Abstract
In Vietnam, bacterial wilt, caused by R. solanacearum, is a major threat to many important field crops. Disease management strategies suggested in the past have remained ineffective. Phylotype analysis and genetic diversity among Vietnamese R. solanacearum is unknown. The present study was conducted to investigate the relationship among isolates of R. solanacearum in the tomato production areas in Vietnam, to analyze genotypic characteristics, to characterize the phylotypes of the isolates and to determine their aggressiveness. All 207 R. solanacearum strains collected from tomato plants in 12 provinces in Vietnam belonged to Asian Phylotype I. All isolates belong to biovar 3, except for isolate Rs86 collected from Ninh Thuan, which belonged to biovar 4. A total of 52 isolates representing 12 origins (provinces) and two Taiwanese strains were selected to investigate genetic diversity and their genetic relationship by using RAPD markers. Based on the unweighted pair group method of arithmetic means endrogram and STRUCTURE results, 54 isolates were grouped into three main clusters. Cluster I includes 3 isolates obtained from Lam Dong province and 1 isolate collected from Tien Giang province. Cluster II had wide distribution from the northern to the southern part of Vietnam. This cluster could be divided into 3 sub-clusters and these sub-clusters were largely separated according to geographical distribution. Sub-cluster IIa consisted of five isolates from Lam Dong province. One isolate from Thai Binh and two isolates from Hai Duong grouped in sub-cluster IIb. Cluster IIc was found only in Hai Phong. Cluster III had highest variation with two sub-clusters. Eight isolates from Southern part, 5 isolates from Nghe An and 2 reference isolates from Taiwan were grouped in sub-cluster IIIa, while 14 isolates mainly from Northern part (Ha Noi, Nam Dinh, and Thai Binh) were grouped in sub-cluster IIIb. Isolates collected in Hai Duong, Hai Phong and Nam Dinh showed higher virulence than those on tomato. The results demonstrated that R. solanacearum isolates even though similar in genetic background do not necessarily cause similar virulence. This is the first report on genetic diversity and aggressiveness of Vietnamese R. solanacearum isolates in tomatoes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial wilt caused by Ralstonia solanacearum is one of the most damaging soilborne diseases of many important crops worldwide. The host range of the pathogen is very wide and includes more than 450 plant species representing over 50 families of plants (Hayward 1991). R. solanacearum is a complex species with exceptional diversity among strains regarding host range, geographical distribution, pathogenicity, epidemiological relationships, and physiological properties (Hayward 1991). Traditionally, this complex species has been subdivided into five races on the basis of differences in host range (Buddenhagen et al. 1962) and six biovars on the basis of carbohydrate utilization (Hayward 1991, 1994). These classification schemes have been applied to describe strains worldwide; however, there are several limitations of each system such as more time consuming, inability to discriminate at sub-specific level etc.
In order to improve understanding of the genetic relatedness among R. solanacearum strains, DNA-based analyses such as restriction fragment length polymorphisms (RFLP), polymerase chain reaction (PCR)-RFLP, 16S rRNA gene sequence analysis, repetitive extragenic palindromic (REP)-PCR, and amplified fragment length polymorphism (AFLP) have been used (Cook et al. 1989; Li et al. 1993; Cook and Sequeira 1994; Fegan et al. 1998; Poussier et al. 2000; Seal et al. 1992; Horita and Tsuchiya 2001; Rodrigues et al. 2012). Fegan and Prior (2005) had proposed phylotype system. The pathogen was classified into four phylotypes. Strains belonging to phylotype III were found only in Africa, while strains belonging to phylotype IV were exclusively found originating from Indonesia. In contrast, the strains belonging to phylotype I and II are present on several continents. Nevertheless, the phylotype I was demonstrated to have the highest evolutionary potential as well as the highest virulence diversity, with a worldwide prevalence and spread (Lebeau et al. 2011; Wicker et al. 2012). This classification scheme was recently confirmed by using a whole-genome microarray approach (Guidot et al. 2007).
Bacterial wilt has been one of the most important diseases of tomato, pepper, groundnut, and potato in Vietnam for a long time. Outbreak zones of this pathogen are still expanding, threatening the production of potato, pepper and tomato in different parts of the country. Several studies on R. solanacearum causing bacterial wilt in groundnut and potato in Northern Vietnam have been reported (Nguyen and Phan 1993; Nguyen et al. 2002; Do 2009; Nguyen 2012). Dinh et al. (2008) used RAPD to investigate genetic diversity of R. solanacearum isolated from peanut collected in the Northern Vietnam. However, phylotype analysis and genetic diversity among Vietnamese R. solanacearum is unknown. The present study was conducted to investigate the relationship among isolates of R. solanacearum in the tomato production areas in Vietnam, to analyze genotypic characteristics, to characterize the phylotypes of the isolates and to determine their aggressiveness.
Materials and methods
Sample collection, bacterial isolates, isolation and conditions
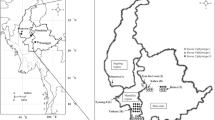
Stem pieces (5–6 cm long) of wilted plants were collected from fields in the areas of tomato production during 2014 from different provinces in Vietnam such as Hai Duong, Hai Phong, Ha Noi, Thai Binh, Nam Dinh, Nghe An, Lam Dong, Can Tho, Ho Chi Minh, Vinh Long, Tien Giang, and Ninh Thuan (Fig. 1). The plant samples were brought to the laboratory for further study. They were first surface disinfected with 70% ethanol, then peeled. The sub samples of these plants were macerated in sterile distilled water. The oozes were then streaked on Kelman’s Triphenyl tetrazolium chloride (TTC) agar medium (Kelman 1954). Plates were incubated at 28 ± 2 °C for 48–72 h. R. solanacearum colonies were again streaked onto fresh TTC medium for purification. Two single colonies were then transferred in 2 ml of double distilled sterile water and the cultures were stored at 20 ± 2 °C.
Biovar determination
The strains were differentiated based on their ability to utilize disaccharides (sucrose, lactose and maltose) and sugar alcohols (mannitol, sorbitol and dulcitol) as described previously by Hayward (1964). 1.0 ml Hayward’s medium was dispensed into 2.0 ml Eppendorf tube containing sucrose, lactose, maltose, mannitol, sorbitol and dulcitol (1% wt/vol) and to which 0.5 ml of bacterial suspension (108 CFU/ml) was also added. The tubes containing the mixture were then incubated at 28 °C. They were then examined after 3 days of incubation for changing pH, which was indicated by the change of color.
DNA extraction from bacterial strains
Total genomic DNA was extracted following a protocol of Bacteriology Unit, AVRDC-The World Vegetable Center, Taiwan (Jaw-Fen WANG, personal communication). A single colony was suspended in a 1.5 ml eppendorf tube contained 50 µl TE buffer. The bacterial suspension was then boiled for 10 min and centrifuged at 13,000 rpm 4 °C for 6 min. The supernatant was transferred into new autoclaved 1.5 ml—eppendorf tube and stored at − 20 °C. DNA quality was checked by electrophoresis of 50 ng aliquots on a 0.8% agarose gel stained with SYBR Green I (Invitrogen, Eugene, OR, USA) and exposed under ultraviolet light.
Confirmation of R. solanacearum strains using specific primers
Ralstonia solanacearum strains were again confirmed by using specific primers AU759: 5′-GTCGCCGTCAACTCACTTTCC-3′; AU760: 5′-GTCGCCGTCAGCAATGCGGAATCG-3′ (Opina et al. 1997). The PCR reactions were performed in Mygenie96 Thermal Block (Bioneer). The 25 µl reaction volume included 5 × buffer containing MgCl2 (Bioscience, England), 2,5 mM deoxyribonucleotide triphosphate mix (Bioscience, England), 2 U of Taq DNA polymerase (Bioscience, England), and 1 pmol of each primer P759/P760 and 15–20 ng of genomic DNA. The amplification reactions were carried out using the following thermal profile: 94 °C for 3 min, 53 °C for 1 min, 72 °C for 1.5 min (1 cycles); 94 °C for 0.3 min, 60 °C for 0.3 min, 72 °C for 0.3 min (30 cycles); 72 °C for 5 min (1 cycle) and stored at 4 °C. Amplified products were incubated with a 1:10,000 dilution of the SYBR Green I nucleic acid gel stain (Invitrogen, USA) for 20 min and separated on a 2% agarose gels using 0.5 × TBE buffer for 2 h at 120 V and photographed under UV light. A 100 bp ladder was used as a molecular weight marker.
Phylotype analysis
Phylotype identification of each strain was conducted using specific primers (Table 1) as described in literature (Fegan and Prior 2005; Prior and Fegan 2005). Phylotype specific multiplex PCR was carried out in 25 µl final volume of reaction mixture, containing 5U Taq Polymerase (Bioscience, England), 5 × buffer contained MgCl2 (Bioscience, England), 6 pmols of the primer Nmult:21:1F, Nmult:21:2F, Nmult:22:InF, Nmult:23:AF, Nmult:22:RR and 4 pmols of the primer AU759 and AU760 (Opina et al. 1997). The following cycling program was used in a thermal cycler (Mygenie96 Thermal Block, Bioneer): 96 °C for 5 min and then cycled through 30 cycles of 94 °C for 15 s, 59 °C for 30 s and 72 °C for 30 s, followed by a final extension period of 10 min at 72 °C and stored at 4 °C. Amplified products were incubated with a 1:10,000 dilution of the SYBR Green I nucleic acid gel stain (Invitrogen, USA) for 20 min and separated on a 2% agarose gels using 0.5 × TBE buffer for 3 h at 120 V and photographed under UV light. A 100 bp ladder was used as a molecular weight marker.
RAPD analysis
A total of 100 UBC (University of British Columbia) RAPD primers (synthesized by Bioneer, Korea) were pre-screened on 12 selected isolates representative for 12 origins. Polymorphic primers were then used for confirmation of generated-fragment stability. Selected polymorphic primers were used to genotype R. solanacearum for genetic diversity analysis. The PCR reactions were performed in Mygenie96 Thermal Block (Bioneer). The 15 µl reaction volume included 2.5 mM MgCl2 (Bioscience, England), 200 µM deoxyribonucleotide triphosphate mix (Bioscience, England), 10 × PCR buffer, 1 U of Taq DNA polymerase (Bioscience, England), and 0.25 µM of random primer and 15–20 ng of genomic DNA. The amplification reactions were carried out using the following thermal profile: 94 °C for 3 min (1 cycle); 94 °C for 1 min, 37 °C for 1 min, 72 °C for 2 min (40 cycles); 72 °C for 7 min (1 cycle). Amplified products were incubated with a 1:10,000 dilution of the SYBR Green I nucleic acid gel stain (Invitrogen, USA) for 20 min and separated on a 1% agarose gels using 0.5 × TBE buffer for three and half hours at 120 V and photographed under UV light. A 100 bp ladder was used as a molecular weight marker.
Bacterial aggressiveness assays
Two tomato genotypes, one resistant to R. solanacearum (Hawaii 7996 (H7996), S. lycopersicum) and the other susceptible (West Virginia 700 (WVa700), S. pimpinellifolium) (Wang et al. 1998), which were provided by AVRDC-The World Vegetable Center and one F1 hybrid (TN52) commonly used in Vietnam were used. Before sowing, seeds were treated by soaking at 4X diluted Chlorox (6% sodium hypochlorite) for 5 min and then rinsed under running water for 15 min and sown immediately in 2-inch pots. The steam-sterilized potting mixture used consisted of sand, soil, rice husk and compost in the ratio of 1:3:1:1. A total of 12 isolates were used for inoculation. The experiment was laid out in split-plot design following with ‘strain’ as the main-plot and ‘plant materials’ as the subplot with three replications and 5 plants per replication.
A suspension of each isolate was made in sterile distilled water and adjusted to 108 cfu/mL. Seedlings with four fully expanded true leaves (about three-week old) were inoculated by pouring 20 ml of the above suspension on the soil surface of each pot. Inoculated plants were rated at 4, 7, 14, 21 and 28 days after inoculation (DAI) using a 0–5 scale, where 0 means no symptom; 1 means one leaf partially wilted; 2 means 2–3 leaves wilted; 3 means 4 or more wilted leaves; 4 means all leaves wilted; 5 means death of the plant (Winstead and Kelman 1952). Percentages of wilted plants (PWP) at 4 weeks after inoculation were calculated following the formulas of PWP = (NW/NT) × 100, where NT is number of total plants and NW is number of wilted plants. Strain aggressiveness was estimated by using the relative area under the disease progress curve (RAUDPC) (Fry 1978), which was calculated in the following manner: First, PWP was used to calculate AUDPC, which expresses the dynamics of disease development according to Shaner and Finney (1977), following the formula: AUDPC = ∑i=1 to n−1 [(Yi+1 + Yi)/2] × [Xi+1 − Xi], where Yi is PWP at the ith observation (i = 1 being the first observation point), and Xi is time at the ith observation, and n is total number of observations. Second, RAUDPC was then divided by the number of days from inoculation to the end of observation period.
Data analysis
All percentage data were transformed by arcsine square-root transformation for the analyses of variance. The data were analyzed using the Statistic 10. The entry mean comparison was performed under each strain when the entry × experiment or entry × strain was significant. The significant differences were determined at P < 0.05 by LSD. Marker bands of RAPD markers were scored as 1 for presence and 0 for absence. From these data input, a dendrogram was constructed based on the Nei’s genetic distances using the Phylip program with the unweighted pair group method by arithmetic averages (UPGMA) cluster analysis. Population structure was analyzed using a model-based approach available in the software STRUCTURE 2.3.4 (Pritchard et al. 2000). In this analysis, we used the no-admixture model and evaluated 1–20 genetic clusters (K) with 5 permutations for each K value. Each run was implemented with a burn-in period of 25,000 steps followed by 500,000 Monte Carlo Markov (MCM) chain. The estimated log-probability of data [LnP(D)] and ΔK value from STRUCTURE overestimates the number of subpopulations. After grouping, genotypic diversity was evaluated using GenAlEx 6.5.
Results
Collection and identification of bacterial strains
A total of 306 stem pieces from wilted plant were collected from fields in the areas of tomato production during 2014 from 12 provinces in Vietnam. Of these, 207 samples produced bacterial ooze (Supplementary Table 1). The bacteria were cultured on TTC medium and incubated at 28 ± 2 °C for 48 h to produce different colonies. Basically, all bacterial strains yielded typical virulent type colonies, which were cream colored, irregular shaped, highly fluidal with pink and dark pink pigmentation in the center. These characters were consistent with R. solanacearum as described by Kelman (1954) on TTC agar medium.
A rapid detection of bacterial isolates using specific universal primer pair 759/760 (Opina et al. 1997) was conducted. Two Taiwanese strains such as Pss4, which belonged to race I/phylotype I and biovar 3, and Pss186, which belonged to race I/phylotype I and biovar 1 (Lin et al. 2014), provided by Dr. Jaw-Fen WANG (AVRDC-The World Vegetable Center—personal communication), were used as control check. The result showed that all of 207 isolates produced a specific DNA band about 280 bp that was similar with amplified products of two reference strains from Taiwan (Fig. 2).
Biovar determination
The isolates were determined as described by Hayward (1964). The biovar test indicated that 206 strains belonged to biovar 3, only one strain Rs86 collected from Ninh Thuan was determined as biovar 4.
Phylotype identification
Based on multiplex PCR analysis, all of 207 isolates from Vietnam and two reference strains from Taiwan belonged to Phylotype I as 144-bp amplicon was observed in all the isolates and together with 280-bp amplicon of R. solanacearum species when PCR products of the strains were subjected to electrophoresis on 2% agarose gel (Fig. 3).
Electrophoresis of PCR products using multiplex PCR primers for phylotype analysis of R. solanacearum strains on 2% agarose. Lanes M: 100 bp DNA ladder, 1: Rs4, 2: Rs5, 3: Rs6, 4: Rs10, 5: Rs11, 6: Rs15, 7: Rs18, 8: Rs19, 9: Rs24, 10: Rs25, 11: Rs26, 12: Rs27, 13: Rs28; 14: Rs29; 15: Rs32; 16: Rs38; 17: Pss4, 18: Pss190
Genetic diversity analysis
RAPD method has been widely used in genetic diversity analysis because it is highly efficient, easy and rapid. In this study, RAPD technique was used as tool to assess polymorphism of 8 R. solanacearum isolates, which were representative 8 origins such as Rs11 (Lam Dong), Rs47 (Ho Chi Minh city), Rs66 (Can Tho), Rs86 (Ninh Thuan), Rs120 (Hai Phong), Rs226 (Ha Noi), Rs300 (Thai Binh), and Rs367 (Nghe An). Each isolate was selected randomly among origin’s isolates. A total of 100 RAPD University of British Colombia (UBC) primers were used to primarily screen (Supplementary Table 2). Of these, 41 UBC primers were polymorphic among the selected isolates (Fig. 4). In order to confirm consistency of polymorphic primers, the polymorphic primers were rescreened on the 12 selected isolates, which were selected representative 12 origins randomly. The result showed that the same band patterns (data not shown). However, only 21 primers (Table 2), which generated 108 DNA fragments polymorphic among isolates and easy to score, were used to genotype 54 R. solanacearum isolates (Supplementary Table 3) that included 52 isolates representing 12 origins in Vietnam and two strains from Taiwan (Pss4 and Pss190) (Fig. 5). The UPGMA dendrogram conducted from Nei’s genetic distance and population structure among 54 isolates can be seen in Fig. 6. The ΔK values and log-probability of data indicate that the best number of optimal groups were occurred at K = 3 and another peak of ΔK could be seen at K = 6 (Supplementary Fig. 1). Therefore, 54 isolates could be confidently grouped into 3 main genetic clusters. The cluster I consisted of 4 isolates from Lam Dong and Tien Giang provinces. Cluster II had 22 isolates from wide geographical origins. This cluster further could be divided into 3 sub-clusters, the distribution of isolates in sub-cluster IIa and sub-cluster IIc obtained from Lam Dong (The southern highland province) and Hai Duong (the northern lowland province), respectively, likewise, sub-cluster IIc consists one isolate of Thai Binh and two isolates of Hai Duong (northern province). Cluster III were quite complex, however, the isolates in this cluster could grouped into 2 major sub-clusters, IIIa and IIIb, correlated with their geographical origin. Sub-cluster IIIa included 8 southern isolates and 5 isolates of Nghe An province, 2 reference isolates from Taiwan were also assembled in this sub-cluster, while 14 isolates from the northern part (Ha Noi, Nam Dinh, Thai Binh) were grouped in sub-cluster IIIb.
The value of Nei’s genetic distance among 6 sub-clusters revealed the closest relationship of cluster IIa and cluster IIc although they belong two distinct geographical regions (Southern highland and Northern lowland), this is partly explained based on their lowest gene diversity and seem to be homologous than other II sub-clusters (IIb). Conversely, in cluster II, the both originating isolates group from the northern (sub-cluster IIa and IIb) posed the highest genetic distance (Tables 3, 4).
Evaluation of aggressiveness of R. solanacearum isolates
Disease incidence was assessed at seedling stage of H7996 (Resistant control), WVa700 (Susceptible control) and TN52 (F 1 hybrid) using 12 isolates, Rs26, Rs38, Rs51, Rs57, Rs66, Rs86, Rs101, Rs127, Rs238, Rs319, Rs341, and Rs373, which were representative of each province. Severe wilting of WVa700, the susceptible control, indicated the environment and the inoculation were suitable for the disease development with all 12 isolates (Fig. 7). TN52, a F1 hybrid commonly planted in tomato production areas in Vietnam, was highly susceptible to all isolates; however, the aggressiveness caused by Southern isolates (Rs26, Rs38, Rs51, Rs57, Rs66, and Rs86) on TN52 was slightly lower than that (or equal) on WVa700. In contrast, the severity caused by Northern isolates (Rs101, Rs127, Rs249, Rs319, Rs341 and Rs373) on WVa700 was slightly lower than or equal to TN52. Disease progressed most rapidly between 7 and 21 days after inoculation. At 21 DAI, nearly 100% of two genotypes (WVa700 and TN52) displayed near maximum wilting. About 90% (inoculated with isolate Rs86) and 80% (inoculated with isolates Rs26, Rs38, Rs51, Rs57, Rs66, Rs249, Rs319, and Rs373) plants of H7996 remained healthy at 28 DAI. Meanwhile, the resistant line H7996 had only 26.7% survival when inoculated with Rs101 and Rs127 and 33.3% with Rs341. Thus, resistance in H7996 was stable to all isolates except for isolates Rs101, Rs127, and Rs341.
Combined analysis showed a significant interaction between isolate and entry, indicating individual entries had different reactions against different strains (Table 5). Genotypes, WVa700 and TN52, had similar reaction to all isolates based on visual wilting symptom. Genotype H7996 was the most resistant to isolates Rs26, Rs38, Rs51, Rs57, Rs66, Rs86, Rs249, Rs319, and Rs373, but not to isolates Rs101, Rs127, and Rs341. Therefore, differences in symptom development of the 12 isolates suggest variation in their virulence. Overall, isolates collected in Hai Phong, which are distributed in cluster C, and from Hai Duong and Nam Dinh, which are distributed in sub-cluster A2 and A4, respectively, had caused more severe disease than those collected in other provinces in Vietnam (Fig. 8). Thus, low level of similarity among isolates collected from different geographical areas indicated that these isolates were lineage of multiple virulent strains. This is the first report on genetic diversity and aggressiveness of Vietnamese R. solanacearum isolates.
Discussion
In Vietnam, bacterial wilt, caused by R. solanacearum, is a major threat to many important crops such as peanut, tomato, pepper, potato, banana, ginger, tobacco (Doan 1995; Le 1997; Do 2009). The infection level fluctuates depending on plant species, ecological zones, season, and weather conditions (Tung et al. 1992; Do 2009). Several management strategies advocated for the control of the disease had very limited success. The genetic diversity of the pathogen might be one of the reasons for the difficulties encountered in disease management. Thus, in this study, we explored the genetic diversity of the pathogen using RAPD marker in order to provide insight into host plant- and site-dependent distribution patterns of this pathogen in Vietnam.
Following the method of Hayward (1964), biovar grouping of 207 isolates of R. solanacearum causing bacterial wilt of tomato and collected from 12 origins (provinces) of Vietnam, was done. Only one isolate from Ninh Thuan, Southern Central of Vietnam, belonged to biovar 4, and the remaining isolates belonged to biovar 3. Several studies on biovar determination of R. solanacearum strains collected from Peanut in Vietnam have been conducted and the strains were found to belong to race 1 biovar 3 and 4 (Nguyen et al. 1997). This is the first study on biovar determination of R. solanacearum from tomato.
All the isolates were evaluated using the hierarchical classification scheme proposed by Fegan and Prior (2005). Multiplex PCR with phylotype-specific primers grouped all of these strains into phylotype I of the R. solanaceraum species complex, supporting the finding of Nguyen et al. (1997) that R. solanacearum isolates collected from different hosts and locations in Vietnam belonged to phylotype I. Ivey et al. (2007) also reported that all R. solanacearum strains collected from eggplant in Philippines belonged to race 1, biovar 3 or 4 and phylotype I. According to phylotype analysis of R. Solanacearum strains collected from wilted potato in India, Sagar et al. (2014) also indicated that all of fifteen bacterial strains belonged to phylotype I. Xue et al. (2011) also reported that majority of R. solanacearum strains in China belonged to Asian phylotype I. Thus, the result in this study is in agreement with finding of Fegan and Prior (2005) and other previous studies.
Several techniques were chosen to analyze genetic diversity of Asian R. solanacearum isolates such PCR–RFLP (Kumar et al. 2004), fliC-RFLP (Xue et al. 2011), AFLP (Jeong et al. 2007), and RAPD (James et al. 2003; Chen et al. 2005; Prasannakumar et al. 2012; Dinh et al. 2008). In this study, highly polymorphic level and genetic variation were detected among different strains using RAPD markers. Fifty-two Vietnamese R. solanacearum strains and two Taiwanese strains were devided into 3 clusters using 21 RAPD polymorphic primers. From the output of RAPD markers, UPGMA cluster analysis tended to group most close geographically isolates together, while genetic structure analysis using STRUCTURE software showed that three gene pools are represented in the overall set of 54 isolates. Together, 52 Vietnamese R. solanacearum strains and two Taiwanese strains were divided into 6 sub-clusters with geographical affiliation using 21 RAPD polymorphic primers. Of the 6 sub-clusters, cluster I, sub-cluster IIIa, and subcluster IIa were the southern isolates, whereas, sub-cluster IIb, IIc and IIIb were represented to the northern strains. Two Taiwan strains grouped in the same cluster with southern Vietnamese isolates. Isolates from Lam Dong were seperated in two different clusters (I and IIa). Poussier et al. (2000) used PCR–RFLP analysis based on the hrp gene to classify African strains, but the resulting classification system was not fully consistent with the results of Fegan et al. (1998) using subgroup specific primers that designed using the sequences of the 16S-23S intergenic spacer region of the rRNA gene, the polygalacturonase gene, and the endoglucanase gene. Horita and Tsuchiya (2001) used REP-PCR analysis using different primer sets with 16S rRNA gene sequence analysis yielded the genomic fingerprints of several Japanese strains; however, the grouping obtained failed to reflect the diverse hosts and geographical origins from which the isolates were obtained. Rodrigues et al. (2012) also found that there is no correlation concerning race, biovar or geographical origin among Brazilian strains.
Symptom expression of tomato bacterial wilt could be affected by pathogen strain, inoculation method, and environmental factors such as temperature (Hayward 1991). Further, resistance to bacterial wilt in tomato is reported to be location-specific (Wang et al. 1998, 2013) and strains-specific (Truong et al. 2008; Wang et al. 2013). Thus, breeding a tomato variety that could be resistant to all known tomato-pathogenic R. solanacearum strains seems unrealistic. Twelve isolates representative of 12 provinces were selected to assess their aggressiveness on H7996 (a resistant line), WVa700 (a susceptible line) and TN52 (a F1 hybrid commonly used in Vietnam tomato production). H7996 was fully susceptible to isolates Rs101 (Hai Duong), Rs127 (Hai Phong), and Rs341 (Nam Dinh), but resistant to isolates Rs26 (Lam Dong), Rs38 (Ho Chi Minh), Rs51 (Tien Giang), Rs57 (Vinh Long), Rs66 (Can Tho), Rs86 (Ninh Thuan), Rs249 (Ha Noi), Rs319 (Thai Binh) and Rs373 (Nghe An). The result is in agreement with a previous study that reported H7996 as highly susceptible to an isolate from Hai Duong, but resistant to isolates from Ha Noi, Quang Ninh, Ha Nam and Phu Tho (Truong et al. 2015). Isolates Rs341 (Nam Dinh) and Rs249 (Ha Noi) belonged to cluster A4 but caused different virulence. In this study, we found that Pss4 and Pss190 had high similarity in genetic diversity but differed in aggressiveness (Truong et al. 2008). Thus, the results demonstrated that R. solanacearum isolates even though similar in genetic background do not necessarily cause similar virulence.
‘Hawaii 7996’ (H7996) was identified as the tomato cultivar with the most stable resistance to R. solanacearum based on field evaluations conducted worldwide (WANG et al. 1998). However, Truong et al. (2008) reported that H7996 was fully susceptible to Pss190 but highly resistant to Pss4. The finding in this study also supported an evidence of the presence of strain-specific resistance in H7996.
References
Buddenhagen I, Sequeira L, Kelman A (1962) Designation of races in Pseudomonas solanacearum (Abstr.). Phytopathology 52:726
Chen Y, He L, Xu J (2005) Detection of bacterial wilt infection in potato using PCR. Acta Phytophylacica Sin 32(2):129–132
Cook D, Sequeira L (1994) Strain differentiation of Pseudomonas solanacearum by molecular genetics methods. In: Hayward AC, Hartman GL (eds) Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, pp 77–93
Cook D, Barlow E, Sequeira L (1989) Genetic diversity of Pseudomonas solanacearum: detection of restriction fragment length polymorphisms with DNA probes that specify virulence and hypersensitive response. Mol Plant Microbe Interact 2:113–121
Dinh PT, Do PT, Nguyen YT (2008) Genetic diversity of Pseudomonas solanacearum isolates caused bacterial wilt in peanut using RADP. J Sci Technol 46(6):43–50 (in Vietnamese)
Do DT (2009) Commonality of bacterial wilt from different hosts in Ha Noi. J Plant Prod 2:38–42 (in Vietnamese)
Doan TT (1995). Study on Pseudomonas solanacearum on different hosts in Northern Vietnam. Ph.D. Dissertation, Ha Noi, Vietnam, Ha Noi Agriculture University (in Vietnamese)
Fegan M, Prior P (2005) How complex is the Ralstonia solanacearum species complex. In: Allen C, Prior P, Hayward AC (eds) Bacterial wilt disease and the Ralstonia solanacearum species complex, 1st edn. APS Press, Minnesota
Fegan M, Taghavi M, Sly LI, Hayward AC (1998) Phylogeny diversity and molecular diagnostics of Ralstonia solanacearum. In: Allen C, Prior P, Elphinstone J (eds) Bacterial wilt disease: molecular and ecological aspects. Springer, Berlin, pp 19–33
Fry WE (1978) Quantification of general resistance of potato cultivars and fungicide effects for integrated control of potato late blight. Phytopathology 68:1650–1655
Guidot A, Prior P, Schoenfeld J, Carrere S, Genin S, Boucher C (2007) Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum inferred from gene distribution analysis. J Bacteriol 189:377–387
Hayward AC (1964) Characteristics of Pseudomonas solanacearum. J Appl Bacteriol 27:265–277
Hayward AC (1991) Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 29:65–87
Hayward AC (1994) The hosts of Pseudomonas solanacearum. In: Hayward AC, Hartman GL (eds) Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford
Horita M, Tsuchiya K (2001) Genetic diversity of Japanese strains of Ralstonia solanacearum. Phytopathology 91:399–407
Ivey MLL, McP Garderner B, Opina N, Miller SA (2007) Diversity of Ralstonia solanacearum infecting eggplant in the Philippines. Phytopathology 97:1467–1475
James D, Girija D, Mathew SK, Nazeem PA, Balu TD, Varma SA (2003) Detection of Ralstonia solanacearum race 3 causing bacterial wilt of solanaceous vegetables in Kerala, using random amplified polymorphic DNA (RAPD) analysis. J Trop Agric 41:33–37
Jeong YH, Kim J, Kang YS, Lee SD, Hwang IY (2007) Genetic diversity and distribution of Korean isolates of Ralstonia solanacearum. Plant Dis 91:1277–1287
Kelman A (1954) The relationship of pathogenicity of Pseudomonas solanacearumto colony appearance in Tetrazolium medium. Phytopathology 51:158–161
Kumar A, Sarma YR, Anandaraj M (2004) Evaluation of genetic diversity of Ralstonia solanacearum causing bacterial wilt of ginger using REP–PCR and PCR–RFLP. Curr Sci 87:1555–1561
Le TL (1997) Influence of different ecology factors on bacterial wilt in peanut in Northern Vietnam. J Plant Prod 4:5–8 (in Vietnamese)
Lebeau A, Daunay MC, Frary A, Palloix A, Wang JF, Dintinger J, Chiroleu F, Wicker E, Prior P (2011) Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resource respond to diverse strain in the Ralstonia solanacearum species complex. Phytopathology 101:154–165
Li X, Dorsch M, Del Dot T, Sly LI, Stackebrandt E, Hayward AC (1993) Phylogenetic studies of the rRNA group II pseudomonads based on 16S rRNA gene sequences. J Appl Bacteriol 74:324–329
Lin CH, Tsai KC, Prior P, Wang JF (2014) Phylogenetic relationship structure of Ralstonia solanacearum isolated from diverse origins in Taiwan. Plant Pathol 63:1395–1403
Nguyen TT (2012) Study on bacterial wilt caused by Ralstonia solanacearum smith in peanut and Potato in Ha Noi and control strategies. Ph.Dissertation, Ha Noi, Vietnam, Ha Noi Agriculture University (in Vietnamese)
Nguyen LT, Phan TB (1993) Bacterial wilt in peanut in Northern Vietnam. In: Proceedings of plant production, pp 15–16 (in Vietnamese)
Nguyen HX, Nguyen YT, Nguyen LV (1997) Results of study on distribution and damage of R. solanacearum in Nothern Vietnam. J Plant Prod 6:27–31 (in Vietnamese)
Nguyen YT, Nguyen VV, Dang LTP (2002) Race and biovar of Ralstonia solanacearum caused bacterial wilt in different hosts. In: Proceeding of plant disease and biotechnology, Ha Noi, Vietnam, pp 64–66 (in Vietnamese)
Opina N, Tavner F, Hollway G, Wang JF, Li TH, Maghirang R, Fegan M, Hayward AC, Krishnapillai V, Hong WF, Holloway BW, Timmis JN (1997) A novel method for development of species and strain-specific DNA probes and PCR primers for identifying Burkholderia solanacearum (Formerly Pseudomonas solanacearum). Asia Pac J Mol Biol Biotechnol 5:19–30
Poussier S, Trigalet-Demery D, Vanderwalle P, Goffinet B, Luisetti J, Trigalet A (2000) Genetic diversity of Ralstonia solanacearum as assessed by PCR-RFLP of the hrp gene region, AFLP and 16S rRNA sequence analysis, and identification of an African subdivision. Microbiology 146:1679–1692
Prasannakumar MK, Chandrashekara KN, Deepa M, Vani A, Khan ANA (2012) Finger printing of Ralstoniasolanacearum isolates by Rep-PCR and RAPD. Pest Man Hortic Ecosyst 18:179–187
Prior P, Fegan M (2005) Recent developments in the phylogeny and classification of Ralstonia solanacearum. Acta Hortic 695:127–136
Pritchard JK, Jonathan K, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Rodrigues LMR, Destéfano SAL, da Silva MJ, Costa GGL, Maringoni AC (2012) Characterization of Ralstonia solanacearum strains from Brazil using molecular methods and pathogenicity tests. J Plant Pathol 94(3):505–516
Sagar V, Gurjar MS, Arjunan J, Bakade RR, Chakrabarti SK, Arora RK, Sharma S (2014) Phylotype analysis of Ralstonia solanacearum strains causing potato bacterial wilt in Karnataka in India. Afr J Microbiol Res 8:1277–1281
Seal SE, Jackson LA, Daniels MJ (1992) Use of tRNA consensus primers to indicate subgroups of Pseudomonas solanacearum by polymerase chain reaction amplification. Appl Environ Microbiol 58:3759–3761
Shaner G, Finney RE (1977) The effect of nitrogen fertilization in the expression of slow-mildewing resistance in Knox wheat. Phytopathology 67:1051–1056
Truong HTH, Wang JF, Esch E (2008) Resistance to Taiwanese race 1 strains of Ralstonia solanacearum in wild tomato germplasm. Eur J Plant Pathol 122:471–479
Truong HTH, Tran TV, Nguyen TTT, Pham BT, Tran HN (2015) Aggressiveness assessment of bacterial Ralstonia solanacearum isolates in the Nothern region. Sci Technol J Agric Rur Dev 260:19–25 (in Vietnamese)
Tung PX, Vander Zaag P, Schemiediche P (1992) Effects of resistance genes, heat tolerance genes and cytoplasm on expression of resistance to Pseudomonas solanacearum in potato. Euphitica 60:127–138
Wang JF, Hanson PM, Barnes JA (1998) Worldwide evaluation of an international set of resistance sources to bacterial wilt in tomato. In: Prior P, Allen C, Elphinstone J (eds) Bacterial wilt disease: molecular and ecological aspects. Springer, Berlin, pp 269–275
Wang JF, Fang IH, Truong HTH, Huang SM, Balareto CH, Dittapongpitch V, Hidayati N (2013) Identification of major QTLs associated with stable resistance of tomato cultivar ‘Hawaii 7996’ to Ralstonia solanacearum. Euphytica 190:241–252
Wicker E, Lefeuvre P, Cambiaire JC, Lamaire C, Poussier S, Proir P (2012) Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum interred from MLSA. Int Soc Microb Ecol J 6:1–14
Winstead NN, Kelman A (1952) Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathology 42:628–634
Xue QY, Yin YN, Yang W, Heuer H, Prior P, Guo JH, Smalla K (2011) Genetic diversity of Ralstonia solanacearum strains from China asessed by PCR-based fingerprints to unravel host plant- and site-dependent distribution patterns. FEMS Microbiol Ecol 75:507–519
Acknowledgements
We are grateful to Vietnam National Foundation for Science and Technology Development (NAFOSTED) for supporting research Grant No 106-NN.99-2013.05. Many thanks to Dr. Jaw-Fen Wang for providing DNA of Taiwanese R. solanacearum strains.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Truong, H.T.H., Duong, TT., Nguyen, T.T.H. et al. Aggressiveness and genetic diversity of Ralstonia solanacearum strains from tomato in Vietnam. Indian Phytopathology 71, 599–610 (2018). https://doi.org/10.1007/s42360-018-0084-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-018-0084-1