Abstract
Recent increasing attention to environmental issues and the shortage of oil resources have spurred political and industrial interest in the development of environmental friendly and cost-effective processes for the production of bio-based chemicals from renewable resources. Thus, microbial production of commercially important chemicals is viewed as a desirable way to replace current petrochemical production. Corynebacterium glutamicum, a Gram-positive soil bacterium, is one of the most important industrial microorganisms as a platform for the production of various amino acids. Recent research has explored the use of C. glutamicum as a potential cell factory for producing organic acids such as lactate and succinate, both of which are commercially important bulk chemicals. Here, we summarize current understanding in this field and recent metabolic engineering efforts to develop C. glutamicum strains that efficiently produce l- and d-lactate, and succinate from renewable resources.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Starch and lignocellulose are the most abundant renewable resources on earth. Environmental concerns and the depletion of oil reserves have resulted in governmental actions to establish greater energy independence by promoting research into environmentally benign and sustainable production of bio-based chemicals and fuels from those renewable resources. For example, the US Department of Energy selected 30 top chemical candidates that could serve as building blocks for the production of a variety of useful chemicals and polymers from renewable resources [77]. Microbial production of chemicals will help reduce CO2 accumulation through reduced emissions and through recycling of the CO2 that is released when bio-based chemicals are combusted. The development of suitable strains of microorganisms is, therefore, critically important for the efficient and cost-effective production of target chemicals from biomass resources. In the post-genomic sequencing era, microbial strains tailored for the production of target compounds can be rationally designed through genetic/metabolic engineering rather than through classical strategies based on mutagenesis and screening. Over the past decade, recombinant strains capable of hydrolyzing starchy and lignocellulosic materials have been developed through genetic/metabolic engineering approaches and used to produce bio-based chemicals and fuels [18, 73].
Corynebacterium glutamicum is a Gram-positive, facultative anaerobic soil bacterium with generally regarded as safe (GRAS) status. This organism was first isolated as a glutamate-producing bacterium in 1956 in Japan [33, 72]. Since that time, the bacterium has been used for industrial production of various amino acids including glutamate and lysine under aerobic conditions [34]. In the first decade of the twenty-first century, the genomic sequences of two C. glutamicum strains, ATCC13032 and R, were revealed [21, 27, 88]. This opened the door for systems metabolic engineering of C. glutamicum for rationally designing strains with high productivity of target compounds by combining with knowledge on metabolism and pathway regulation. In addition to its use in the production of various amino acids, C. glutamicum has recently attracted attention as a potentially versatile platform for producing bio-based chemicals and fuels.
In 1993, Dominguez et al. [10] showed that C. glutamicum produces lactate, succinate and acetate as fermentation products when the oxygen is limited. In 2004, a milestone study was achieved by Inui and colleagues showing that C. glutamicum retains its metabolic activity under oxygen deprived conditions when its growth is arrested, and produces l-lactate and succinate at large scale from glucose in minimal medium under oxygen deprivation realized by high-cell density cultivation [23]. The high rate of glucose consumption under oxygen deprivation has been observed only in C. glutamicum R, ATCC13032, and the closely related C. efficiens YS-314 among Corynebacterium relatives tested [80]. After development of the system for producing l-lactate and succinate under oxygen deprivation, the range of target compounds produced by the organism has been expanded by metabolic engineering to include l-valine, l-alanine, and non-natural fermentation products such as ethanol, isobutanol, 1,2-propanediol, and xylitol [4, 16, 17, 24, 26, 45, 59, 62, 81, 82]. Strain optimization for the production of l- and d-lactate and succinate has also been reported (reviewed in this article).
To date, two informative review articles summarizing lactate and succinate production by C. glutamicum have been published [75, 79]. After publication of these articles, several interesting studies have been reported regarding the metabolic engineering of C. glutamicum for the production of lactate and succinate. The present review focuses on recent advances in the metabolic engineering of C. glutamicum for the purpose of optimizing strains to produce l- and d-lactate and succinate from renewable resources.
l- and d-lactate production from glucose
Lactate (lactic acid) is currently used in the food, cosmetic, and pharmaceutical industries, and has attracted considerable attention as a renewable resource material for producing next generation plastics. Lactic acid was selected by the US Department of Energy as one of the top 30 candidates for potential use as sugar-derived building blocks to produce a variety of bulk chemicals and commercially important polymers [77]. However, poly-l-lactic acid, which is a commonly used synthetic polymer, has a markedly lower melting temperature than petroleum-based polymers, thus limiting its practical application [28]. Stereocomplex poly-lactic acid (scPLA), which is a 1:1 mixture of polymerized l- and d-lactic acids, is a promising alternative candidate due to its high heat resistance [12, 20]. However, to produce high-quality scPLA, stereoisomers of both l- and d-lactic acid that are of high optical purity are required. Unlike chemical synthesis, microbial production of lactic acid is, therefore, ideal because either optically pure isomer can be synthesized depending on the chiral-specific l- or d-lactate dehydrogenase (LDH) enzyme expressed by the microorganism. Lactic acid bacteria are used exclusively for lactate production because of their high lactate productivity [48]. However, these bacteria require a nutrient-rich medium for growth, which increases both the production and downstream purification costs [67]. Moreover, the presence of d-lactate is essential for the growth of some lactic acid bacteria, which spoils the optical purity [15]. Other microorganisms, such as metabolically engineered Escherichia coli, have been developed for large-scale and cost-effective l- and d-lactate production in minimal medium [47, 90].
Corynebacterium glutamicum is a facultative anaerobic bacterium and thus can grow aerobically or anaerobically using either oxygen or nitrate, respectively, as the external electron acceptor [46, 63]. However, Inui et al. [23] showed that even when its growth is arrested in the absence of oxygen or nitrate, C. glutamicum maintains its metabolic activity to produce l-lactate as a major product and succinate and acetate as minor products. l-lactate is produced from pyruvate by l-LDH (encoded by the L-ldhA gene), and succinate is produced mainly through an anaplerotic pathway catalyzed by phosphoenolpyruvate carboxylase (PEPC) and through the reductive branch of the tricarboxylic acid (TCA) cycle via oxaloacetate, malate, and fumarate [23] (Fig. 1). Inui et al. [23] established a C. glutamicum two-stage production system consisting of a biomass-production phase under aerobic conditions followed by a production phase under oxygen deprived conditions in minimal medium with a high density of cells in a sealed bottle in which the cells serve as a “biocatalyst”. Anaerobic conditions under high cell density are immediately achieved (<5 s), with a dissolved oxygen concentration of <0.01 ppm [19, 49]. Without genetic modification, the wild-type C. glutamicum strain R produced 574 mM (51.7 g/L) l-lactate in minimal medium after 8 h, with a yield of 1.79 mol/mol glucose when a concentration of 30 g of dry cells/L was used [49] (Table 1). A linear correlation between cell concentration (up to 60 g of dry cells/L) and the rates of lactate and succinate production was observed. When sodium bicarbonate was added at the beginning of the cultivation, the glucose consumption rate increased. The rate was maximized to 68 mM/h when 100 mM sodium bicarbonate was added [49] due to stimulation of the carbon dioxide-fixing reaction catalyzed by PEPC. Indeed, the succinate yield increased from 0.09 to 0.37 mol/mol glucose. The drawback was that the yield of lactate decreased from 1.79 to 1.56 mol/mol glucose. When 400 mM sodium bicarbonate was added, 1,061 mM (95.6 g/L) l-lactate was produced after 6 h, as mixed organic acids [49] (Table 1). The titer was less advantageous, but the productivity was comparable to metabolically engineered E. coli 090B3, which produced 142.2 g/L of l-lactate within 30.5 h, with a yield of 1.74 mol/mol glucose [47] (Table 1).
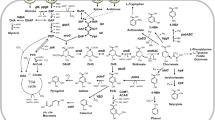
The central metabolic pathways of C. glutamicum including pathways for the assimilation of carbon sources (glucose, xylose, arabinose, cellobiose) used for the production of either l- or d-lactate. When producing d-lactate, native l-LDH encoding gene is deleted. Gray arrows represent pathways not present due to inactivation of genes encoding the enzymes marked with an X, or not activated under anaerobic conditions. Bold lines represent pathways that are useful for increasing d-lactate production by overexpressing the corresponding genes, or that can be exploited for utilizing new carbon sources by introducing assimilating genes. 6PGDH 6-phosphogluconate dehydrogenase, AK acetate kinase, AmyA α-amylase, AraA l-arabinose isomerase, AraB l-ribulokinase; AraD l-ribulose-5-phosphate 4-epimerase, AraE l-arabinose transporter, BglA phospho-β-glucosidases, BglF V317A mutated PTS permease enabling d-cellobiose import, CS citrate synthase, CtfA acetyl CoA:CoA transferase, FBA fructose bisphosphate aldolase, FDH formate dehydrogenase, Fum fumarase, GAPDH glyceraldehyde phosphate dehydrogenase, G6PDH glucose-6-phosphate dehydrogenase, GLK glucokinase, ICD isocitrate dehydrogenase, ICL isocitrate lyase, IolT inositol permease, L-LDH l-Lactate dehydrogenase, D-LDH d-Lactate dehydrogenase, MalE malic enzyme, MDH malate dehydrogenase, MS malate synthase, ODHC 2-oxoglutarate dehydrogenase complex, PC pyruvate carboxylase, PDHC pyruvate dehydrogenase complex, PEPC phosphoenolpyruvate carboxylase, PFK phosphofructokinase, PGI phosphoglucose isomerase, PQO pyruvate/quinone oxidoreductase, PTA phosphotransacetylase, PTS Glu phosphotransferase system for glucose import, PYK pyruvate kinase, SDH succinate dehydrogenase, TPI triosephosphate isomerase, XylA xylose isomerase, XylB xylulokinase
As mentioned above, because optically pure l- and d-lactate are necessary to synthesize scPLA, large-scale production of d-lactate under oxygen deprivation has also been examined. Interestingly, C. glutamicum R does not possess a d-lactate dehydrogenase (Dld)-encoding gene, which enables to grow on d-lactate in C. glutamicum ATCC13032 [29]. To produce optically pure d-lactate using C. glutamicum R, the native l-LDH-encoding gene L-ldhA was deleted, and the d-LDH-encoding D-ldhA gene from Lactobacillus delbrueckii under the control of the C. glutamicum R ldhA promoter was introduced using a plasmid. The resultant strain (ΔldhA/pCRB204) produced 1,336 mM (120.3 g/L) d-lactate with >99.9 % optical purity within 30 h when using 60 g of dry cells/L [50] (Table 1). The yield of d-lactate was 1.73 mol/mol glucose. In this study, the pathway for succinate production was untouched to maintain a high glucose consumption rate, with the result that the succinate yield remained high (0.19 mol/mol glucose). To minimize the succinate yield, the ppc gene was deleted in the subsequent study. As expected, the yield of succinate from the resultant strain (CRZ2/pCRB203) decreased to 0.05 mol/mol glucose [69]. However, the d-lactate productivity also decreased (to 14.5 mM/h) when using 100 g of wet cells/L (corresponding 25 g of dry cells/L). Replacement of the promoter controlling expression of D-ldhA from ldhA to the promoter for gapA (encoding glyceraldehyde phosphate dehydrogenase [GAPDH]) (CRZ2/pCRB215) increased d-lactate productivity to 19.1 mM/h. Further dramatic improvement in d-lactate productivity was achieved by overexpressing glycolytic genes. Recently, it was shown that overexpression of glycolytic genes is an effective strategy for increasing the production of fermentation products in C. glutamicum R under oxygen deprivation. In an alanine-producing strain of C. glutamicum R in which the ldhA and ppc genes were deleted and the alanine dehydrogenase-encoding gene (alaD) from Lysinibacillus sphaericus was overexpressed, overexpression of the GAPDH-encoding gene increased alanine production by 2.7-fold under oxygen deprivation [26]. Moreover, simultaneous overexpression of the genes encoding GAPDH, phosphoglucose isomerase, phosphofructokinase (PFK), and pyruvate kinase was shown to result in a stepwise 6.4-fold increase in alanine production compared with the control strain [82]. In d-lactate-producing C. glutamicum (CRZ2/pCRB215), individual chromosomal overexpression of the 10 glycolytic genes showed that overexpression of the genes encoding glucokinase (GLK), GAPDH, PFK, triosephosphate isomerase (TPI), and fructose bisphosphate aldolase (FBA) increased d-lactate productivity by 98, 39, 15, 13, and 10 %, respectively, compared with the CRZ2/pCRB215 parent strain [69] (Fig. 1). Overexpression of the glycolytic genes also affected the yield of by-products. Overexpression of the GAPDH-encoding gene was shown to increase succinate yield by 59 %. Overexpression of the GLK-encoding gene was shown to dramatically increase the yields of dihydroxyacetone and glycerol, by 9.0- and 6.8-fold, respectively. Upon simultaneous overexpression of the five genes, the strain (LPglc279/pCRB215) produced 2,169 mM (195.4 g/L) d-lactate, with a yield of 1.80 mol/mol glucose after 80 h of cultivation under oxygen deprivation when using 100 g of wet cells/L (corresponding 25 g of dry cells/L) (Tsuge and Inui, Unpublished result; Table 1). The optical purity of the d-lactate produced under these circumstances was >99.9 %. These values are similar to those obtained when using metabolically engineered E. coli B0013-070B (122.8 g/L of d-lactate within 28 h, with a yield of 1.69 mol/mol glucose [90] [Table 1]), as strain LPglc279/pCRB215 produced 1,394 mM (125.6 g/L) d-lactate within 32 h.
Anaerobic/microaerobic succinate production from glucose
Succinate (Succinic acid), an intermediate of the TCA cycle, is selected by the U.S. Department of Energy as one of the top 12 candidate sugar-derived building blocks [77]. Bio-based succinic acid can be used as a building block for the production of a variety of bulk chemicals including 1,4-butanediol, tetrahydrofuran, and gamma-butyrolactone, that are currently produced from n-butane/butadiene via maleic anhydride using chemical processes [41]. Succinic acid is also useful as a building block for producing commercially important polymers, such as polybutylene succinate adipate [60, 89]. Although the current market for succinic acid is remained in the order of 30–50 kton/year based predominantly on petrochemical-based production, bio-based succinic acid could potentially lead to a market in the order of one to several megatons [25]. The potential for using a microbial bio-based process to replace chemical-based succinic acid production has been extensively studied. Several microorganisms, including Anaerobiospirillum succiniciproducens and Actinobacillus succinogenes have been found to produce succinate as the end product of anaerobic fermentation [14, 43]. Metabolically engineered E. coli and Saccharomyces cerevisiae are also used to produce succinate [5, 54, 84]. Two companies, BioAmber and DSM-Roquette, currently produce succinic acid on a large scale using recombinant E. coli and S. cerevisiae, respectively [7].
Inui et al. [23] reported that C. glutamicum R produces succinate under oxygen deprivation, mainly through an anaplerotic pathway catalyzed by PEPC and through the reductive branch of the TCA cycle via oxaloacetate, malate, and fumarate (Fig. 2). Addition of sodium bicarbonate at the beginning of the process enhances the reaction catalyzed by PEPC and accelerates the rate of succinate production in proportion to the concentration, up to 400 mM [49]. Inui et al. later showed that C. glutamicum holds great potential as a host strain for producing succinate by metabolic engineering with the use of the two-stage production system. They tailored C. glutamicum R for producing succinate by deleting the L-ldhA gene to shut off lactate production and by overexpressing the pyruvate carboxylase (PC)-encoding gene (pyc) (ΔldhA-pCRA717). Overexpression of the gene encoding another anaplerotic enzyme, PEPC had no dramatic effect on succinate production. The engineered strain produced 1,240 mM (146.3 g/L) succinate from glucose within 46 h, with a yield of 1.40 mol/mol glucose, when using 50 g of dry cells/L and 400 mM sodium bicarbonate [51] (Table 1). The strain did not produce any lactate. However, a large amount of acetate was still produced as a by-product with a yield of 0.29 mol/mol glucose. Acetate is produced mainly through three pathways catalyzed by phosphotransacetylase (pta) plus acetate kinase (ackA), pyruvate/quinone oxidoreductase (pqo), and acetyl coenzyme A (CoA):CoA transferase (cat) [85] (Fig. 2). Litsanov and colleagues further engineered C. glutamicum ATCC13032 to minimize acetate production by deleting these four genes. They also chromosomally overexpressed the mutant pyc gene encoding PC possessing a P458S amino acid exchange, that reportedly enhanced the reaction [22] (Fig. 2). To increase the reducing equivalents, the NAD+-coupled formate dehydrogenase-encoding gene (fdh) from Mycobacterium vaccae was also chromosomally overexpressed (Fig. 2). Last, the GAPDH-encoding gene was overexpressed by introducing plasmid pAN6-gap, with the aim of increasing glucose consumption, as was observed with alanine production [26]. The resultant strain, BOL-3/pAN6-gap, produced 1,134 mM (133.8 g/L) succinate within 53 h from glucose and formate, with a yield of 1.67 mol/mol glucose when using a culture of cells at an OD600 of 50 (approximately 20 g of dry cells/L) and 250 mM sodium bicarbonate [38] (Table 1). Unexpectedly, overexpression of the GAPDH-encoding gene did not increase glucose consumption but instead increased the succinate yield by 10 %, as was observed with d-lactate production (described above).
The central metabolic pathways of C. glutamicum including pathways for the assimilation of carbon sources (glucose, xylose, arabinose, cellobiose) used for the production of succinate under anaerobic conditions. Gray arrows represent pathways not present due to inactivation of genes encoding the enzymes marked with an X, or not activated under anaerobic conditions. Bold lines represent pathways that are useful for increasing succinate production by overexpressing the corresponding genes, or that can be exploited for utilizing new carbon sources by introducing assimilating genes
Because C. glutamicum is cultured at neutral pH (6.9–7.5) under oxygen deprived conditions, the dissociated form of succinic acid is the primary product. This indicates that succinate is exported through a protein exporter under these conditions. Two research groups independently indentified the succinate exporter in C. glutamicum. Using a comparative genomic analysis approach, Huhn et al. [19] identified the exporter, designated SucE. Later, Fukui et al. [11] identified the same exporter (designated SucE1) through comprehensive transcriptional analysis of C. glutamicum under microaerobic conditions. They also demonstrated that overexpression of the SucE1-encoding gene increases succinate production by 1.5-fold at an Eppendorf-tube scale under anaerobic conditions. Zhu et al. [92] investigated the effect of overexpressing the SucE1-encoding gene on succinate production at bioreactor scale and found that succinate productivity and the rate of glucose consumption increased by 19 and 8 %, respectively (Fig. 2). The parent strain they used was SA1 with deletion of ldhA, pta, ackA, pqo, and cat genes and with plasmid-based overexpression of the native pyc gene. They also overexpressed genes encoding isocitrate lyase (aceA), malate synthase (aceB), and citrate synthase (gltA) from a plasmid (strain SA4), with the aim of converting more carbon to succinate through the oxidative TCA cycle and glyoxylate pathway, as well as the reductive branch of the TCA cycle through the anaplerotic pathway (Fig. 2). Overexpression of aceA, aceB, and gltA resulted in a 12.8 ± 0.8 % carbon flux toward the glyoxylate pathway. Strain SA5, in which the succinate transporter-encoding gene (sucE) was overexpressed using a plasmid in SA4, produced 926 mM (109.4 g/L) succinate within 98 h, with a yield of 1.32 mol/mol glucose when using 27.5 g of dry cells/L and 230 mM sodium bicarbonate [92] (Table 1). This level of production was highly comparable to that obtained with metabolically engineered S. cerevisiae PMCFfg (which produced 13.0 g/L of succinate within 120 h, with a yield of 0.21 mol/mol glucose), even though the process was carried out at pH 3.8, which considerably reduces the cost of downstream processes [84] (Table 1).
The above-mentioned studies involved a two-stage bioprocess consisting of aerobic growth in complex or minimal medium followed by anaerobic succinate production in a sealed bottle or a fermenter after harvesting and resuspension of the cells in fresh minimal medium at high cell density. To overcome the practical disadvantages of such two-stage processes, a one-stage fermentation process in a single bioreactor was demonstrated by Wieschalka et al. This process includes three phases: (1) an aerobic growth phase on glucose and acetate; (2) a self-induced microaerobic phase at the end of the exponential growth phase with minimal aeration to assist in physiological adaptation from the aerobic to subsequent anaerobic conditions; and (3) an anaerobic production phase, achieved by supplying the fermenter with CO2 gas. They used the pyruvate-overproducing strain ELB-P, in which aceE (encoding the E1p subunit of the pyruvate dehydrogenase complex), ldhA, pqo, the last 249 bp of the C-terminal domain of ilvN (encoding the small subunit of acetohydroxyacid synthase), alaT (encoding the alanine aminotransferase), and avtA (encoding the valine:pyruvate aminotransferase) were deleted [78]. This growth-decoupled one-stage bioprocess produced 330 mM (39.0 g/L), succinate with a yield of 1.02 mol/mol glucose [79] (Table 1).
Aerobic succinate production from glucose
One-stage bioprocesses involving aerobic growth and anaerobic succinate production require delicate regulation of the bioreactor. Moreover, in anaerobic cultivation, by-products must be produced to generate reducing equivalents. To circumvent these problems, aerobic succinate production was considered. Because succinate must be produced through the oxidative TCA cycle under aerobic conditions, the key target of metabolic engineering for aerobic succinate production is deletion of the succinate dehydrogenase-encoding genes (sdhCAB) (Fig. 3). Litsanov and colleagues showed that although the growth rate and biomass formation of a ΔsdhCAB strain were 9 and 28 % lower than the wild type, the ΔsdhCAB strain accumulated 40 mM succinate after 22.5 h of aerobic cultivation [39]. Because the major by-product formed during cultivation of the ΔsdhCAB strain was acetate (125 mM after 22.5 h), the four genes responsible for acetate production (pqo, pta, ackA, cat; see above) were also deleted to minimize production of this by-product (Strain BL-1). Acetate accumulation decreased to 22 mM in BL-1, whereas succinate accumulation increased to 66 mM. Overexpression of the anaplerotic enzymes-encoding genes pyc P458S and ppc (BL-1/pAN6-pyc P458S ppc) to increase the availability of oxaloacetate as a substrate for citrate synthase resulted in production of 82 mM (9.7 g/L) succinate after 22.5 h aerobic cultivation, with a yield of 0.36 mol/mol glucose [39] (Fig. 3; Table 1). Interestingly, overexpression of the pyc P458S gene increased the succinate titer by only 3 %, whereas overexpression of the ppc gene increased the titer by 21 %. In contrast, during anaerobic cultivation, overexpression of the pyc gene resulted in a twofold increase in succinate production, whereas overexpression of the ppc gene had no significant effect [23]. Previous studies showed that PC-catalyzed pathway from pyruvate to oxaloacetate is responsible for 91 % of the oxaloacetate replenishment during aerobic growth of C. glutamicum on glucose [53], whereas the PEPC-catalyzed pathway from phosphoenolpyruvate to oxaloacetate is primarily responsible for producing succinate under oxygen deprived conditions [23]. These results demonstrate that the genes encoding anaplerotic enzymes that play a minor role in oxaloacetate replenishment under both conditions (PEPC under aerobic and PC under anaerobic) can increase the availability of oxaloacetate when overexpressed. Low theoretical yield is the primary disadvantage of aerobic succinate production, because a major proportion of the carbon source is converted into biomass and CO2. Therefore, Litsanov et al. tried to restrict growth by decreasing the level of the nitrogen source, which is essential for growth but not for succinate formation. As a result, the maximal biomass concentration under nitrogen-limiting conditions was 45 % lower than that under conditions of nitrogen excess, but succinate production was 43 % higher under nitrogen-limiting conditions than under conditions of nitrogen excess. As expected, the succinate yield under nitrogen-limiting conditions increased to 0.45 mol/mol glucose [39]. Although all known genes involved in acetate production were deleted, a relatively large amount of acetate was still produced (15 mM). Zhu and colleagues tried to solve this problem by introducing the gene encoding acetyl-CoA synthase (acsA) from Bacillus subtilis, which converts acetate to acetyl-CoA through acetyladenylate (Fig. 3). Plasmid-based expression of acsA in a strain in which the sdhCAB, ldhA, pqo, cat, and pta genes were deleted and in which the native pyc and ppc genes were overexpressed (strain ZX1) resulted in elimination of acetate production and an increase in the yield of succinate to 0.5 mol/mol glucose [91]. Overexpression of the gene encoding citrate synthase (gltA) as well (strain ZX1/pEacsAgltA) increased the succinate concentration by 23 % and increased the yield to 0.61 mol/mol glucose (Fig. 3). In contrast to anaerobic conditions, under aerobic conditions, the overexpression of succinate exporter-encoding gene had no effect on succinate productivity. In fed-batch cultivation, strain ZX1/pEacsAgltA produced approximately 238 mM (28.1 g/L) succinate within 67 h, with a yield of 0.63 mol/mol glucose [91] (Table 1).
The central metabolic pathways of C. glutamicum, including pathways for the assimilation of carbon sources (glucose, arabinose, glycerol) used for the production of succinate under aerobic conditions. Gray arrows represent pathways not present due to inactivation of genes encoding the enzymes marked with an X. Bold lines represent pathways that are useful for increasing succinate production by overexpressing the corresponding genes or that can be exploited for utilizing new carbon sources by introducing assimilating genes. ACS acetyl-CoA synthase, GlpD glycerol-3-phosphate dehydrogenase, GlpF glycerol facilitator, GlpK glycerol kinase
Production of lactate and succinate from alternative resources
So far, we have focused on studies examining the production of lactate and succinate primarily from purified glucose, as this is the simplest way to examine the effect of metabolic engineering. In terms of production costs, economically feasible microbial production of chemicals depends heavily on the use of low-cost carbon sources, especially when the products are commodity chemicals. Because wild-type C. glutamicum cannot directly utilize low-cost carbon sources such as lignocellulose, glycerol, and starch, considerable research has focused on metabolically engineering strains to expand the substrate spectrum. A particularly promising carbon source for microbial production of commodity chemicals is lignocellulosic biomass, which consists largely of cellulose, hemicellulose, and lignin, since it is the most abundant renewable organic resource on earth [3]. Cellulose, a polysaccharide consisting of a linear chain of glucose molecules, becomes a suitable carbon source following depolymerization by chemical or enzymatic processes. Hemicellulose, which consists primarily of glucose and the C5 sugars xylose and arabinose, can also be utilized as a carbon source after the pretreatment. While some microorganisms, such as E. coli, naturally utilize xylose and arabinose, C. glutamicum must be engineered to assimilate these C5 sugars.
The first C. glutamicum strain capable of utilizing xylose was developed by introducing the plasmid pCRA811, which carries the xylose isomerase (xylA) and xylulokinase (xylB) genes from E. coli [30] (Figs. 1, 2). The resultant strain, CRX2, grew on xylose as a sole carbon source. Interestingly, when organic acids were produced under oxygen deprivation, the l-lactate yield was higher when using glucose (1.36 mol/mol glucose) than when using xylose (1.06 mol/mol glucose), whereas the succinate yield was lower when using glucose (0.28 mol/mol glucose) than when using xylose (0.50 mol/mol glucose). When using a mixture of glucose and xylose at a mass ratio of 2:1, which is typical of lignocellulosic hydrolysates [3], xylose was consumed only after the glucose was depleted under aerobic growth conditions, showing apparent carbon catabolite repression [8]. Under oxygen deprivation, the carbon catabolite repression was partially deregulated, but the rate of xylose consumption doubled once the glucose was exhausted, showing that metabolism of xylose is still affected by the presence of glucose [30]. Carbon catabolite repression under anaerobic conditions was later fully deregulated when five copies of the xylA and xylB genes were integrated into the chromosome of C. glutamicum R (strain X5C1). Strain X5C1 consumed xylose at a constant rate, regardless of the presence or absence of glucose [57]. The strain also carried one copy each of the bglF V317A and bglA genes (encoding the β-glucoside-specific enzyme IIBCA component of the phosphotransferase system and phospho-β-glucosidase, respectively), which enabled C. glutamicum R to utilize cellobiose [35] (Figs. 1, 2). Strain X5C1 produced approximately 460 mM (41.4 g/L) l-lactate and 125 mM (14.8 g/L) succinate within 12 h from a mixture of 222 mM (40 g/L) glucose, 134 mM (20 g/L) xylose, and 29.3 mM (10 g/L) cellobiose [58] (Table 1). Another C5 sugar contained in hemicellulose, arabinose, could be utilized by C. glutamicum R upon introduction of plasmid pCRA820, which carried the ara operon, consisting of the araB, araA, and araD genes (encoding l-ribulokinase, l-arabinose isomerase, and l-ribulose-5-phosphate 4-epimerase, respectively) from E. coli (Figs. 1, 2, 3; strain CRA1) [31]. Around the same time, Kawaguchi and colleagues reported that of 10 C. glutamicum strains tested, only strain ATCC31831 could utilize arabinose as a sole carbon source [32]. The strain possessed arabinose-utilization genes (araB, araD, araA) analogous to those in E. coli, forming an operon with a different gene order than E. coli. The strain also possessed an arabinose transporter gene (araE) located upstream of the araBDA operon. Unexpectedly, chromosomal integration of araE increased the rate of both xylose and arabinose consumption by 2.7- and 2.8-fold, respectively, under oxygen deprivation [58] (Figs. 1, 2). Strain X5C1 containing an integrated araE gene (strain ACX-araE) completely consumed xylose (17.5 g/L), arabinose (7 g/L), and cellobiose (7 g/L) simultaneously with glucose (35 g/L) within 14 h under oxygen deprivation (the amount of organic acids produced is unknown) [58]. Aerobic succinate production from arabinose was recently demonstrated by Chen et al. [6], who introduced plasmid pXaraBAD (which carries the E. coli araBAD operon) into strain ZX1/pEacsAgltA (see above), and the resultant strain produced 74.4 mM (8.8 g/L) succinate within 84 h, with a yield of 0.58 mol/mol arabinose (Fig. 3; Table 1).
The production of l-lactate and succinate from C5 sugars in the studies described above involved the use of purified sugars, which is not industrially feasible. Lignocellulosic biomass requires dilute acid pretreatment to accelerate the accessibility of enzymes and subsequent hydrolysis of the polysaccharide [13]. During acid pretreatment, some amount of side products, such as furfural, 5-hydroxymethyfurfural, vanillic acid, and acetic and formic acids are released, which usually inhibits microbial growth and fermentation [2, 42]. Although C. glutamicum is highly tolerant to such fermentation inhibitors under anaerobic conditions when they are individually added to the medium [56, 71], the production of organic acids using lignocellulose hydrolysates under these conditions has not been demonstrated. Recently, Wang et al. [74] demonstrated succinate production from hydrolysates of corn cob, which is a significant lignocellulosic by-product of industrial corn production. The corn cob hydrolysates were found to contain toxic compounds, such as 5-hydroxymethyfurfural, vanillic acid, and p-hydroxybenzoic acid. Succinate production was indeed inhibited when using corn cob hydrolysates, but the inhibition could be reversed by addition of activated charcoal to absorb the toxins. Strain NC-2, in which the ldhA gene was deleted and the xylA and xylB genes from E. coli were introduced, produced 40.8 g/L of succinate within 48 h, with a yield of 0.69 g/g total sugar under anaerobic conditions in two-stage fermentation [74] (Table 1). Metabolically engineered E. coli BA305 produced 83 g/L of succinate within 36 h from sugarcane bagasse hydrolysates, with a yield of 0.87 g/g total sugar [37] (Table 1). Although C. glutamicum produced less succinate from hydrolysates than did E. coli, further metabolic engineering to accelerate succinate production (as described above) could make C. glutamicum a superior host strain for producing succinate from lignocellulose hydrolysates (Table 1).
Glycerol is another attractive carbon source for microbial production of fermentation products, because it is produced in abundance as a major by-product of biodiesel production [86]. Another advantage is that glycerol requires no complicated pretreatment before use as a raw material for fermentation. Although C. glutamicum cannot naturally utilize glycerol as a carbon source, introduction of genes of the E. coli glpFKD operon, which encode glycerol facilitator, glycerol kinase, and glycerol-3-phosphate dehydrogenase, respectively, enabled C. glutamicum to grow on glycerol as a sole carbon source [55] (Fig. 3). A plasmid carrying the glpFKD genes was introduced into strain BL-1 (see above) for aerobic succinate production (BL-1/pVWEx1-glpFKD). The strain produced 79 mM (9.3 g/L) succinate under aerobic conditions, with a yield of 0.21 mol/mol glycerol [40] (Table 1), a level of production comparable to that obtained when using glucose. Thus, glycerol must be considered a useful resource for succinate production.
Hydrolyzed starch is widely used as a carbon source for microbial production of fermentation products. Although C. glutamicum cannot naturally utilize starch, several strains have been developed that are capable of directly utilizing starch by secretion or cell-surface display of α-amylase encoded by the amyA gene from Streptococcus bovis 148 or Streptomyces griseus [61, 65, 66] (Figs. 1, 2, 3). Using AmyA-displaying C. glutamicum ATCC13032 (ATCC13032/pCC-pgsA-amyA), a mixture of 88.9 g/L of l-lactate and 10.4 g/L of succinate was directly produced from 128.5 g/L of soluble starch through consolidated bioprocessing after five cycles of anaerobic cultivation using 60 g of dry cells/L [70] (Table 1). Studies of succinate production from starch have advanced through the use of E. coli. Metabolically engineered E. coli NZN111 produced 127.1 g/L of succinate within 39 h from cassava starch by simultaneous saccharification and fermentation [5] (Table 1). Recently, microalgae have attracted attention as a host strain for producing bio-fuels, gas, and chemicals because these organisms grow and produce fermentation products using only CO2 [9]. Microalgae have also attracted attention as a potential feedstock for microbial production after acid hydrolysis or even without pretreatment because they contain large amounts of carbohydrates such as starch or glycogen [1, 44]. To test the utility of using microalgal biomass as a carbon source for C. glutamicum, Lee et al. [36] used Chlamydomonas reinhardtii as a feedstock for CO2-derived succinate production by metabolically engineered C. glutamicum after simply disrupting the cells by glass bead beating. Strain BL-1 (see above) harboring a plasmid carrying the amyA gene from S. bovis (BL-1/pSbAmyA) produced 0.5 g/L of succinate from microalgal starch without the need for complicated pretreatments, with a yield of 0.28 g/g starch under aerobic conditions (Table 1).
Conclusions and perspectives
Driven by metabolic engineering, C. glutamicum is now an attractive platform for producing lactate and succinate with industrial feasibility because of its high productivity in minimal medium, especially under anaerobic conditions. Although further strain optimization to improve the titer and yield of lactate and succinate is desirable, research is now moving to the next stage and focusing on areas, such as the optimization of bioreactor conditions, or the use of lignocellulosic hydrolysates rather than on the use of purified sugars.
An as yet unexploited target for constructing an optimized strain, especially for succinate production, is optimizing the redox state. A well-balanced redox state is critically important for the production of fermentation products under anaerobic conditions. Recently, it was reported that overexpression of the E. coli transhydrogenase-encoding gene pntAB in C. glutamicum ATCC13032 (the aim of which was to increase the amount of NADH available to accelerate succinate production) increased the NADH/NAD+ ratio and decreased the NADPH/NADP+ ratio during prolonged aerobic growth. Although the growth rate was unchanged, the succinate productivity increased 2.2-fold in the transition phase, which was 16–24 h after growth was initiated [83]. Optimizing the redox state through simple genetic engineering could further improve succinate production under anaerobic conditions. Another expectation for improving process productivity derives from recently revealed research on the global regulators governing the expression of genes involved in sugar uptake, glycolysis, the pentose phosphate pathway, and the TCA cycle [64, 68, 76]. On l-isoleucine production, overexpression of the gene encoding the global regulator Lrp (which regulates the expression of several operons involved in the metabolism of branched-chain amino acid) was shown to increase the productivity [87].
In addition to metabolic engineering to increase process productivity, physiological alteration of C. glutamicum, such as render the organism highly tolerant to low pH, high temperatures, and fermentation inhibitors, is an ideal strategy for industrial production. Since C. glutamicum is cultivated at neutral pH under both aerobic and anaerobic conditions, altering the organism so that it would exhibit a high tolerance to low pH would be beneficial. Purifying organic acids from low-pH fermentation broth is usually significantly more cost efficient than purifying these compounds from neutral-pH broth [7]. This makes yeast an attractive platform for the production of organic acids due to its high acid tolerance. In addition, cultivation at low pH reduces the risk of contamination. Tolerance to high temperatures is also beneficial for reducing the risk of contamination. Moreover, engineering strains to be high-temperature tolerant could reduce the costs associated with cooling during fermentation. Lastly, even though C. glutamicum has high tolerance to fermentation inhibitors under anaerobic conditions, it is expected that the organism would acquire complete tolerance to fermentation inhibitors contained in lignocellulosic hydrolysates through genetic engineering of, for example, sigma factors which regulate the expression of genes involved in tolerance to various stresses [52]. Such acquired phenotypes can be advantageous for producing other fermentation products as well as the organic acids.
Overall, C. glutamicum is now a major candidate microorganism for use as a platform for industrial production of lactate and succinate, and the potential exists for the use of C. glutamicum as a cell factory for producing fine, value-added chemicals as well as bulk chemicals.
References
Aikawa S, Joseph A, Yamada R, Izumi Y, Yamagishi T, Matsuda F, Kawai H, Chang JS, Hasunuma T, Kondo A (2013) Direct conversion of Spirulina to ethanol without pretreatment or enzymatic hydrolysis processes. Energy Environ Sci 6:1844–1849
Almeida JR, Bertilsson M, Gorwa-Grauslund MF, Gorsich S, Lidén G (2009) Metabolic effects of furaldehydes and impacts on biotechnological processes. Appl Microbiol Biotechnol 82(4):625–638
Aristidou A, Penttilä M (2000) Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol 11(2):187–198
Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77(10):3300–3310
Chen C, Ding S, Wang D, Li Z, Ye Q (2014) Simultaneous saccharification and fermentation of cassava to succinic acid by Escherichia coli NZN111. Bioresour Technol 163:100–105
Chen T, Zhu N, Xia H (2014) Aerobic production of succinate from arabinose by metabolically engineered Corynebacterium glutamicum. Bioresour Technol 151:411–414
Cok B, Tsiropoulos I, Roes AL, Patel MK (2014) Succinic acid production derived from carbohydrates: an energy and greenhouse gas assessment of a platform chemical toward a bio-based economy. Biofuel Bioprod Bioref 18(1):16–29
Deutscher J (2008) The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol 11(2):87–93
Desai SH, Atsumi S (2013) Photosynthetic approaches to chemical biotechnology. Curr Opin Biotechnol 24(6):1031–1036
Dominguez H, Nezondet C, Lindley ND, Cocaign M (1993) Modified carbon flux during oxygen limited growth of Corynebacterium glutamicum and the consequences for amino acid overproduction. Biotechnol Lett 15(5):449–454
Fukui K, Koseki C, Yamamoto Y, Nakamura J, Sasahara A, Yuji R, Hashiguchi K, Usuda Y, Matsui K, Kojima H, Abe K (2011) Identification of succinate exporter in Corynebacterium glutamicum and its physiological roles under anaerobic conditions. J Biotechnol 154(1):25–34
Fukushima K, Chang YH, Kimura Y (2007) Enhanced stereocomplex formation of poly(l-lactic acid) and poly (d-lactic acid) in the presence of stereoblock poly (lactic acid). Macromol Biosci 7(6):829–835
Geddes CC, Peterson JJ, Mullinnix MT, Svoronos SA, Shanmugam KT, Ingram LO (2010) Optimizing cellulase usage for improved mixing and rheological properties of acid-pretreated sugarcane bagasse. Bioresour Technol 101(23):9128–9136
Guettler MV, Rumler D, Jain MK (1999) Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int J Syst Bacteriol 49(1):207–216
Goffin P, Deghorain M, Mainardi JL, Tytgat I, Champomier-Vergès MC, Kleerebezem M, Hols P (2005) Lactate racemization as a rescue pathway for supplying d-lactate to the cell wall biosynthesis machinery in Lactobacillus plantarum. J Bacteriol 187(19):6750–6761
Hasegawa S, Uematsu K, Natsuma Y, Suda M, Hiraga K, Jojima T, Inui M, Yukawa H (2012) Improvement of the redox balance increases l-valine production by Corynebacterium glutamicum under oxygen deprivation conditions. Appl Environ Microbiol 78(3):865–875
Hasegawa S, Suda M, Uematsu K, Natsuma Y, Hiraga K, Jojima T, Inui M, Yukawa H (2013) Engineering of Corynebacterium glutamicum for high-yield l-valine production under oxygen deprivation conditions. Appl Environ Microbiol 79(4):1250–1257
Huang GL, Anderson TD, Clubb RT (2014) Engineering microbial surfaces to degrade lignocellulosic biomass. Bioengineered 5(2):96–106
Huhn S, Jolkver E, Krämer R, Marin K (2011) Identification of the membrane protein SucE and its role in succinate transport in Corynebacterium glutamicum. Appl Microbiol Biotechnol 89(2):327–335
Ikada Y, Jamshidi K, Tsuji H, Hyon S (1987) Stereocomplex formation between enantiomeric poly(lactides). Macromolecules 20(4):904–906
Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62(2–3):99–109
Ikeda M, Ohnishi J, Hayashi M, Mitsuhashi S (2006) A genome-based approach to create a minimally mutated Corynebacterium glutamicum strain for efficient l-lysine production. J Ind Microbiol Biotechnol 33(7):610–615
Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H (2004) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7(4):182–196
Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H (2004) Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol 8(4):243–254
Jansen ML, van Gulik WM (2014) Towards large scale fermentative production of succinic acid. Curr Opin Biotechnol 30:190–197
Jojima T, Fujii M, Mori E, Inui M, Yukawa H (2010) Engineering of sugar metabolism of Corynebacterium glutamicum for production of amino acid l-alanine under oxygen deprivation. Appl Microbiol Biotechnol 87(1):159–165
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol 104(1–3):5–25
Karjomaa S, Suortti T, Lempiainen R, Selin J, Itavaara M (1998) Microbial degradation of poly-(l-lactic acid) oligomers. Polym Degrad Stab 59(1–3):333–336
Kato O, Youn JW, Stansen KC, Matsui D, Oikawa T, Wendisch VF (2010) Quinone-dependent d-lactate dehydrogenase Dld (Cg1027) is essential for growth of Corynebacterium glutamicum on d-lactate. BMC Microbiol 10:321
Kawaguchi H, Vertès AA, Okino S, Inui M, Yukawa H (2006) Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol 72(5):3418–3428
Kawaguchi H, Sasaki M, Vertès AA, Inui M, Yukawa H (2008) Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Appl Microbiol Biotechnol 77(5):1053–1062
Kawaguchi H, Sasaki M, Vertès AA, Inui M, Yukawa H (2009) Identification and functional analysis of the gene cluster for l-arabinose utilization in Corynebacterium glutamicum. Appl Environ Microbiol 75(11):3419–3429
Kinoshita S, Udaka S, Shimono M (1957) Studies on the amino acid fermentation part 1. Production of l-glutamic acid by various microorganisms. J Gen Appl Microbiol 3(3):193–205
Kinoshita S (1985) Glutamic acid bacteria. In: Demain AL, Solomon NA (eds) Biology of industrial microorganisms. Benjamin Cummings, London, pp 115–146
Kotrba P, Inui M, Yukawa H (2003) A single V317A or V317M substitution in enzyme II of a newly identified beta-glucoside phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiology 149(6):1569–1580
Lee J, Sim SJ, Bott M, Um Y, Oh MK, Woo HM (2014) Succinate production from CO2-grown microalgal biomass as carbon source using engineered Corynebacterium glutamicum through consolidated bioprocessing. Sci Rep 4:5819
Liang L, Liu R, Li F, Wu M, Chen K, Ma J, Jiang M, Wei P, Ouyang P (2013) Repetitive succinic acid production from lignocellulose hydrolysates by enhancement of ATP supply in metabolically engineered Escherichia coli. Bioresour Technol 143:405–412
Litsanov B, Brocker M, Bott M (2012) Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78(9):3325–3337
Litsanov B, Kabus A, Brocker M, Bott M (2012) Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb Biotechnol 5(1):116–128
Litsanov B, Brocker M, Bott M (2013) Glycerol as a substrate for aerobic succinate production in minimal medium with Corynebacterium glutamicum. Microb Biotechnol 6(2):189–195
McKinlay JB, Vieille C, Zeikus JG (2007) Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol 76(4):727–740
Mills TY, Sandoval NR, Gill RT (2009) Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol Biofuels 2:26
Nghiem NP, Davison BH, Suttle BE, Richardson GR (1997) Production of succinic acid by Anaerobiospirillum succiniciproducens. Appl Biochem Biotechnol 63–65:565–576
Nguyen MT, Choi SP, Lee J, Lee JH, Sim SJ (2009) Hydrothermal acid pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. J Microbiol Biotechnol 19(2):161–166
Niimi S, Suzuki N, Inui M, Yukawa H (2011) Metabolic engineering of 1,2-propanediol pathways in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90(5):1721–1729
Nishimura T, Vertès AA, Shinoda Y, Inui M, Yukawa H (2007) Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl Microbiol Biotechnol 75(4):889–897
Niu D, Tian K, Prior BA, Wang M, Wang Z, Lu F, Singh S (2014) Highly efficient l-lactate production using engineered Escherichia coli with dissimilar temperature optima for l-lactate formation and cell growth. Microb Cell Fact 13:78
Okano K, Tanaka T, Ogino C, Fukuda H, Kondo A (2010) Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl Microbiol Biotechnol 85(3):413–423
Okino S, Inui M, Yukawa H (2005) Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 68(4):475–480
Okino S, Suda M, Fujikura K, Inui M, Yukawa H (2008) Production of d-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 78(3):449–454
Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H (2008) An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol 81(3):459–464
Pátek M, Nešvera J (2011) Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol 154(2–3):101–113
Petersen S, de Graaf AA, Eggeling L, Möllney M, Wiechert W, Sahm H (2000) In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J Biol Chem 275(46):35932–35941
Raab AM, Gebhardt G, Bolotina N, Weuster-Botz D, Lang C (2010) Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab Eng 12(6):518–525
Rittmann D, Lindner SN, Wendisch VF (2008) Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl Environ Microbiol 74(20):6216–6222
Sakai S, Tsuchida Y, Nakamoto H, Okino S, Ichihashi O, Kawaguchi H, Watanabe T, Inui M, Yukawa H (2007) Effect of lignocellulose-derived inhibitors on growth of and ethanol production by growth-arrested Corynebacterium glutamicum R. Appl Environ Microbiol 73(7):2349–2353
Sasaki M, Jojima T, Inui M, Yukawa H (2008) Simultaneous utilization of d-cellobiose, d-glucose, and d-xylose by recombinant Corynebacterium glutamicum under oxygen-deprived conditions. Appl Microbiol Biotechnol 81(4):691–699
Sasaki M, Jojima T, Kawaguchi H, Inui M, Yukawa H (2009) Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl Microbiol Biotechnol 85(1):105–115
Sasaki M, Jojima T, Inui M, Yukawa H (2010) Xylitol production by recombinant Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 86(4):1057–1066
Sauer M, Porro D, Mattanovich D, Branduardi P (2008) Microbial production of organic acids: expanding the markets. Trends Biotechnol 26(2):100–108
Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ (2006) Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J Biotechnol 124(2):381–391
Smith KM, Cho KM, Liao JC (2010) Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol 87(3):1045–1055
Takeno S, Ohnishi J, Komatsu T, Masaki T, Sen K, Ikeda M (2007) Anaerobic growth and potential for amino acid production by nitrate respiration in Corynebacterium glutamicum. Appl Microbiol Biotechnol 75(5):1173–1182
Tanaka Y, Takemoto N, Ito T, Teramoto H, Yukawa H, Inui M (2014) Genome-wide analysis of the role of global transcriptional regulator GntR1 in Corynebacterium glutamicum. J Bacteriol 196(18):3249–3258
Tateno T, Fukuda H, Kondo A (2007) Production of l-Lysine from starch by Corynebacterium glutamicum displaying alpha-amylase on its cell surface. Appl Microbiol Biotechnol 74(6):1213–1220
Tateno T, Fukuda H, Kondo A (2007) Direct production of l-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl Microbiol Biotechnol 77(3):533–541
Tejayadi S, Cheryan M (1995) Lactic acid from cheese whey permeate. Productivity and economics of a continuous membrane bioreactor. Appl Microbiol Biotechnol 43(2):242–248
Teramoto H, Inui M, Yukawa H (2011) Transcriptional regulators of multiple genes involved in carbon metabolism in Corynebacterium glutamicum. J Biotechnol 154(2–3):114–125
Tsuge Y, Yamamoto S, Suda M, Inui M, Yukawa H (2013) Reactions upstream of glycerate-1,3-bisphosphate drive Corynebacterium glutamicum (d)-lactate productivity under oxygen deprivation. Appl Microbiol Biotechnol 97(15):6693–6703
Tsuge Y, Tateno T, Sasaki K, Hasunuma T, Tanaka T, Kondo A (2013) Direct production of organic acids from starch by cell surface-engineered Corynebacterium glutamicum in anaerobic conditions. AMB Express 3(1):72
Tsuge Y, Hori Y, Kudou M, Ishii J, Hasunuma T, Kondo A (2014) Detoxification of furfural in Corynebacterium glutamicum under aerobic and anaerobic conditions. Appl Microbiol Biotechnol 98(20):8675–8683
Udaka S (1960) Screening method for microorganisms accumulating metabolites and its use in the isolation of Hicrococcus alatamicus. J Bactariol 79(5):754–755
Van Vleet JH, Jeffries TW (2009) Yeast metabolic engineering for hemicellulosic ethanol production. Curr Opin Biotechnol 20(3):300–306
Wang C, Zhang H, Cai H, Zhou Z, Chen Y, Chen Y, Ouyang P (2014) Succinic acid production from corn cob hydrolysates by genetically engineered Corynebacterium glutamicum. Appl Biochem Biotechnol 172(1):340–350
Wendisch VF, Bott M, Eikmanns BJ (2006) Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol 9(3):268–274
Wennerhold J, Krug A, Bott M (2005) The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J Biol Chem 280(49):40500–40508
Werpy T, Petersen G (2004) Top value added chemicals from biomass. Volume 1: results of screening for potential candidates from sugars and synthesis gas. U.S. Department of Energy, Oak Ridge TN, USA, p 76
Wieschalka S, Blombach B, Eikmanns BJ (2012) Engineering Corynebacterium glutamicum for the production of pyruvate. Appl Microbiol Biotechnol 94(2):449–459
Wieschalka S, Blombach B, Bott M, Eikmanns BJ (2013) Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol 6(2):87–102
Yamamoto S, Sakai M, Inui M, Yukawa H (2011) Diversity of metabolic shift in response to oxygen deprivation in Corynebacterium glutamicum and its close relatives. Appl Microbiol Biotechnol 90(3):1051–1061
Yamamoto S, Gunji W, Suzuki H, Toda H, Suda M, Jojima T, Inui M, Yukawa H (2012) Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl Environ Microbiol 78(12):4447–4457
Yamamoto S, Suda M, Niimi S, Inui M, Yukawa H (2013) Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol Bioeng 110(11):2938–2948
Yamauchi Y, Hirasawa T, Nishii M, Furusawa C, Shimizu H (2014) Enhanced acetic acid and succinic acid production under microaerobic conditions by Corynebacterium glutamicum harboring Escherichia coli transhydrogenase gene pntAB. J Gen Appl Microbiol 60(3):112–118
Yan D, Wang C, Zhou J, Liu Y, Yang M, Xing J (2014) Construction of reductive pathway in Saccharomyces cerevisiae for effective succinic acid fermentation at low pH value. Bioresour Technol 156:232–239
Yasuda K, Jojima T, Suda M, Okino S, Inui M, Yukawa H (2007) Analyses of the acetate-producing pathways in Corynebacterium glutamicum under oxygen-deprived conditions. Appl Microbiol Biotechnol 77(4):853–860
Yazdani SS, Gonzalez R (2007) Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr Opin Biotechnol 18(3):213–219
Yin L, Shi F, Hu X, Chen C, Wang X (2013) Increasing l-isoleucine production in Corynebacterium glutamicum by overexpressing global regulator Lrp and two-component export system BrnFE. J Appl Microbiol 114(5):1369–1377
Yukawa H, Omumasaba CA, Nonaka H, Kos P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153(4):1042–1058
Zeikus JG, Jain MK, Elankovan P (1999) Biotechnology of succinic acid production and markets for derived industrial products. Appl Microbiol Biotechnol 51(5):545–552
Zhou L, Niu DD, Tian KM, Chen XZ, Prior BA, Shen W, Shi GY, Singh S, Wang ZX (2012) Genetically switched d-lactate production in Escherichia coli. Metab Eng 14(5):560–568
Zhu N, Xia H, Wang Z, Zhao X, Chen T (2013) Engineering of acetate recycling and citrate synthase to improve aerobic succinate production in Corynebacterium glutamicum. PLoS ONE 8(4):e60659
Zhu N, Xia H, Yang J, Zhao X, Chen T (2014) Improved succinate production in Corynebacterium glutamicum by engineering glyoxylate pathway and succinate export system. Biotechnol Lett 36(3):553–560
Acknowledgments
We are grateful to Dr. Masayuki Inui for permission to present unpublished results. This work was supported by the Commission for Development of Artificial Gene Synthesis Technology for Creating Innovative Biomaterial from the Ministry of Economy, Trade and Industry (METI), Japan, and Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction, Kobe). This work was also supported by a Grant-in-Aid for Young Scientists (B) to YT from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special Issue: Metabolic Engineering.
Rights and permissions
About this article
Cite this article
Tsuge, Y., Hasunuma, T. & Kondo, A. Recent advances in the metabolic engineering of Corynebacterium glutamicum for the production of lactate and succinate from renewable resources. J Ind Microbiol Biotechnol 42, 375–389 (2015). https://doi.org/10.1007/s10295-014-1538-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-014-1538-9