Abstract
Causing potato brown rot, Ralstonia solanacearum (R. solanacearum) strains are reported as one of the most destructive bacteria to potato (Solanum tuberosum L.) in China. In this study, 113 strains were isolated from potato, collected in the four major agroecological zones in China. The study showed that 102 strains belonged to the phylotype IIB sequevar 1 (race 3 biovar 2). The 11 remaining strains belonged to the phylotype I, sequevar 13, 17, 18, 16 or 14 M, a new sequevar closely related to sequevar 14. Thirty-four strains were further characterized according to their virulence at low temperature on three wild potato species. IIB-1 strains all belonged to high and moderate virulence, while others belonged to the low virulence group, which had limited pathogenicity.
Resumen
Causante de la pudrición café, se reporta a las variantes de Ralstonia solanacearum (R. solanacearum) como una de las bacterias más destructivas en papa (Solanum tuberosum L.) en China. En este estudio, se aislaron 113 cepas en papa, que se colectaron en las cuatro zonas agroecológicas principales en China. El estudio mostró que 102 variantes pertenecían al filotipo IIB sequevar 1 (raza 3, biovar 2). Las 11 variantes restantes pertenecían al filotipo I, sequevar 13, 17, 18, 16 o 14 M, un nuevo sequevar estrechamente relacionado con el sequevar 14. Posteriormente se caracterizaron 34 cepas de acuerdo con su virulencia a baja temperatura en tres especies silvestres de papa. Todas las cepas IIB-1 pertenecieron a los grupos de virulencia alta y moderada, mientras que otras pertenecieron al grupo de baja virulencia, el cual tuvo patogenicidad limitada.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato (Solanum tuberosum L.) is considered as one of the most important food and vegetable crops worldwide, especially in China. The cultivated area in China in 2014 was 5.6 million ha producing 96 million tons with an average yield of 17 T/ha (FAO 2014). Causing brown rot of potato, Ralstonia solanacearum (R. solanacearum) strains are reported as the second most destructive disease after late blight, both nationally and globally. It causes considerable amount of damage and crop loss in tropical, subtropical and warm temperate regions with an estimated production loss evaluated at more than $950 million per year (Patil et al. 2012). In China, R. solanacearum strains were first isolated from peanut in 1930s. As reported, they have spread from 42°N (Guyuan County, Hebei Province) to 20°N (Hainan Province), and affected more than 44 host species of 22 plant families, being especially serious on potato and tobacco. A recent increase in the occurrence of bacterial wilt has been reported in China, where large outbreaks were reported in 16 provinces including Hubei (Li et al. 2015, 2016; Lin et al. 2014; Xu et al. 2009, 2011). Brown rot has only been reported on potato in a few provinces so far, including Guizhou, Yunnan, and Hubei. A comprehensive and systematic investigation of this economically important pathogen is currently lacking (He et al. 1999; Lin et al. 2004; Zheng et al. 2014).
R. solanacearum is a complex species, which can attack approximately 450 species in 54 families (Hayward 1991). Historically, R. solanacearum has been divided into five races (Buddenhagen et al. 1962), which is based on their host range, and six biovars (Hayward 1964) according to their metabolism of three dextrose sugars and three hexose alcohols. These phenotypic classification approaches are frequently used. Though these are the most classical methods to classify R. solanacearum strains, there are many drawbacks. For example, they are laborious, and time-consuming (Villa et al. 2005), and neither can reflect differences on the geographical basis nor are phylogenetically meaningful.
Using the hierarchical classification scheme which is based on the partial sequences of the ITS region (between 16S–23S rRNA) and the aggressive gene Endoglucanase (egl), R. solanacearum strains are partitioned into four phylotypes, corresponding to four genetic clusters associated with strains geographical origin (I: Asian, II: American, III: African, and IV: Indonesian), and fifty-two sequevars, reflecting the current known genetic diversity (Fegan and Prior 2005). The Brown rot ecotype clusters strains that belong to the phylotypes IIB sequevar 1 (IIB-1) and phylotype IIB sequevar 2 (IIB-2) (Wicker et al. 2012). However, under laboratory conditions, most of sequevars belonging to four phylotypes can affect potato, except for sequevar10 that causes banana blood disease (Phylotype IV), at temperatures over 28 °C (Cellier and Prior 2010). It is widely known that strains of R. solanacearum are tropical or subtropical pathogens. As “cold-tolerant” strains (Milling et al. 2009; Cellier and Prior 2010), IIB-1 strains may become more and more popular in the Northern territories, previously thought as relatively free of R. solanacearum. Therefore, it is imperative to inspect and study this type of strains. In China, two out of five races (race 1 and 3), two biovars (bv. 2 and 3) and four out of 52 sequevars (1, 13, 17 and 18) belonging to phylotype I and II were detected and can cause brown rot (Lin et al. 2014; Xu et al. 2009, 2011).
Because of the large number of strains exhibiting complex genetic and phenotypic characteristics, the diversity and distribution of potato bacterial wilt strains in China is currently poorly understood especially for the main groups of strains.
The objectives of this study were to (i) assess the distribution of potato bacterial wilt in Chinese major agro-ecological zones, (ii) ascertain the biovar and estimate the genetic diversity of R. solanacearum using the hierarchical classification scheme, and (iii) assess the virulence at cool temperature and the genetic diversity of R. solanacearum strains.
Materials and Methods
Bacterial Isolation and Preservation
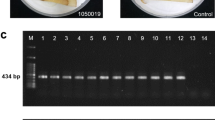
In 2014 and 2015, potato plants with wilt symptoms or potato tubers with brown rot symptoms were collected from a total of 54 sites in 21 provinces in Chinese major agro-ecological zones with the help of local potato growers and agricultural agencies. From the tubers or stems of wilted plants, 113 R. solanacearum strains were isolated on tetrazolium chloride agar (TZC) medium and incubated for 48 to 72 h at 28 °C (Kelman 1954). Typical colonies with a pink center and milky edge, were purified on TZC medium. Using the specific primer pair 759/760 (Table 1), R. solanacearum strains were confirmed by PCR, which would produce a single 280-bp fragment as PCR product (Opina et al. 1997).
Biovar Determination
The biovar of strains can be determined by their ability to utilize three disaccharides and three hexose alcohols in 96 well plates, as previously outlined (Hayward 1964). Briefly, identification was carried out on basal medium composed as below: 1.0 g of yeast extract; 0.2 g of KCl,; 0.2 g of MgSO4.7H2O; 1.0 g of NH4H2PO4; 15 g of agar; 6 mL of 1% aqueous solution of bromothymol blue; 1 L of distilled water, and adjusted to pH 7.2. 5 mL of 10% solutions of lactose, cellobiose, maltose, mannitol, dulcite, and sorbitol, which have been pre-filtered by using 0.22 μm, MF-Millipore MCF membrane individually, were mixed with 45 mL of molten basal medium, separately. 200 μL of these media was then added to plates of 96-well micro-titer. The positive and negative controls were Hayward’s medium without a carbon source or un-inoculated wells. All wells were inoculated with 2 μL of OD600 = 0.1 (≈1 × 108 CFU mL−1) fresh cell suspension. These plates, whose color was recorded after the interval of 24 h, were incubated at 28 °C for approximately 3 weeks until the color of positive cultures changed from green to yellow. Each test was replicated three times.
DNA Extraction and Phylotype Identification
Using TaKaRa MiniBEST Bacterial Genomic DNA Extraction Kit Ver.3.0 (Beijing Tiangen Biotechnology Co., Ltd., China), the total DNA of all strains was extracted. Phylotype identification was conducted on all strains using specific multiplex PCR (Pmx-PCR) with phylotype-specific primers targeting the spacer region 16S–23S r RNA (ITS) (Table 1). The PCR reaction was carried out according to Fegan and Prior (2005), using the species-specific primer 759/760. Strains, which were classified as phylotype II, were further tested using the specific PCR primer pair 630-F / 631-R (Table 1), which should amplify a 278-bp band from strains of phylotype IIB-1 (Fegan et al. 1998).
Identification of Sequevar of R. solanacearum
According to Poussier et al. (2000), by sequencing the partial egl gene, sequevar of all strains were identified through the PCR application with primer pair Endo-F and Endo-R (Table 1), The reaction mixture (total volume of 50 μL) contained 25 μL of 2× UTaq PCR Master Mix (Beijing Zoman Biotechnology Co., Ltd., China), 20 μmol of each primer, 100 ng of DNA as template, and the rest was deionized water. PCR was performed as followings: initial run, 96 °C for 5 min, followed by 30 cycles of 95 °C for 30s, 64 °C for 30s and 72 °C for 2 min, and the last extending cycle, 72 °C for 10 min. Samples (3 μL) of reaction mixtures were examined and the remained PCR products were purified and sequenced by Sangon Biotech (Shanghai Sangon Biotech Co., LTD., China). Using the algorithm of Jukes and Cantor (1969), the dendrogram was generated by MEGA 6.0 (ImageMagick Studio LLC) with 1000 bootstrap resamplings.
Pathogenicity Assays
In order to determine the virulence of collected strains, 34 R. solanacearum strains, representing a range of sequevars from different provinces, were used in pathogenicity assays, performing the root injury inoculation technique (Cellier and Prior 2010). Potato lines C9701 (from species S. chacoense), W5337.4X (from the hybridization F1 progeny of species S. phureja × S. tuberosum) and CT7-6 (from species S. bulbocastanum), were used for this assay. These lines were chosen because they exhibited varying compatibility with R. solanacearum strain PeaHuB4 and potato cultivars show no resistance in previous study in our lab (data not shown). About 10 μL volume of tested strains were streaked on TTC medium plate, separately, cultivated at 28 °C. For each strain, an isolated clone was selected to be restreaked and cultured for 24 to 38 h at 28 °C on the same medium. A bacterial suspension in 10 mM Tris (pH 7.1) was made from the restreaked culture and adjusted to OD600 = 0.1 (≈108 CFU mL −1). Four week-old seedlings at 5–7 leaf stage were inoculated by adding 10 mL of above prepared suspension on lateral roots that had been cut by a scalpel. Five seedlings of every potato line were inoculated for each strain in a greenhouse, with temperature as 20 ± 2 °C and RH as 75%. And then their symptoms were recorded every 2 days for continuous 3 weeks after inoculation, using the following scales: 0 = plant without visible symptoms; 1 = 0 to 25% leaves wilted; 2 = 26% to 50% wilted; 3 = 51% to 75%; 4 = 76% to 100%. This experiment was repeated three times.
The virulence of strains was evaluated using the Disease Index (DI) and Area under Disease Progress Curve (AUDPC). DI was calculated as followings: DI = (n1 × 1 + n2 × 2 + n3 × 3 + n4 × 4) / (n0 + n1 + n2 + n3 + n4) × 4, n0, n1, n2, n3, n4 represented as the number of plants with symptoms of 0, 1, 2, 3 and 4, respectively. The resistance reactions of the three potato materials to each strain were evaluated based on DI on the 21th day after inoculation as follows: (i) DI values of 0.0 were considered as no disease symptom (N); (ii) DI values of 0.1 to 1.5 was considered resistant (R); (iii) 1.6 to 3.0 as medium resistant (MR); (iv) 3.1 to 4.0 as susceptible (S), according to Horita and Tsuchiya (2001).
Area under Disease Progress Curve (AUDPC) was calculated by the following formula according to the trapezoidal integration of the DI from 0 to 21 days after inoculation (Wicker et al. 2007):
Where X is the disease index, n is the number of individual plant evaluated, and (t i+1 − t i ) is the time interval days between two consecutive evaluations. The type of virulence was classified into 3 groups based on the AUDPC values of three potato materials, using SPSS version 19.0 hierarchical cluster analysis marked them as follows: L as low virulence, M as medium virulence, H high virulence.
DI and AUDPC values were estimated using the method of variance (ANOVA) by SPSS. Means were separated using Least Significant Difference (p = 0.05), where significant treatment effects were identified.
Results
The Collection and Traditional Identification of R. solanacearum Strains
There are four major agroecological zones in China (Fig. 1), including Zone1: North single-cropping region, Zone II: Central double-cropping, Zone III: South winter-cropping, and Zone IV: Southwest mixed-cropping zones (Jansky et al. 2009). All regions were surveyed and potato samples were taken from a total of 54 sites located in 21 provinces. A total of 113 R. solanacearum isolates from 24 of 54 sites in 11 of 21 provinces were isolated. None could be isolated from the other 30 sites (Fig. 1). A total of 12 strains were obtained from other hosts, and were also subjected to the same tests as tested strains.
Biovar Characteristics
Strains isolated from potato were identified as biovar 2 (n = 102), biovar 3 (n = 8) and biovar 4 (n = 3). Biovar 3 and 4 were found in zones II and IV, while biovar 2 was distributed in all four zones. Comparatively, 12 strains from other hosts were identified as biovar 3 (Table 2).
Phylotype Identification
Using the Pmx-PCR method, 2 phylotypes were identified: phylotype I (Asian) and phylotype II (American). Of the 113 strains, 90% (102 strains) belonged to phylotype II and 10% (11 strains) to phylotype I. All phylotype II strains were tested by 630/631, which is the specific primer pair of brown rot pathogen. One band of 278-bp R3bv2-specific fragment was amplified in all of the 102 strains. The 12 strains from other hosts were identified as phylotype I (Table 2).
Phylogenetic Analysis
Partial egl gene sequences of all the 125 strains and 23 reference strains, which could help to assign the phylogenetic position of the strains, were used to produce the phylogenetic trees (Fig. 2). The phylogenetic position in egl-based tree was completely consistent with their phylotype determination identified by Pmx-PCR and 630/631-PCR. All the tested strains were divided into two phylotypes (I and IIB) clustered by the phylogenetic tree based on sequence of egl gene. In phylotype IIB, all 102 strains clustered with reference strains IPO1609, UW551 and JT516, which belong to sequevar 1. Only few strains were grouped into previously known sequevars (such as 13, 14, 15, 16, 17, 18 and 34). However, there are four strains, PeaHuB4, PeaFJ1, HZAU089 and HZAU091, that did not cluster with any of the reference strains. We assigned these to a new sequevar (i.e., 14 M) because they are mostly related to sequevar 14 type.
Phylogenetic neighbor-joining tree based on the partial endoglucanase (egl) gene sequence of R. solanacearum infecting potato from China. The dendrogram was generated by MEGA 6.0 using the algorithm of Jukes and Cantor (1969) with 1000 bootstrap resamplings. Numbers at branch points indicate percent bootstrap support for 1000 iterations. ★ Present reference sequevars; ● present new sequevar 14 M
Pathogenicity Assessment
Thirty-four representative strains were tested for their pathogenicity by inoculating three wild potato lines C9701, W5337.4X and CT7-6. The potato lines showed different resistance to R. solanacearum strains 21 days post inoculation (DPI) (Table 3). CT7-6 was the most susceptible with a final DI mean of 2.86 ± 0.71 and AUDPC of 25.94 ± 8.63, and exhibited medium resistance (MR) and susceptibility (S) to 79% of all strains tested and no wilting symptoms (N) to 12% of the strains. The respective values of C9701 as a moderately susceptible species were 2.46 ± 0.89 and 21.20 ± 9.31 for DI and AUDPC value, respectively, and this species was susceptible (S) to 56% of the strains and showed no symptoms (N) to 27% of the strains. W5337.4X was the most resistant line, with DI = 1.76 ± 0.92 and AUDPC =16.59 ± 9.11, showing no symptom (N) to 44% and susceptibility (S) to 38% of the strains.
The virulence of the 34 strains was estimated by the cluster analysis based on AUDPC values from the three wild potato species. Phenotypes were divided into three types, including type H (high virulence), type M (moderate virulence) and type L (low virulence) (Fig. 3). Only IIB-1 strains, whether isolated from potato or not, belonged to type M and H, indicating strong virulence to potato at low temperature. However, there were four IIB-1 strains that belonged to type L, exhibiting weak virulence (Fig. 4). The virulence of strains may have slight correlation with region, because ignoring sampling numbers (n = 2), strains isolated from Zone I all belonged to high virulence, while the virulence of strains isolated from Zone II and IV had three types.
Virulence analysis of R. solanacearum at low temperature. The dendrogram was generated by SPSS 22 through weighed Euclidean distance classification. a, Red represents strains belonged to I/13; b, Purple represents I/14; c, Yellow represents I/14 M; d, Orange represents I/15; e, Green represents I/16; f, Blue represents I/17; g, Pink represents I/34; h, Black represents IIB/1
IIB-1 strains, with different virulence types, exhibited varied aggressiveness characteristics (Fig. 5). At 21 days, L, H and M type strains were equally pathogenic (P < 0.01), with an average DI of 4.00. This indicates that the three wild potato lines should be considered susceptible or highly susceptible to these strains. However, for strains belonging to type L, the pathogenesis was significantly lower (P < 0.01) than type H and type M, which showed almost no virulence on W5337.4X, with a mean DI of 0.40 ± 0.40 and AUDPC 4.42 ± 4.42. These strains showed the highest virulence on CT7-6 among the three lines evaluated, only with a mean DI of 2.25 ± 0.75, and AUDPC value 10.45 ± 4.91. The plant phenotypes reflected different virulence types of different strains (Fig. 4).
The area under disease progress curve (AUDPC) values and disease index (DI) for three potato species, inoculated with R. solanacearum strains of different virulence type. The mean of AUDPC value was assessed from 0 to 21 days after inoculation. And the mean of disease index was assessed at 21 days after inoculation. The vertical lines at the top of each bar represent the standard error of the mean. Values with the same capital letters for the same material were not significant different at P < 0.01, with the same lower-case letters not significant different at P < 0.05 according to Least Significant Difference
Discussion
This study characterized R. solanacearum strains affecting potato, through an extensive collection of strains collected throughout China. The type IIB-1 represented 90% of all strains isolated and it was distributed in all four zones. The results demonstrate that potato bacterial wilt, or brown rot is most likely caused by type IIB-1 strains of R. solanacearum, which is being the dominant pathogen across the four local major potato agroecological zones. One would expect such results because of the frequent movement of potentially-infected symptomless plant material from one region into another. Type IIB-1 strains can live longer in potato tubers than in water under cool temperature, and longer than other type strains (Milling et al. 2009). Moreover, most seed potatoes are transported from North to South in China. We have isolated R. solanacearum from potato tubers from the North single-cropping region, which is the main region for cultivating seed potatoes and maintains a cold temperature in winter. Other factors such as the widespread use of monoculture and suitable climatic conditions could also contribute to the transmission of this pathogen.
Our study showed that potato brown rot is more serious in the central double-cropping and southwest mixed-cropping zones which have favorable climatic conditions, and consequently disease incidence and prevalence of bacterial wilt. In contrast, R. solanacearum strains were difficult to be isolated in cool regions such as the North single-cropping zone which includes the following provinces: Heilongjiang, Gansu, Jilin, Inner Mongolia, Ningxia, Gansu and part of provinces as Liaoning, Hebei, Shanxi, Qinghai, and Xinjiang. Factors that may have influenced such distribution is the low temperatures (−4 ~ 10 °C), and annual rainfall (50 ~ 1000 mm) with significant fluctuation in night and day temperatures that the region typically experience. IIB-1 strains are sensitive to fluctuations in temperature, leading to loss of virulence and in some cases death (Scherf et al. 2010). The other reason why it is difficult to isolate R. solanacearum strains from the North single-cropping zone might be the stricter or better environmental condition for potato growth including using virus-free potato seeds, systemic disease quarantine and so on, because the North single-cropping zone is an important seed potato production base. In this survey, potato brown rot was found in all four of potato major agroecological zones. The geographical distribution of R. solanacearum of potato in this study was in accordance with the previous report (Xu et al. 2009) which had isolated pathogen from various hosts in China. Of the 113 potato R. solanacearum strains, 102 strains belonged to biovar 2 (90%), which is the most prevalent, 11 belonged to biovar 3, and 3 to biovar 4. Previous reports showed that strains belonging to biovars 1, 2, 2 T, 3 and 4 can infect potato worldwide. While Biovar 2 were the most prevalent strains, causing potato brown rot in most of these countries, which implied that biovar 2 have a wide distribution (Cruz et al. 2012; Jeong et al. 2007; Khoodoo et al. 2010; Ustun et al. 2008).
The results of Pmx-PCR were in accordance with the results of phylogenetic analysis based on sequences of partial egl genes. The Chinese potato strains of biovar 3 and 4 all belonged to phylotype I, and which is thought to be of Asian origin, and all the biovar 2 strains belonged to phylotype II, originated from America (Fegan and Prior 2005). Historically, strains from all the phylotypes can infect potato, with phylotype II strains found worldwide. For example, R. solanacearum isolated from natural infected potatoes belonged to: phylotype I, II and III in India (Sagar et al. 2014), I and IV in Japan (Horita et al. 2014), II from Uruguay (Siri et al. 2011), Mauritius (Khoodoo et al. 2010), Iran (Nouri et al. 2009) and the United States (Hong et al. 2012). Remarkably, the Chinese strains, belonging to phylotype II, exhibit close genetic relationship with the reference strains isolated from American, African and European, which indicates that the genetic background of the Chinese strains may have the same as those of non-Chinese strains. However, strains that belonged to phylotype I exhibited distant genetic relationships with the reference strains, indicating their different background. This supports the close relationship of phylotype of R. solanacearum to geographic origin as previously found by Fegan and Prior (2005).
Cluster analysis of the partial egl gene partitioned Chinese strains of phylotype I from potato into six different sequevars, including five different named sequevars and a single unidentified sequevar, which we designated as 14 M because of its close relation to sequevar 14. Sequevar 1, 13, 17, 18 were previously described on potato, and sequevar 16 was previously described infecting other plants (Xu et al. 2009). Furthermore, sequevar 1, composed of a large number of strains, was widely distributed in the four Chinese regions. The previously unidentified sequevar 14 M may be a new sequevar mutated from sequevar 14, since its placement was close to sequevar 14. The genetic diversity of sequevar 14 M requires further study.
IIB-1 strains originated in South America, in the 1920s it was first reported in Italy, Egypt and Spain, but now strains of type IIB-1 are found all over the world (Clarke et al. 2015). One characteristic is their ability to cause potato wilt disease at cold temperatures, while other types cannot (Aundy et al. 2014; Cellier and Prior 2010; Clarke et al. 2015). Many methods, such as microarray analysis and multi-gene sequence analysis and SNP analysis, were used to explore IIB-1 strains’ origin and route of transmission (Cellier et al. 2012; Clarke et al. 2015; Wicker et al. 2012). The results showed that, IIB-1 strains originated in South America, where they may have evolved with potato or other Solanaceae hosts, and eventually disseminated across the world through the Mediterranean, Northern Europe, Africa, and the Indian Ocean region (Clarke et al. 2015). It is still not known how IIB-1 strains were introduced in China.
Some studies on the virulence of R. solanacearum at low temperature demonstrated that only IIB-1 strains have strong virulence (Milling et al. 2009; Cellier and Prior 2010; Huerta et al. 2015). So, the variation in virulence at low temperature among Chinese strains of R. solanacearum was evaluated using wild potato wild lines with different resistance to R. solanacearum strain PeaHub4 at 28 °C (Data not shown). The results were similar to previous studies, with only strains belonging to IIB-1 having strong virulence (Cellier and Prior 2010). However, there are also four IIB-1 strains that exhibited weak virulence, having almost no wilt symptom or few leaves wilted at a later period. In order to understand the mechanism of R. solanacearum interaction with potato at low temperature, further research is warranted.
These findings provide the first comprehensive analyses of the populations of R. solanacearum affecting potato in China, which can eventually bring about the development of geographically targeted and pathogen-targeted management practices, and aid potato breeding programs in China which would mainly focus on resistance to IIB-1 strains.
References
Aundy, K., P.P. Thekken, and P. Birendranath. 2014. Genetic characterization of an Indian isolate of Ralstonia solanacearum race 3/ biovar 2/ phylotype IIB from potato. Indian Phytopathology 67 (4): 346–352.
Buddenhagen, I., L. Sequeira, and A. Kelman. 1962. Designation of races in Pseudomonas solanacearum. Phytopathology 52: 726.
Cellier, G., and P. Prior. 2010. Deciphering phenotypic diversity of Ralstonia solanacearum strains pathogenic to potato. Phytopathology 100 (11): 1250–1261.
Cellier, G., B. Remenant, F. Chiroleu, P. Lefeuvre, and P. Prior. 2012. Phylogeny and population structure of brown rot-and Moko disease-causing strains of Ralstonia solanacearum phylotype II. Applied and Environmental Microbiology 78 (7): 2367–2375.
Clarke, C.R., D.J. Studholme, B. Hayes, B. Runde, A. Weisberg, R. Cai, T. Wroblewski, M.C. Daunay, E. Wicker, J.A. Castillo, and B.A. Vinatzer. 2015. Genome-enabled phylogeographic investigation of the quarantine pathogen Ralstonia solanacearum race 3 biovar 2 and screening for sources of resistance against its core effectors. Phytopathology 105 (5): 597–607.
Cruz, L., M. Eloy, F. Quirino, H. Oliveira, and R. Tenreiro. 2012. Molecular epidemiology of Ralstonia solanacearum strains from plants and environmental sources in Portugal. European Journal of Plant Pathology 133 (3): 687–706.
FAO. 2014. Website: http://faostat.fao.org/.
Fegan, M., and P. Prior. 2005. How complex is the ‘Ralstonia solanacearum species complex’. In Bacterial wilt disease and the Ralstonia solanacearum species complex, ed. C. Allen, P. Prior, and A.C. Hayward, 449–462. Madison: APS Press.
Fegan, M., and P. Prior. 2006. Diverse members of the Ralstonia solanacearum species complex cause bacterial wilts of banana. Australasian Plant Pathology 2006 (35): 93–101.
Fegan M, M. Taghavi, L.I. Sly and A.C. Hayward. 1998. Phylogeny, diversity and molecular diagnostics of Ralstonia solanacearum. In Bacterial Wilt Disease: Molecular and Ecological Aspects, eds. P. Prior, C. Allen and J. Elphinstone, 19–33. Berlin: Springer.
Hayward, A.C. 1964. Characteristics of Pseudomonas solanacearum. Journal of Applied Bacteriology 27: 265–277.
Hayward, A.C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annual Review of Phytopathology 29: 65–87.
He, Y.K., Z.J. Zhao, Y.H. Li, Z.K. Zhang, X.P. Li, and J.X. Deng. 1999. A study on biotype of strains of Pseudomonas solanacearum of potato in Yunnan. Southwest China Journal of Agricultural Sciences 04: 78–81.
Hong, J.C., D.J. Norman, D.L. Reed, M.T. Momol, and J.B. Jones. 2012. Diversity among Ralstonia solanacearum strains isolated from the southeastern United States. Phytopathology 102 (10): 924–936.F.
Horita, M., and K. Tsuchiya. 2001. Genetic diversity of Japanese strains of Ralstonia solanacearum. Phytopathology 91 (4): 399–407.
Horita, M., K. Tsuchiya, Y. Suga, K. Yano, T. Waki, D. Kurose, and N. Furuya. 2014. Current classification of Ralstonia solanacearum and genetic diversity of the strains in Japan. Journal of General Plant Pathology 80 (6): 455–465.
Huerta, A.I., A. Milling, and C. Allen. 2015. Tropical strains of Ralstonia solanacearum outcompete race 3 biovar 2 strains at lowland tropical temperatures. Applied and Environmental Microbiology 81 (10): 3542–3551.
Jansky, S.H., L.P. Jin, K.Y. Xie, C.H. Xie, and D.M. Spooner. 2009. Potato production and breeding in China. Potato Research 52 (1): 57–65.
Jeong, Y., J. Kim, Y. Kang, S. Lee, and I. Hwang. 2007. Genetic diversity and distribution of Korean isolates of Ralstonia solanacearum. Plant Disease 91 (10): 1277–1287.
Jukes, T.H., and C.R. Cantor. 1969. Evolution of protein molecules. In Mammalian protein metabolism, ed. H.N. Munro, 121–132. New York: Academic.
Kelman, A. 1954. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium chloride medium. Phytopathology 44: 693–695.
Khoodoo, M.H.R., E.S. Ganoo, and A.S. Saumtally. 2010. Molecular characterization and epidemiology of Ralstonia solanacearum race 3 biovar 2 causing brown rot of potato in Mauritius. Phytopathology 158 (7–8): 503–512.
Li, Y.Y., H.L. Liu, L. Zheng, J.B. Huang and X.H. Li. 2015. Research progress on genetic diversity of Ralstonia solanacearum in China. Journal of Anhui Agriculture Sciences 14: 107–110, 112.
Li, Y.Y., F. Ji, H.L. Wang, H. Tom, X.H. Li, and J.B. Huang. 2016. Genetic diversity and pathogenicity of Ralstonia solanacearum causing tobacco bacterial wilt in China. Plant Disease 100: 1–9.
Lin, J.J., Q. Huang, F.R. Gui, M.F. Zheng, Z.Y. Li, and Y.X. Sun. 2004. Comparison of pathogenicity for Ralstonia solanacearum of potao in Yunnan province. Southwest China Journal of Agricultural Sciences 06: 738–740.
Lin, C.H., K.C. Tsai, P. Prior, and J.F. Wang. 2014. Phylogenetic relationships and population structure of Ralstonia solanacearum isolated from diverse origins in Taiwan. Plant Pathology 10 (1111): 12209.
Milling, A., F.H. Meng, T.P. Denny, and C. Allen. 2009. Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology 99 (10): 1127–1134.
Nouri, S., M. Bahar, and M. Fegan. 2009. Diversity of Ralstonia solanacearum causing potato bacterial wilt in Iran and the first record of phylotype II/biovar 2T strains outside South America. Plant Pathology 58: 243–249.
Opina, N., F. Tavner, G. Hollway, J.F. Wang, T.H. Li, and R. Maghirang. 1997. A novel method for development of species and strain-specific DNA probes and PCR primers for identifying Burkholderia solanacearum. Asia-Pacific Journal of Molecular Biology and Biotechnology 5: 19–30.
Patil, V.U., J. Gopal, and B.P. Singh. 2012. Improvement for bacterial wilt resistance in potato by conventional and biotechnological approaches. Agriculture Research 1 (4): 299–316.
Poussier, S., P. Prior, J. Luisetti, A.C. Hayward, and M. Fegan. 2000. Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Systematic and Applied Microbiology 23 (4): 479–486.
Sagar, V., A. Jeevalatha, S. Mian, S.K. Chakrabarti, M.S. Gurjar, R.K. Arora, S. Sharma, R.R. Bakade, and B.P. Singh. 2014. Potato bacterial wilt in India caused by strains of phylotype I, II and IV of Ralstonia solanacearum. European Journal of Plant Pathology 138 (1): 51–65.
Scherf, J.M., A. Milling, and C. Allen. 2010. Moderate temperature fluctuations rapidly reduce the viability of Ralstonia solanacearum race 3, biovar 2, in infected geranium, tomato and potato plants. Applied and Environmental Microbiology 76 (21): 7061–7067.
Siri, M.I., A. Sanabria, and M.J. Pianzzola. 2011. Genetic diversity and aggressiveness of Ralstonia solanacearum strains causing bacterial wilt of potato in Uruguay. Plant Disease 95 (10): 1292–1301.
Toukam, G.M.S., G. Cellier, E. Wicker, C. Guilbaud, R. Kahane, C. Allen, and P. Prior. 2009. Broad diversity of Ralstonia solanacearum strains in Cameroon. Plant Disease 93 (11): 1123–1130.
Ustun, N., M. Ozakman, and A. Karahan. 2008. Outbreak of Ralstonia solanacearum biovar 2 causing brown rot on potato in the Aegean region of Turkey. Plant Disease 92 (6): 973–973.
Villa, J.E., K. Tsuchiya, M. Horita, M. Natural, N. Opina, and M. Hyakumachi. 2005. Phylogenetic relationships of Ralstonia solanacearum species complex strains from Asia and other continents based on 16S rDNA, endoglucanase and hrpB gene sequences. Journal of General Plant Pathology 71 (1): 39–46.
Wicker, E., L. Grassart, R. Coranson-Beaudu, D. Mian, C. Guilbaud, M. Fegan, and P. Prior. 2007. Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Applied and Environmental Microbiology 73 (21): 6790–6801.
Wicker, E., P. Lefeuvre, J.C. de Cambiaire, C. Lemaire, S. Poussier, and P. Prior. 2012. Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. Isme Journal Multidisciplinary Journal of Microbial Ecology 6 (5): 961–974.
Xu, J., Z.C. Pan, P. Prior, J.S. Xu, Z. Zhang, H. Zhang, L.Q. Zhang, L.Y. He, and J. Feng. 2009. Genetic diversity of Ralstonia solanacearum strains from China. European Journal of Plant Pathology 125 (4): 641–653.
Xue, Q.Y., Y.N. Yin, W. Yang, H. Heuer, P. Prior, J.H. Guo, and K. Smalla. 2011. Genetic diversity of Ralstonia solanacearum strains from China assessed by PCR-based fingerprints to unravel host plant- and site-dependent distribution patterns. FEMS Microbiology Ecology 22 (2): 507–519.
Zheng, X.A., B.T. Song, X.D. Tan, and H.L. Chen. 2014. Identification of potato bacterial wilt pathogens. Chinese Potato Journal 02: 83–89.
Acknowledgements
This Project was supported by the Fundamental Research Funds for the Central Universities (Grant No. 2013PY080), the Initial Project Fund from Huazhong Agricultural University (Grant No. 12012) and the Promoting Scientific Research Cooperation and High Level Personnel Training Project in American and Canadian Areas (Grant No. 2012-1738). We thank all of those who supplied the wilted potato plants and tubers for the isolation of R. solanacearum strains and who kindly offered us the reference strains. We also thank all of the faculty members of the Potato Research Team in Huazhong Agricultural University for their support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, L., Wang, B., Zhao, G. et al. Genetic and Pathogenic Diversity of Ralstonia solanacearum Causing Potato Brown Rot in China. Am. J. Potato Res. 94, 403–416 (2017). https://doi.org/10.1007/s12230-017-9576-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12230-017-9576-2