strains, using whole cell fatty acid analysis. Syst Appl Microbiol 14:335–345
Janse JD (1996) Potato brown rot in Western Europe—History, presence occurrence and some remarks on possible origin, epidemiology and control strategies. EPPO Bull 26:679–985
Jeong Y, Kim J, Kang Y, Lee S, Hwang I (2007) Genetic diversity and distribution of Korean isolates of Ralstonia solanacearum. Plant Dis 91:1277–1287
Ji P, Allen C, Sanchez-Perez A, Yao J, Elphinstone JG, Jones JB, Momol MT (2007) New diversity of Ralstonia solanacearum strains associated with vegetable and ornamental crops in Florida. Plant Dis 91:195–203
Kelman A (1953) The bacterial wilt caused by Pseudomonas solanacearum. A literary review and bibliography. Tech Bull North Carol Agric Exp Stn 99:1–194
Kelman A (1954) The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium chloride medium. Phytopathology 44:693–695
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lewis Ivey ML, McSpadden Gardener BB, Opina N, Miller SA (2007) Diversity of Ralstonia solanacearum infecting eggplant in the Philippines. Phytopathology 97:1467–1475
Liu Y, Kanda Y, Yano K, Kiba A, Hikichi Y, Aino M, Kawaguchi A, Mizoguchi S, Nakaho K, Shiomi H, Takikawa Y, Ohnishi K (2009) Molecular typing of Japanese strains of Ralstonia solanacearum in relation to the ability to induce a hypersensitive reaction in tobacco. J Gen Plant Pathol 75:369–380
Lwin M (2006) Biological control of bacterial wilt disease through antagonistic microbial populations. Doctor of Technical Science dissertation, Asian Institute Technology, Bangkok, Thailand
Mahbou Somo Toukam G, Cellier G, Wicker E, Guilhaud C, Kahane R, Allen C, Prior P (2009) Broad diversity of Ralstonia solanacearum strains in Cameroon. Plant Dis 93:1123–1130
Marin JE, El-Nashaar HM (1993) Pathogenicity and new phenotypes of Pseudomonas solanacearum from Peru. In: Hartman GL, Hayward AC (eds) Bacterial wilt, vol 45. ACIAR Proceedings, Canberra, pp 78–84
MAS (Myanmar Agricultural Services) and the International Potato Center (CIP) (1990) Potato research and development in Union of Myanmar (1983–1989), Rangoon and Manilla, pp 1–47
MOAI (Ministry of Agriculture and Irrigation) (2014) Myanmar agriculture at a glance. Department of Agricultural Planning, Myanmar, pp 6–7
Myint MM (2001) Potato late blight in Myanmar. Proceedings of the East and Southeast Asia Linkage Group, Baoding, China. J Agric Univ Hebei 24:24–26
Nouri S, Bahar M, Fegan M (2009) Diversity of Ralstonia solanacearum causing potato bacterial wilt in Iran and the first record of phylotype II/biovar 2T strains outside South America. Plant Pathol 58:243–249
Opina N, Tavner F, Hollway G, Wang JF, Li TH, Maghirang R, Fegan M, Hayward AC, Krishnapillai V, Hong WF, Holloway BW, Timmins J (1997) A novel method for development of species and strain-specific DNA probes and PCR primers for identifying Burkholderia solancearum (formerly Pseudomonas solancearum). Asia Pac J Mol Biol Biotechnol 5:19–30
Pegg KG, Moffett ML (1971) Host range of the ginger strain of Pseudomonas solanacearumin Queensland. Aust J Exp Agric Anim Husb 11:696–698
Poussier S, Prior P, Luisetti J, Hayward C, Fegan M (2000) Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst Appl Microbiol 23:479–486
Prior P, Fegan M (2005) Recent developments in the phylogeny and classification of Ralstonia solanacearum. Acta Hort 695:127–136
Sagar V, Jeevalatha A, Mian S, Chakrabarti SK, Gurjar MS, Arora RK, Sharma S, Bakade RR, Singh BP (2013) Potato bacterial wilt in India caused by strains of phylotype I, II and IV of Ralstonia solanacearum. Eur J Plant Pathol 138:51–65
Smith JJ, Offord LC, Holderness M, Saddler GS (1995) Genetic diversity of Burkholderia solanacearum (synonym Pseudomonas solanacearum) race 3. Kenya. Appl Environ Microbiol 61:4263–4268
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tsuchiya K (2014) Genetic diversity of Ralstonia solanacearum and disease management strategy. J Gen Plant Pathol 80:504–509
Villa JE, Tsuchiya K, Horita M, Natural M, Opina N, Hyakumachi M (2005) Phylogenetic relationships of Ralstonia solanacearum species complex strains from Asia and other continents based on 16 S rDNA, endoglucanase, and hrpB gene sequences. J Gen Plant Pathol 71:39–46
Wicker E, Lefeuvre P, de Cambiaire J-C, Lemaire C, Poussier S, Prior P (2012) Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J 6:961–974
Winstead NN, Kelman A (1952) Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathology 42:628–634
Xu J, Pan ZC, Prior P, Xu JS, Zhang Z, Zhang H, Zhang LQ, He LY, Feng J (2009) Genetic diversity of Ralstonia solanacearum strains from China. Eur J Plant Pathol 125:641–653
Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y (1995) Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov: proposal for Ralstonia pickettii, Ralstonia solanacearum and Ralstonia eutropha. Microbiol Immunol 39:897–904
Zaw MN (2010) Occurrence of bacterial wilt in Kalaw Township and its control measure by effective microorganisms. MS thesis, Yezin Agricultural University, Yezin
Acknowledgements
We are grateful to the Ministry of Education, Culture, Sports, Science and Technology of Japan, Laboratory of Plant Pathology, Kyushu University of Japan, and Yezin Agricultural University of Myanmar to fulfill this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kyaw, H.W.W., Tsuchiya, K., Matsumoto, M. et al. Genetic diversity of Ralstonia solanacearum strains causing bacterial wilt of solanaceous crops in Myanmar. J Gen Plant Pathol 83, 216–225 (2017). https://doi.org/10.1007/s10327-017-0720-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-017-0720-0