Abstract
Mining activities generate large quantities of wastes that significantly alter the biogeochemistry and ecological structure of entire river basins. Microbial communities that develop in these areas present a variety of survival and adaptation mechanisms. Knowing this diversity at the molecular level is strategic both for understanding adaptive processes and for identifying genomes with potential use in bioremediation and bioprospecting. In this work, prokaryotic and eukaryotic communities were evaluated by meta-taxonomics (16S and 18S amplicons) in sediments and water bodies impacted by acid mine drainage in an important coal mining area in southern Brazil. Five sampling stations were defined on a gradient of impacts (pH 2.7–4.25). Taxon diversity was directly proportional to pH, being greater in sediments than in water. The dominant prokaryotic phyla in the samples were Proteobacteria, Actinobacteria, Acidobacteria, OD1, Nitrospirae, and Euryarchaeota, and among the eukaryotes, algae (Ochrophyta, Chlorophyta, Cryptophyceae), fungi (Basidiomycota, Ascomycota, and Cryptomycota), and protists (Ciliophora, Heterolobosea, Cercozoa). The prokaryotic genera Leptospirillum, Acidithiobacillus, Acidiphilium, Thiomonas, Thermogymnomonas, and Acidobacterium, and the eukaryotic genera Pterocystis and Poteriospumella were associated with more acidic conditions and higher metal concentrations, while the prokaryotic genera Sediminibacterium, Gallionella Geothrix, and Geobacter were more abundant in transitional environments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mining activities have impacts on large areas, altering the biogeochemistry and ecology of terrestrial and aquatic environments. One of the main recognized impacts is acid mine drainage (AMD), a strongly acidic effluent with high concentrations of metals and sulfates, capable of impacting entire river basins and even coastal areas (Simate and Ndlovu 2014; Rambabu et al. 2020).
AMD is generated by the oxidation of sulfide minerals, the most common being pyrite (FeS2). Oxidation can occur naturally from the exposure of minerals to oxygen, water, and microorganisms, but it is accelerated by mining activities that increase the exposure of these minerals (Carlier et al. 2020; Wu et al. 2021). The rate of chemical oxidation of ferrous iron is negligible below pH 4; therefore, the activity of iron-oxidizing acidophilic bacteria plays a crucial role in the generation of AMD (Hallberg 2010). AMD composition can differ drastically from one region to another due to local geology, microclimate, group of microorganisms, and water source (Simate and Ndlovu 2014; Rambabu et al. 2020). Its generation can occur in active or abandoned mining sites, especially those that did not have deactivation planning (Neff et al. 2021).
A survey carried out in 2018 (Maus et al. 2020), places Brazil in 14th position in the world in active mining area. The southern state of Santa Catarina is one of the largest coal mining centers in the country, with more than 140 years of activities (Silva et al. 2011; Rangel et al. 2016). Intensive mining activity has produced considerable amounts of waste and, consequently, a serious environmental deterioration. The hydrographic basins of the Tubarão, Urussanga and Araranguá rivers are part of the Santa Catarina Carboniferous Basin (SCCB). This area receives AMD from 134 open-pit mine sites covering a total area of 2964 ha, with 115 waste deposit areas (2734 ha), 77 acid lakes (58 ha), and hundreds of underground mines (Silva et al. 2013). It is estimated that there are more than 1000 abandoned mines in the SCCB (Lattuada et al. 2009). In 1980, this region was designated as a “National Critical Area for Pollution Control and Environmental Conservation”. In subsequent years, actions were taken by the General Attorney of the Republic of Brazil demanding that mining companies recover impacted areas (Silva et al. 2013).
Although SCCB has a high-alert environmental liability, most of the existing studies at the site are of physical–chemical characterization. The highest concentrations of metals, especially Fe, Al, Cu, Zn, Pb, and Mn, together with the lowest pH values (pH = 2.4–3.8) were documented in samples from water bodies close to mines (Silva et al. 2011, 2013). A few studies address biology, related to the resistance of microorganisms to heavy metals (Castro-Silva et al. 2003), the use of fish as bioindicators (Osório et al. 2014), and the evaluation of microbiology used to assist the plant growth in the phytoremediation process (Pille da Silva et al. 2019). Studies describing the behavior of microbial communities in AMD areas in SCCB have not been carried out to date. To access these microbial communities, it is important to apply meta-taxonomic methods based on total DNA extraction from environmental samples (Brumfield et al. 2020).
The use of sequencing meta-taxonomy methods allows the identification and comparison of entire microbial communities, including non-cultivable organisms. This technique has great relevance in the evaluation of AMD environments, which generally have characteristics so extreme that they make it impossible to cultivate most of the organisms that live there. With these methods, it is possible to carry out microbial ecology studies and discover how the dynamics of these communities respond to AMD. Several studies have demonstrated that geochemical gradients substantially alter the microbial community (Xu et al. 2020; She et al. 2021). This makes it possible to check species that may be indicators of AMD conditions.
The main objective of this work was to characterize by meta-taxonomy the prokaryotic and eukaryotic (16S and 18S amplicons) communities in the sediments and water bodies of different areas impacted by AMD in the SCCB, correlating their composition with physicochemical variables, in an attempt to describe microbiota response patterns to different degrees of environmental change.
Materials and methods
Study area and sampling
The study was carried out in five different locations at SCCB, located in the State of Santa Catrina, Southern Brazil (Fig. 1). The sampling points were distributed across three different river basins, namely: Araranguá, Urussanga, and Tubarão. At the intersection of these three river basins, there is an area of approximately 1,625 km2 impacted by mining activity.
The identification of samples and a brief descriptions of them are summarized in Table 1. Three of them (#1, #2 and #3) were in mine mouths with intense AMD generation (pH 2.7–3.6), one located at the mouth of a mine (#4) with less acidic characteristics (pH above 4.25), and the last one located at the junction of two rivers (#5) that receive AMD and domestic sewage from different points in the river basin.
Sediment samples from the surface layer (~ 5 cm deep) were collected at five points for each collection station, in a water depth of about 0.3 m, using a sterilized PVC (Polyvinyl chloride) tube with a diameter of 40 mm. The five fractions were mixed in a sterile bag and a compound fraction was subsequently transferred to a sterile falcon tube (50 mL volume). The entire process was carried out using sterile vinyl gloves. The samples were refrigerated to the laboratory in a thermal box. In the laboratory, a fraction of the sediment was placed in 5 mL tubes and submerged with RNA Later until DNA extraction, which occurred 5 days after collection. The remainder of the sediment present in the falcon tube was sent for analysis of metals and nutrients.
Running water samples were collected until reaching a volume of 5 L (5 L sterile gallons) for each station. The gallons were stored in a thermal box for subsequent chemical and genetic meta-taxonomy analyses.
Sterilization of materials was carried out in an autoclave for heat-resistant materials and by bathing in sodium hypochlorite, followed by sterilized distilled water for heat-sensitive materials. All etherized materials were packaged airtight until use.
Geochemical analyses
Water and sediment samples from the five stations were collected in 50 mL polyethylene tubes and stored in thermal boxes in the dark until arrival at the laboratory. Samples for determining metals in water were acidified to pH 2.0 with concentrated sulfuric acid. The concentrations of the main metal components of the regional geochemical matrix—Fe, Mn, Zn, Cu, Ni—(Freitas et al. 2011) were determined in water and sediment samples. The concentration of total organic carbon (TOC), total nitrogen (TN), and total phosphorus (TP) was also determined in the sediment samples. The analyses were carried out in the LAQUA analytical laboratory (Chemical and Environmental Analysis Laboratory) belonging to the Santa Catarina Carbon Industry Charitable Association (SATC).
Total concentrations of Cu, Fe, Zn, Mn, and Ni in water and sediment were determined by inductively coupled plasma mass spectrometry (ICP-MS), following standard methodologies for sediments (U.S. EPA 1996) and for water (APHA 2018a).
TOC was determined on a total organic carbon analyzer following the SMWW 5310 D standard methodology (APHA 2018b). This method provides characteristics of the organic content of the sample, independent of the oxidation state of the organic matter and does not measure other bound elements. For this analysis, the sample is acidified to remove inorganic carbon and oxidized with persulfate at temperatures of 116 to 130 °C. The resulting carbon dioxide is measured by non-dispersive infrared spectrometry.
PT determination for sediment samples was carried out by underwent microwave-assisted acid digestion and subsequent analysis by ICP-MS, in accordance with the EPA 3051A (U.S. EPA 2007) and SMWW 3120 B (APHA 2018a) methodologies. NT determination in sediments was carried out by alkaline oxidation at 100 to 110 °C in digestion block, with the digestate subsequently analyzed spectrophotometrically as nitrate according to the SMWW 4500-N method (APHA 2018c).
DNA extraction
A composite sediment sample from each sampling station was homogenized and subjected to DNA extraction using the DNeasy PowerSoil kit (Qiagen®) following the manufacturer’s instructions. Water samples were filtered through a 0.22 µm cellulose membrane (~ 3 Liter) and then DNA was extracted with the DNeasy PowerWater extraction kit (Qiagen®), following the manufacturer’s instructions. The concentration and quality of eluted DNA were determined using a spectrophotometer (NanoDrop3300, Thermo Fisher Scientific, USA).
Amplicon library and sequencing
The extracted purified DNA was used for single-end library construction based on an Illumina protocol. The V3-V4 regions of 16S rRNA genes were amplified by polymerase chain reaction (PCR) using primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R 5′-GGACTACNNGGGTATCTAAT-3′ (Takahashi et al. 2014). The V9 regions of the 18S rRNA genes were amplified by PCR using primers Euk1391F (5′-GTACACACCCGCCCGTC) and EukBR e (5′-TGATCCTTCTGCAGGTTCACCTAC) (Amaral-Zettler et al. 2009). All PCR reactions were performed with Phusion® High-Fidelity PCR Master Mix (New England Biolabs). The amplicon libraries were purified using the standard protocol for Agencourt Ampure XP Bead (Beckman Coulter, USA) and their DNA concentration measured using the Qubit™ HS DNA Assay kit (Thermo Fisher Scientific, USA). Sequencing libraries were prepared from the purified amplicon libraries using a second PCR. Each PCR reaction (25 μL) contained PCRBIO HiFi buffer (PCRBiosystems, UK), PCRBIO HiFi Polymerase (1U) (PCRBiosystems, UK), adaptation mix (400 nM each forward and reverse), and up to 10 ng of amplicon library model. Library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and the Agilent Bioanalyzer 2100 system. Finally, purified sequencing libraries were pooled at equimolar concentrations and diluted to 2 nM, and samples were sequenced (301 bp) at a MiSeq (Illumina) using a MiSeq Reagent v3 kit (Illumina, USA) following standard guidelines for preparation and loading samples into the MiSeq. The Phix control library was used as a run-time control (20% spikedin) for monitoring run quality to overcome the low complexity issue that is often observed with amplicon samples.
DNA data processing
The QIIME2 package (Bolyen et al. 2019) was used to perform quality control, amplicon sequence variant (ASV) calling, and taxonomic classification of the sequencing reads. The primer sequences were removed prior to calling ASVs (17 to 20 base pairs removed at the 5' end of the reads). The quality filtering and ASV calling steps were performed using the DADA2 denoise-paired pipeline (Callahan et al. 2016), with the parameters maxN = 0; truncQ = 2; minOverlap = 10. Representative sequences resulting from ASVs calling were classified with Naïve Bayes classifiers implemented in the q2-feature-classifier plugin, trained using sequences extracted from the SILVA 138 SSU database (Quast et al. 2013) according to the primers 16S and 18S used in the sequencing process, through the fit-classifier-naive-bayes command. The result was a set of taxonomic tables with relative abundances for each sample. The raw data is available in the NCBI Sequence Read Archive (SRA) under accession number PRJNA1015584.
Downstream analyses
Alpha-diversity indices were estimated for all samples using the Python package scikit-bio 0.5.5 (http://scikit-bio.org/). Subsequent downstream analyses were done using the Microeco R package (Liu et al. 2021), where relative abundances at the genus and phylum levels were calculated using the trans_abund class method. To highlight probable relationships between abiotic data and the composition of organisms, multivariate canonical correspondence analyses were carried out using the Past 3.10 software (v. nov/2015), where environmental (except pH) and ASV abundance data were normalized by logarithmic transformation. For these analyses, the 20 most abundant taxonomic groups classified at the genus level were considered.
Results
Physical–chemical and chemical variables
The samples showed considerable differences in pH values, concentration of metals, phosphorus, and nitrogen (Table 2). Stations #1, #2, #3 showed AMD typical of a pyrite-rich waste leachate. Water pH between 2.70 and 3.20 and iron concentrations are indicative of intense pyrite weathering in these stations. Most of the water parameters at station #4 are also typical of AMD, although they were less acidic and contained lower concentrations of some metallic ions, which suggests the effect of contact with alkaline rocks. The waters that arrive at station #5 are the result of the junction of the Mãe Luzia and Sangão rivers, and also have a low pH (3.8); however, the concentration of metals is much lower when compared to the other collected stations, demonstrating processes of dilution or even physical–chemical and biological removal.
Station #1 presented the highest concentrations of Fe in water and sediment, although station #3 presented higher concentrations of other metals (Mn, Zn, Cu, Ni) in water. In the sediments, the highest concentrations of Mn, Zn, and Cu occurred at station #2 and the highest concentration of Ni at station #5. The amount of iron in the sediment from station #1 represents a fraction of 26.8% of the sample weight, which represents an extremely high value for a sediment.
The TOC in the sediments showed the highest content at station #3, followed by stations #1 and #2. In relation to the TN, the differences between stations were low, with a higher concentration in station #2, followed by station #1. The highest concentration of TP in the sediment was found at station #5, which receives the load of other effluents along the Mãe Luzia and Sangão rivers. Stations #1 and #4 showed concentrations below the analytical detection limit.
Microbiome biodiversity
A total of 525,136 and 687,620 qualified sequences were identified for sequencing the 16S and 18S rRNA genes, respectively. To make samples comparable and avoid bias due to differences in sequencing depth, sequences were leveled based on the sample with the lowest number of sequences (29,149 for 16S and 38,404 for 18S). The number of ASVs obtained for each sample ranged from 16 to 51 for 16S and from 18 to 100 for 18S, in 10 samples for each marker gene (Supplementary Material, Table S1 and S2).
The alpha-diversity assessment using the Shannon index showed a greater biodiversity of prokaryotes in the samples collected in the sediment (Fig. 2), with higher values for stations #4 and #5. In relation to water, the highest alpha-diversity of prokaryotes was found at station #5 (junction of rivers), while station #2 presented the lowest value. Regarding the alpha-diversity of eukaryotic organisms, the greatest diversity occurred in sample #5 for water and in sample #4 for sediment, and the lowest values were found in the mine mouth waters of stations #1, #2, and #3. Considering the biodiversity of both prokaryotic and eukaryotic targets in water and sediment, the highest values were verified for stations #4 and #5, which consist of a mine with a lower degree of contamination and river water, respectively.
Community composition
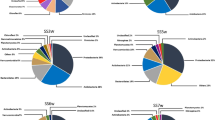
The composition of the microbial community was evaluated at the level of phyla and genera and the results are presented in Fig. 3. The complete list of ASVs is available in the supplementary material (Tables S1 and S2).
The dominant prokaryotic phyla in all samples were Pseudomonadota, Actinomycetota, Acidobacteriota, OD1, Nitrospirota, Euryarchaeota, Chlorobiota, Bacteroidota, and Cyanobacteriota. The phylum Proteobacteria was the most dominant in the water samples, with an abundance greater than 50% in all samples, reaching 90% in the sample from station #2. For sediment samples, the Pseudomonadota phylum was also the most abundant, but with a less expressive proportion (15 to 40%), when compared to water samples. Still for sediment samples, the phyla Acidobacteriota and Actinomycetota also had a prominent abundance. The distribution of taxonomic phyla in sediment samples was more homogeneous and balanced than in water samples. By comparing samples #1, #2, and #3 in the sediment, it was possible to verify similar patterns in the distribution of phyla. However, the water samples presented varied patterns in the abundance of phyla, showing a relevant difference in communities between these compartments.
The genera Acidithiobacillus and Leptospirillum were the most abundant in samples from station #1, #2, and #3 in both water and sediment, but showed low abundance in sample #5 water. The genus Acidiphilium was relatively important in the water and sediment samples from station #3, and also in the sediment from stations #1 and #5. The genus Gallionella was the most abundant in sample #4 water, and also occurred in sample #5 water. The genus Sediminibacterium was present in water and sediment samples at station #4 and in water sample #5. It is also worth highlighting that the genus Thiomonas was the second most frequent in water sample #1.
The distribution of taxonomic phyla of eukaryotic organisms showed very different patterns between sediment and water samples. For the water samples, the dominant phyla were Ochrophyta, Ciliophora, Basidiomycota, Ascomycota and Cryptophyceae. The common fact in all water samples is that the phylum Ochrophyta is among the dominant ones in practically all samples, being in greater proportion in stations #1, #2 and #3. The phylum Ochrophyta includes the microalgae Bacillariophyceae, common in microphytobenthic biofilms, in rivers and even in AMD environments. Samples #2 and #5 water presented Ciliophora as the dominant phylum. For the sediment samples, the dominant phyla were Heterolobosea, Basidiomycota, Ciliophora, Ascomycota, Cryptomycota, Chlorophyta, Cercozoa, and Phragmoplastophyta. Sediment samples #1 and #2 had a greater abundance of the phylum Heterolobosea, while sample #5 had the phylum Cryptomycota as more abundant. Samples #3 and #4 had three phyla with the highest occurrence in each of them, with the phylum Basidiomycota being common for both.
Regarding eukaryotic organisms, chrysophycean alga genus Poteriospumella was the most abundant in sample #3 water, cryptophyceaen alga genus Cryptomonas in sample #1 water, ciliate genus Opisthonecta in sample #2 water, and ameboid family Vampyrellidae in sample #3 sediment. The chrysophycean genus Poterioochromonas was found in all water samples and in sediment samples #1 and #5; its greatest abundance occurred in water sample #4.
The multivariate canonical correspondence analyses (CCA) for prokaryotic organisms (16S) are presented in Fig. 4. It is possible to verify that the sample separation pattern is the same for water and sediments, but the organisms and factors forcing the explainability of variance are distinct. The raw data from the CCAs are available in the supplementary material (Tables S3 and S4).
In the CCA involving water samples (Fig. 4A), where the first and second axes explained, respectively, 48.04% and 21.14% of the variance, samples #4 and #5 showed an association with higher pHs and higher concentration of suspended material, while samples #1, #2, and #3 are characterized by low pH, and higher conductivity and concentrations of Zn, Mn, Ni, Fe, and Cu. Leptospirillum and Acidithiobacillus predominated in these samples, showing a clear association with these last variables. It is also noticeable, although more at the level of the second axis and, therefore, with less variance explainability, a clear separation of two groups of taxa, one associated with sample #4, characterized by slightly higher pHs and the other with sample #5, characterized by slightly lower pHs and higher SPM. In other words, pH was once again the main factor determining the gradients, also positively influencing metal concentrations.
For sediment samples (Fig. 4B), it is observed, at the level of the first axis, which explained 59.61% of the variance, that samples #4 and #5 differ from samples #1, #2, and #3. The main gradient that explains this differentiation is related to pH, Ni, and Cu, which presented higher values in samples #4 and #5, and TOC, Mn, N, and Fe, with higher values in samples #1, #2, and #3. The pH, therefore, is positively related to Ni and Cu and negatively to TOC, Mn, N, and Fe. The taxa most associated with acidity were Deinobacterium, Acidobacterium, Leptospirillum, Acidithiobacillus, Thermogymnomonas, Mycobacterium, and Bdellovibrium. The taxa Candidatus solibacter, Treponema, Geothrix, Desulfomonile, and C. Koribacter were more associated with less acidic pHs. P is associated with the second axis, which explained 19.83% of the variance, being less important in determining taxon composition and showing a certain relationship with the genera Acidocella and Paludibacter. Zn, despite being important in sample #2, did not prove to be an important variable in defining the taxonomic composition, given its low weight in both axes.
The multivariate canonical correspondence analyses for the 20 most abundant eukaryotic organisms (18S) are presented in Fig. 5, and the raw data from these CCAs are presented in the Supplementary Material (Tables S5 and S6). For the water compartment, sample separation showed different trends than previous analyzes (Fig. 5A). At the level of the first axis, which explained 40.78% of the variance, samples #4 and #5 appear in opposite positions, with #5 on the positive side, associated with conductivity, Mn and Zn, and sample #4 on the negative, associated with higher pH. For the second axis, which explained 28.65% of the variance, sample #3 was positioned on the positive side, associated with Ni and Cu, as opposed to samples #1 and #2 on the negative side, associated with Fe and SPM. In relation to organisms, sample #4 was characterized by greater importance of the amoeboid Naegleria and the fungi Gonapodya and Neobulgaria. On the opposite side, related to sample #5, mainly ciliates (Epicarchesium, Vorticella, Choreotrichia), Magnusiophyces fungi and Spumella algae (Chrysophyceae), Chlorophyceae, and Klebsormidiophyceae appear. For the second axis, Poteriospumella (Chrysophyceae), Hypotricha ciliates, and LKM11 fungi appear on the positive side, more associated with sample #3, while Cryptomonas, other Chrysophyceae, Tetramitia amoeboids, and Opisthonecta ciliates appear on the negative side, associated with samples #1 and #2.
In CCA 18S for the sediment samples (Fig. 5B), the relationships between abiotic variables and taxa were more complex, which was expected due to the great diversity of groups of organisms represented, including fungi, algae, ciliates, amoeboids, and plants. However, the separation of samples and abiotic variables was similar to that revealed for 16S. At the level of the first axis, which explained 32.25% of the variance, the variables that most influenced the distribution were Mn, TOC, and N, associated with samples #2 and #3, as opposed to Ni and Zn, associated with samples #2 and #3 and samples #4 and #5. The weight of pH in this analysis was smaller and more associated with the second axis, which explained 28.88% of the variance, together with Cu, as opposed to Fe and P. Sample #1, more associated with the second axis, separated of the others due to their higher P values. In relation to organisms, the algae Poteriospumella and Trebouxiophyceae, together with the amoeboids Vampyrellidae were associated with TOC, Mn, and N and samples #2 and #3, as opposed to algae Klebsormidiophyceae and fungi from the LKM11 group and Magnusiomyces, associated with Zn, Ni, and higher pH and samples #4 and #5. At the level of the second axis, on the negative side, more associated with sample #4, plants (Magnoliophyta), the fungi Gonapodya and Malassezia, the ciliates Nyctotheroides and the alveolates Gregarina appeared. On the positive side of the second axis, the diatom Pinnularia and Hypotricha ciliates stood out.
Discussion
The present study evaluated for the first time the microbial composition of waters and sediments from the SCCB, in southern Brazil, through meta-genomic analyses, relating this information with abiotic data measured in parallel. SCCB, like all areas affected by mining tailings, has aquatic ecosystems that are severely impacted, primarily by broad changes in pH, which exerts strong selective pressure on the structure of biological communities (Novis and Harding 2007). pH, in turn, generates a series of cascading effects on the geochemistry of environments, providing the leaching and mobilization of toxic metals from the geological matrix, the production of high concentrations of sulfates, in addition to affecting the concentration of inorganic and organic nutrients and the availability of gases (Gray 1997). Although the impacts determined a decrease in the diversity of microorganisms, it was possible to detect many taxa adapted to the extreme conditions of the AMD, which suggests a versatile genomic framework.
The sampling procedure performed attempted to cover areas with different characteristics in relation to AMD contamination, to try to highlight the main contrasts in biodiversity, since this is a first meta-genomic approach in the region. The differences were sensitive, with pH and some metals being the main determinants of these differences.
Station #1, also called Língua do Dragão, is the place of greatest environmental concern in the region due to the high flow and high concentration of Fe. This area has already been targeted by other studies that evaluated the geological composition and reported the origin of Fe. In a study using Raman spectroscopy methodology, the highest composition of its sediment was given by goethite (Fe(OH)3), followed by jarosite (KFe3(SO4)2 (OH)6), calcite (CaCO3) (Silva et al. 2011). The values found in the sediment and water were extremely high. For reference issues, the Brazilian national resolution (CONAMA Resolution no. 357/2005) for irrigation, animal watering and landscape harmony provides for a maximum of 5.0 mg/L of Fe and 0.5 mg/L of Mn, while the World Health Organization (WHO) determines that the maximum concentration allowed in drinking water is 0.3 mg/L of Fe and 0.2 mg/L of Mn (Herschy 2012). At station #1, Fe concentration exceeds the CONAMA resolution n°357/2005 by more than 100 times, and more than 38 times for Mn concentration.
The high concentrations of metals and acids contained in AMD are extremely harmful to all ecosystems adjacent to mining areas. These residues are leached into rivers, groundwater, and even marine environments, extending the effects of metal contamination (Oyetibo et al. 2021). This situation can be exemplified by the sample from station #5, which comprises the junction of two rivers that cross the cities of the Araranguá river basin, and still presents very low pH and high concentrations of metals. The concentration of Fe in this station is 25 times higher than that indicated by the WHO and 1.5 times higher than that indicated by CONAMA resolution no. 357/2005, considering the same uses above, a fact that exposes the level of environmental risk in the region and the need for mitigations. A fact that draws attention at this station is that the Fe concentration in the river sediment was one of the highest evaluated, while that in the water was the lowest. This may occur due to the entry of domestic sewage effluents along the river. Some studies have shown that the mixing AMD with sewage promotes a series of biogeochemical reactions. Fe in large quantities acts as a coagulant in sewage waste, causing part of the metals to precipitate together with solid waste, thus promoting an increase in pH, reduction of dissolved metals, and even a decrease in the biochemical oxygen demand and P concentration (Strosnider et al. 2013; Hughes and Gray 2013). The rivers that are found a few meters upstream from station #5 have distinct characteristics. One runs through urbanized areas and receives a greater supply of sewage, and the other has a lower contribution of sewage and remains more acidic due to the influence of the AMD. These processes can at least partially explain the higher P concentration in the sediment at station #5.
The greater alpha-diversity evidenced in almost all sediment samples compared to water samples was also verified in a study of prokaryotic diversity in the São Domingos mine, Portugal (Carlier et al. 2020). The authors’ explanation, which can be extended to the present study, is that the physicochemical conditions are homogeneous in water, while microniches with different physicochemical conditions can be generated in the sediments, favoring greater microbial diversity. Differentiated microlayers of minerals in the bed of water bodies and the accumulation of organics that precipitate can cause the establishment of stratified microbial communities in the sediments close to the water–sediment interface (Carlier et al. 2020). At station #5, eukaryotic alpha-diversity was greater in the water than in the sediment. This fact can be related to the entry of domestic sewage into the river, conditioning the proliferation of organisms from different niches, especially photoautotrophic eukaryotes (microalgae) and heterotrophs (protozoa and fungi).
The acidity and toxicity conditions found in mining environments induce adaptive responses from microorganisms (Ayangbenro et al. 2018). Due to this, the ecological diversity indices found in this study are lower than those found in other environments such as soil, sediment or water in conserved sites (Walters and Martiny 2020; Rambabu et al. 2020). Therefore, only a few specialized taxa such as those mentioned in this work survive in these environments.
The dominance of the phylum Pseudomonadota in AMD environments has been cited in other studies (Kuang et al. 2013; Hua et al. 2014; Ettamimi et al. 2019; Villegas-Plazas et al. 2019; Giddings et al. 2020). Pseudomonadota generally dominate in AMD due to their metabolic plasticity, which includes iron and sulfur oxidants that enable them to grow under low-pH, metal-rich conditions (Giddings et al. 2020). Similarly, Actinobacteria have been reported in other AMD environments (Kuang et al. 2013). Both Pseudomonadota and Actinomycetota thrive in diverse sediments and have developed mechanisms to inhabit metal-rich environments (Giddings et al. 2020). Members of the phylum Nitrospirota participate in the oxidation and fixation of ammonium in low pH and oligotrophic environments and subsequently play an important role in the nitrification process in the sediment of AMD-impacted areas (Giddings et al. 2020). As occurred in the environments studied, information collected from the bacterial communities that dominate several environments polluted by AMD show that Pseudomonadota, Nitrospirota, Acidobacteriota, Chloroflexota, and Actinomycetota are the dominant phyla (Kuang et al. 2013; Méndez-García et al. 2015; Lukhele et al. 2019; Munyai et al. 2021).
The genera reported as most common in AMD environments are Acidithiobacillus (Auld et al. 2013; Wang et al. 2019; Gómez-Villegas et al. 2022), Leptospirillum (Hua et al. 2014; Gómez-Villegas et al. 2022), Acidiphilium (Munyai et al. 2021; Gómez-Villegas et al. 2022), Gallionella (He et al. 2007), Thermogymnomonas (Mesa et al. 2017), and Sediminibacterium (Ettamimi et al. 2019). In the compilation data for this study, these genera represented 37% in water samples and 47% in sediment samples. Within the diversity of microbial communities that inhabit AMD water, acidophilic sulfur and/or iron-oxidizing bacteria are the predominant groups. The acidic nature and high concentration of dissolved metals exert strong selective pressure on microorganisms, favoring genera that develop multiple stress resistance mechanisms to deal with extreme environmental conditions (Mathivanan et al. 2021).
Acidithiobacillus and Leptospirillum were the most occurring genera in all environments evaluated with pH below 3.8. These organisms are obligate or facultative chemolithoautotrophs widely found in AMD environments. The genus Acidithiobacillus is an important representative of sulfur-oxidizing bacteria. The species of this genus are strict autotrophic Gram-negative bacteria, they can fix both carbon and nitrogen from the atmosphere, using reduced sulfur compounds as a primary source of energy and generating the corresponding oxidized sulfur species (Wang et al. 2019). Some species of the genus are also capable of oxidizing ferrous iron to ferric iron or growing anaerobically, using ferric iron as a final electron acceptor (Wang et al. 2019). The most abundant species of this genus in the samples of the present study were A. ferriphilus and A. ferrooxidans which, in addition to sulfides and other sulfur compounds, are capable of oxidizing ferrous iron to ferric, being important contributors to the biogeochemical cycles of Fe and S (Auld et al. 2013; Wang et al. 2019; Gómez-Villegas et al. 2022).
The genus Leptospirillum includes different iron-oxidizing species, whose role in the acidic environment has gained increasing recognition (Hua et al. 2014; Gómez-Villegas et al. 2022). Metabolic reconstructions suggest that Leptospirillum can perform carbon fixation through the novel tricarboxylic acid reductive cycle (Montoya et al. 2012) and has been found to be the main nitrogen fixer in AMD systems (Hua et al. 2014; Villegas-Plazas et al. 2019).
In the present study, the genus Acidiphilium was more frequent in samples with lower pH and showed a positive correlation with Fe and Cu. Acidiphilium is a versatile acidophilic genus generally found in AMD environments (Li et al. 2020; Munyai et al. 2021; Gómez-Villegas et al. 2022). From a metabolic point of view, most Acidiphilium spp. are aerobic heterotrophs that use organic compounds to obtain carbon and energy, although many can obtain some extra energy from the mixotrophic oxidation of ferrous iron (Gómez-Villegas et al. 2022). Furthermore, some members of the genus have been described as capable of reducing Fe3+ under anaerobic or microaerophilic conditions, or carrying out photosynthetic assimilation of CO2 (Kisková et al. 2018). Hua et al. (2014) argued that the presence of the genus Acidiphilium facilitates the dominance of other species by degrading toxic organic substances.
In water samples from stations #4 and #5, which showed lower concentrations of metals and pH 4.25 and 3.8, respectively, the genus Gallionella was highly abundant. Organisms in this genus are neutrophilic and chemolithotrophic Fe oxidizers, which inhabit relatively nutrient-poor environments containing reduced iron (He et al. 2007). In addition, iron oxidation can be carried out mainly by Gallionella spp. in environments with little oxygen (He et al. 2007). Another abundant genus in the water of stations #4 and #5 and in the sediment of station #4 was Sediminibacterium. This genus was also found to be one of the most abundant in a mining transition area with a pH of 6.4 (Ettamimi et al. 2019; Carlier et al. 2020).
Sediment samples from stations #1, #2, and #3 showed an abundance of the genus Thermogymnomonas, an archaea that belongs to the phylum Euryarchaeota. The species is obligately aerobic and heterotrophic. In Los Rueldos, Spain, Thermogymnomonas spp. were abundant in all samples with chemical characteristics compatible with their niche as an obligate heterotrophic aerobe (Mesa et al. 2017).
As highlighted above, the prokaryotic communities of stations #1, #2, and #3 were similar to those of other AMD environments with severe characteristics, where low biodiversity prevails. Furthermore, at these stations, there is a community with taxonomic groups specialized in carrying out the biogeochemical recycling of Fe, S, N, C, and O. Stations #4 and #5 presented characteristics of greater biodiversity and some taxonomic groups normally found in environments with less acidity impact.
The results for the eukaryotic community also indicated a relatively low diversity, as seen for archaea and bacteria, a fact similar to that found in biofilms in Richmond Mine at Iron Mountain, USA (Baker et al. 2009). The association of the Eukarya domain with AMD has been reported to a lesser extent when compared with Bacteria (Oyetibo et al. 2021). Microalgae from the phylum Ochrophyta, such as the benthic diatom Pinnularia (Aguilera 2013), have been previously reported in the waters of Tinto River, Spain, and the Chrysophyceae from the genus Poteriospumella (Gómez-Villegas et al. 2022) were found in phosphogypsum stacks near Tinto River. Species belonging to these genera, especially Pinnularia, are quite widespread in environments with pH values around 3.0 (Aguilera 2013). Of all the environmental variables that affect freshwater diatoms, pH appears to be the most important, and most taxa show a preference for a narrow pH range (Aguilera 2013). This feature was observed in water samples #1, #2, and #3, which showed high dominance of photosynthetic eukaryotes.
Fungi from the phyla Basidiomycota and Ascomycota were also found at the Richmond Mine at Iron Mountain, USA (Baker et al. 2009). The LKM11 clade of environmental fungi belongs to a group located close to the phylogenetic root of the kingdom Fungi, Rozellomycota (Lara et al. 2010). Phylogeny reconstruction indicated that the LKM11 group shared high similarity with Paramicrosporidium fungi, which are endonuclear parasites of free-living amoebae (Mesa et al. 2017).
Gómez-Villegas et al. (2022) described the existence of ciliates (phylum Ciliophora), amoebas and other protist predators in phosphogypsum stacks near Tinto River. Protists from the phylum Heterolobosea were also found in the Richmond Mine at Iron Mountain, USA (Baker et al. 2009). The genus identified in this phylum with the greatest abundance was Naegleria. This eukaryote is reported to be an organism adapted to life in the harsh environment of AMD (Baker et al. 2009).
It is interesting to note the prevalence of photosynthetic organisms, fungi, ciliates, and protists in the samples. The vast majority of photosynthetic organisms were found in water, as expected, since they need light to carry out photosynthesis. While the greatest amount of fungi and heterotrophic protists were found in sediment, where they have a greater amount of organic matter and nutrients and, normally, lower irradiance. Photosynthetic eukaryotes play a fundamental role in the formation of atmospheric oxygen, which could favor the aerobic oxidation of Fe and S (González-Toril et al. 2011). Fungi, largely represented by Ascomycota and Basidiomycota, are mainly found in low-pH subsurface biofilms that thrive in AMD participating in C cycling as the main decomposers of the microbial community Bacteria (Oyetibo et al. 2021). Amoebas act as herbivores in the AMD ecosystem (Volant et al. 2016; Mesa et al. 2017). Ciliates can impact the abundance of bacteria and archaea and, therefore, community composition and function (Mesa et al. 2017).
Despite the small number of samples evaluated, the canonical correspondence analyses carried out clearly and with good explainability of variance showed the main relationships between the taxonomic composition of main organisms and environmental variables, whether in different compartments (water and sediment) or in prokaryotic (16S) and eukaryotes (18S) groups.
It was possible to verify that the majority of the most abundant prokaryotic and eukaryotic organisms were associated with the environmental water of sample #5, which refers to the river with the least contamination by heavy metals and which visibly has a greater sewage load. This is an expected fact, since it is the least hostile environmental condition among those evaluated and, therefore, less toxic for the survival of organisms.
Prokaryotic diversity was quite different among sites with different levels of contamination, highlighting the prevalence of some groups of organisms that can serve as bioindicators of conditions in mining environments. The genera Leptospirillum, Acidithiobacillus, Acidiphilium, Thiomonas, Thermogymnomonas, and Acidobacterium are the most prominent taxonomic groups in impacted environments, correlated with lower pH and higher Fe concentration. Some of these groups have already been associated with a greater AMD contamination such as the genera Acidiphilium, Acidibacter, Acidobacterium, and Acidocella found in low pH environments (pH < 3) in the São Domingos mining area, in southern Portugal (Ettamimi et al. 2019; Carlier et al. 2020). The genera Acidocella, Sediminibacterium, Geothrix, Geobacter, and Gallionella were correlated with milder environments, characterized as transitioning from AMD pollution. The Sediminibacterium and Gallionella groups, as already mentioned, are characteristic of transition areas (He et al. 2007; Ettamimi et al. 2019); Geothrix and Geobacter are anaerobic sulfate-reducing bacteria found in environments with weak acidic conditions in coal mining (Chen et al. 2021).
Among the eukaryotic organisms found in water, Poteriospumella (Chrysophyceae), Hypotricha ciliates, and LKM11 fungi were associated with conditions of greater contamination (lower pH and higher concentration of Cu, Ni, Zn and Mn) of AMD. The Hypotrichia subclass belongs to the ciliate group, and has previously been associated with acidic mining lake environments (Weisse et al. 2013). In the sediment, again the Poteriospumella algae together with the Trebouxiophyceae algae and the Vampyrellidae amoeboids were associated with conditions of greater impact. The correlation of prokaryotic organisms in environments with greater AMD contamination is well established, as previously reported, but for eukaryotic organisms, there are few studies, which shows the relevance of better understanding these groups and even exploring their use for processes in biotechnology and bioremediation.
One of the facts that caught our attention were the groups related to the sediment with the highest concentration of copper, including Tetramitia, which is a heterotrophic protist, Gregarina, a unicellular organism from the phylum Apicomplexa, considered a parasite of many invertebrates, and Malassezia, which is a yeast. Cu is a toxic metal for most eukaryotes; to survive higher concentrations, it is necessary to develop resistance mechanisms. These mechanisms are more widespread in prokaryotes. In eukaryotes, few strategies are found, being more common in fungi and yeasts (Antsotegi-Uskola et al. 2020), mainly comprising Cu sequestration by metallothioneins, extracellular sequestration by EPS and synthesis of chelating agents.
Like Cu, Fe concentration was related to the prevalence of some eukaryotic organisms in the sediment of the evaluated environments. In places like station #1, which has 26% Fe in the sediment, organisms need resistance mechanisms to this metal to survive. Thus, to achieve effective homeostasis, they must balance their need to efficiently eliminate this metal from the environment to ensure adequate supplies are maintained, with careful management of cellular-free Fe levels to protect against toxicity (Touati 2000; Andrews et al. 2003).
The results of the present study proved to be very important for understanding the prokaryotic and eukaryotic populations existing in coal mining environments in SCCB. It is interesting to see that mining environments for the same ore (coal) in a very close geographical location, sharing the same climate, presented different abiotic, biodiversity, and population characteristics. An important additional step to this initial study would be to evaluate functional metagenomics, to better understand the adaptive mechanisms and their biotechnological potentials under these extreme conditions.
Concluding remarks
As a first meta-taxonomic study of prokaryotes and eukaryotes from sediment and water in the SCCB mining area, this work contributed to better understanding the impact of AMD on these ecosystems. It revealed that differences in community composition can distinguish sites highly contaminated with AMD (pH 2.7–3.6) from sites with intermediate levels of contamination (pH 3.8–4.25), indicating possible bioindicators. For the SCCB region, the prokaryotic genera Leptospirillum, Acidithiobacillus, Acidiphilium, Thiomonas, Thermogymnomonas, and Acidobacterium were found as bioindicators of areas highly impacted by AMD (low pH and high concentration of metals); and eukaryotes the alga Poteriospumella (Chrysophyceae), Hypotricha ciliates, and LKM11 fungi. For transitional environments, with weak conditions of acidity and metals, the genera that have a vocation as bioindicators are Sediminibacterium, Gallionella Geothrix, and Geobacter.
The sediment compartment presented a higher concentration of metals than water, which can be explained by sequential dissolution processes, with subsequent sedimentation and concentration. The variation in pH was the factor with the greatest influence on the composition of the prokaryotic community, while for the eukaryotic community, the concentration of nutrients (TOC and N) and metals (Ni, Cu, Mn, and Zn) was more influential in explaining the community composition.
The Pseudomonadota phylum, which houses several sulfur-oxidizing bacteria, was the most abundant in all samples, being a striking characteristic in these AMD environments. Eukaryotic organisms presented a great diversity of photosynthetic phyla (Ochrophyta, Chlorophyta, and Cryptophyta), fungi (Basidiomycota, Ascomycota, and Cryptomycota), ciliates (Ciliophora), and protists (Heterolobosea, Cercozoa).
Data availability
The raw data is available in the NCBI Sequence Read Archive (SRA) under accession number PRJNA1015584.
Abbreviations
- AMD:
-
Acid mine drainage
- SCCB:
-
Santa Catarina Carboniferous Basin
- TOC:
-
Total organic carbon
- TN:
-
Total nitrogen
- TP:
-
Total phosphorus
- ASV:
-
Amplicon sequence variant
- CCA:
-
Canonical correspondence analyses
- SPM:
-
Suspended particulate material
References
Aguilera A (2013) Eukaryotic organisms in extreme acidic environments, the Río Tinto case. Life 3:363–374. https://doi.org/10.3390/LIFE3030363
Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM (2009) A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 4:e6372. https://doi.org/10.1371/journal.pone.0006372
Andrews SC, Robinson AK, Rodríguez-Quiñones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. https://doi.org/10.1016/S0168-6445(03)00055-X
Antsotegi-Uskola M, Markina-Iñarrairaegui A, Ugalde U (2020) New insights into copper homeostasis in filamentous fungi. Int Microbiol 23:65–73. https://doi.org/10.1007/S10123-019-00081-5
APHA (2018a) 3120 metals by plasma emission spectroscopy. In: Lipps WC, Baxter TE, Braun-Howland E (eds) Standard methods for the examination of water and wastewater, 23rd edn. APHA Press, Washington DC
APHA (2018b) 5310 total organic carbon. In: Lipps WC, Baxter TE, Braum-Howland E (eds) Standard methods for the examination of water and wastewater, 23rd edn. APHA Press, Washington, DC
APHA (2018c) 4500-N NITROGEN. In: Lipps WC, Baxter TE, Braum-Howland E (eds) Standard methods for the examination of water and wastewater, 23rd edn. APHA Press, Washington, DC
Auld RR, Myre M, Mykytczuk NCS et al (2013) Characterization of the microbial acid mine drainage microbial community using culturing and direct sequencing techniques. J Microbiol Methods 93:108–115. https://doi.org/10.1016/J.MIMET.2013.01.023
Ayangbenro AS, Olanrewaju OS, Babalola OO (2018) Sulfate-reducing bacteria as an effective tool for sustainable acid mine bioremediation. Front Microbiol. https://doi.org/10.3389/fmicb.2018.01986
Baker BJ, Tyson GW, Goosherst L, Banfield JF (2009) Insights into the diversity of eukaryotes in acid mine drainage biofilm communities. Appl Environ Microbiol 75:2192–2199. https://doi.org/10.1128/AEM.02500-08
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Brumfield KD, Huq A, Colwell RR et al (2020) Microbial resolution of whole genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data. PLoS ONE 15:1–21. https://doi.org/10.1371/journal.pone.0228899
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Carlier JD, Ettamimi S, Cox CJ et al (2020) Prokaryotic diversity in stream sediments affected by acid mine drainage. Extremophiles 24:809–819. https://doi.org/10.1007/s00792-020-01196-8
Castro-Silva MA, De Souza Lima AO, Gerchenski AV et al (2003) Heavy metal resistance of microorganisms isolated from coal mining environments of Santa Catarina. Brazilian J Microbiol 34:45–47. https://doi.org/10.1590/S1517-83822003000500015
Chen D, Feng Q, Li W et al (2021) Effects of acid drainage from abandoned coal mines on the microbial community of Shandi River sediment, Shanxi Province. Int J Coal Sci Technol 8:756–766. https://doi.org/10.1007/s40789-021-00433-5
de Rangel WM, Schneider J, Soares CRFS et al (2016) Phytoremediation, vol 3. Springer International Publishing, Cham
Ettamimi S, Carlier JD, Cox CJ et al (2019) A meta-taxonomic investigation of the prokaryotic diversity of water bodies impacted by acid mine drainage from the São Domingos mine in southern Portugal. Extremophiles 23:821–834. https://doi.org/10.1007/S00792-019-01136-1
Freitas APP, Schneider IAH, Schwartzbold A (2011) Biosorption of heavy metals by algal communities in water streams affected by the acid mine drainage in the coal-mining region of Santa Catarina state, Brazil. Miner Eng 24:1215–1218. https://doi.org/10.1016/J.MINENG.2011.04.013
Giddings LA, Chlipala G, Kunstman K et al (2020) Characterization of an acid rock drainage microbiome and transcriptome at the Ely Copper Mine Superfund site. PLoS ONE 15:1–25. https://doi.org/10.1371/journal.pone.0237599
Gómez-Villegas P, Guerrero JL, Pérez-Rodriguez M et al (2022) Exploring the microbial community inhabiting the phosphogypsum stacks of Huelva (SW SPAIN) by a high throughput 16S/18S rDNA sequencing approach. Aquat Toxicol 245:106103. https://doi.org/10.1016/J.AQUATOX.2022.106103
González-Toril E, Águilera Á, Souza-Egipsy V et al (2011) Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain). Appl Environ Microbiol 77:2685–2694. https://doi.org/10.1128/AEM.02459-10
Gray NF (1997) Environmental impact and remediation of acid mine drainage: a management problem. Environ Geol 30:62–71. https://doi.org/10.1007/S002540050133/METRICS
Hallberg KB (2010) New perspectives in acid mine drainage microbiology. Hydrometallurgy 104:448–453. https://doi.org/10.1016/J.HYDROMET.2009.12.013
He Z, Xiao S, Xie X et al (2007) Molecular diversity of microbial community in acid mine drainages of Yunfu sulfide mine. Extremophiles 11:305–314. https://doi.org/10.1007/S00792-006-0044-Z
Herschy RW (2012) Water quality for drinking: WHO guidelines. World Health Organization
Hua ZS, Han YJ, Chen LX et al (2014) Ecological roles of dominant and rare prokaryotes in acid mine drainage revealed by metagenomics and metatranscriptomics. ISME J 9:1280–1294. https://doi.org/10.1038/ismej.2014.212
Hughes TA, Gray NF (2013) Remoción de acidez y de metales de drenajes ácidos de minas usando aguas residuales municipales y barros activados. Mine Water Environ 32:170–184. https://doi.org/10.1007/s10230-013-0218-8
Kisková J, Perháčová Z, Vlčko L et al (2018) The bacterial population of neutral mine drainage water of Elizabeth’s Shaft (Slovinky, Slovakia). Curr Microbiol 75:988–996. https://doi.org/10.1007/S00284-018-1472-6
Kuang J-L, Huang L-N, Chen L-X et al (2013) Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J 7:1038–1050. https://doi.org/10.1038/ismej.2012.139
Lara E, Moreira D, López-García P (2010) The environmental clade LKM11 and Rozella form the deepest branching clade of fungi. Protist 161:116–121. https://doi.org/10.1016/J.PROTIS.2009.06.005
Lattuada RM, Menezes CTB, Pavei PT et al (2009) Determination of metals by total reflection X-ray fluorescence and evaluation of toxicity of a river impacted by coal mining in the south of Brazil. J Hazard Mater 163:531–537. https://doi.org/10.1016/j.jhazmat.2008.07.003
Li L, Liu Z, Zhang M et al (2020) Insights into the metabolism and evolution of the genus Acidiphilium, a typical acidophile in acid mine drainage. mSystems. https://doi.org/10.1128/MSYSTEMS.00867-20
Liu C, Cui Y, Li X, Yao M (2021) microeco: an R package for data mining in microbial community ecology. FEMS Microbiol Ecol. https://doi.org/10.1093/FEMSEC/FIAA255
Lukhele T, Selvarajan R, Nyoni H et al (2019) Diversity and functional profile of bacterial communities at Lancaster acid mine drainage dam, South Africa as revealed by 16S rRNA gene high-throughput sequencing analysis. Extremophiles 23:719–734. https://doi.org/10.1007/S00792-019-01130-7
Mathivanan K, Chandirika JU, Vinothkanna A et al (2021) Bacterial adaptive strategies to cope with metal toxicity in the contaminated environment—a review. Ecotoxicol Environ Saf 226
Maus V, Giljum S, Gutschlhofer J et al (2020) A global-scale data set of mining areas. Sci Data. https://doi.org/10.1038/s41597-020-00624-w
Méndez-García C, Peláez AI, Mesa V et al (2015) Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol. https://doi.org/10.3389/fmicb.2015.00475
Mesa V, Gallego JLR, González-Gil R et al (2017) Bacterial, Archaeal, and Eukaryotic diversity across distinct microhabitats in an acid mine drainage. Front Microbiol. https://doi.org/10.3389/fmicb.2017.01756
Montoya L, Celis LB, Razo-Flores E, Alpuche-Solís ÁG (2012) Distribution of CO2 fixation and acetate mineralization pathways in microorganisms from extremophilic anaerobic biotopes. Extremophiles 16:805–817. https://doi.org/10.1007/s00792-012-0487-3
Munyai R, Ogola HJO, Modise DM (2021) Microbial community diversity dynamics in acid mine drainage and acid mine drainage-polluted soils: implication on mining water irrigation agricultural sustainability. Front Sustain Food Syst 5:321. https://doi.org/10.3389/FSUFS.2021.701870/BIBTEX
Neff AN, DeNicola DM, Maltman C (2021) Passive treatment for acid mine drainage partially restores microbial community structure in different stream habitats. Water 13:3300. https://doi.org/10.3390/w13223300
Novis PM, Harding JS (2007) Extreme acidophiles. In: Seckbach J (ed) Algae and Cyanobacteria in extreme environments. Springer Netherlands, Dordrecht, pp 443–463
Osório FHT, Silva LFO, Piancini LDS et al (2014) Water quality assessment of the Tubarão River through chemical analysis and biomarkers in the Neotropical fish Geophagus brasiliensis. Environ Sci Pollut Res 21:9145–9160. https://doi.org/10.1007/s11356-013-1512-5
Oyetibo GO, Enahoro JA, Ikwubuzo CA, Ukwuoma CS (2021) Microbiome of highly polluted coal mine drainage from Onyeama, Nigeria, and its potential for sequestrating toxic heavy metals. Sci Rep 11:17496. https://doi.org/10.1038/s41598-021-96899-z
Pille da Silva E, Dutra de Armas R, Ademar Avelar Ferreira P et al (2019) Soil attributes in coal mining areas under recovery with bracatinga (Mimosa scabrella). Lett Appl Microbiol 68:497–504. https://doi.org/10.1111/lam.13153
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Rambabu K, Banat F, Pham QM et al (2020) Biological remediation of acid mine drainage: review of past trends and current outlook. Environ Sci Ecotechnol. https://doi.org/10.1016/j.ese.2020.100024
She Z, Pan X, Wang J et al (2021) Vertical environmental gradient drives prokaryotic microbial community assembly and species coexistence in a stratified acid mine drainage lake. Water Res 206:117739. https://doi.org/10.1016/J.WATRES.2021.117739
Silva LFO, Wollenschlager M, Oliveira MLS (2011) A preliminary study of coal mining drainage and environmental health in the Santa Catarina region, Brazil. Environ Geochem Health 33:55–65. https://doi.org/10.1007/S10653-010-9322-X
Silva LFO, Ortiz F, de Vallejuelo S, Martinez-Arkarazo I et al (2013) Study of environmental pollution and mineralogical characterization of sediment rivers from Brazilian coal mining acid drainage. Sci Total Environ 447:169–178. https://doi.org/10.1016/j.scitotenv.2012.12.013
Simate GS, Ndlovu S (2014) Acid mine drainage: challenges and opportunities. J Environ Chem Eng 2:1785–1803. https://doi.org/10.1016/J.JECE.2014.07.021
Strosnider WHJ, Nairn RW, Peer RAM, Winfrey BK (2013) Passive co-treatment of Zn-rich acid mine drainage and raw municipal wastewater. J Geochemical Explor 125:110–116. https://doi.org/10.1016/j.gexplo.2012.11.015
Takahashi S, Tomita J, Nishioka K et al (2014) Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE. https://doi.org/10.1371/journal.pone.0105592
Touati D (2000) Iron and oxidative stress in Bacteria. Arch Biochem Biophys 373:1–6. https://doi.org/10.1006/ABBI.1999.1518
U.S. EPA (1996) Method 3050B acid digestion of sediments, sludges, and soils 1.0 scope and application, p 12
U.S. EPA (2007) Method 3051A (SW-846): microwave assisted acid digestion of sediments, sludges, and oils. Washington, DC
Villegas-Plazas M, Sanabria J, Junca H (2019) A composite taxonomical and functional framework of microbiomes under acid mine drainage bioremediation systems. J Environ Manage 251:109581. https://doi.org/10.1016/J.JENVMAN.2019.109581
Volant A, Héry M, Desoeuvre A et al (2016) Spatial distribution of Eukaryotic communities using high-throughput sequencing along a pollution gradient in the arsenic-rich creek sediments of Carnoulès Mine, France. Microb Ecol 72:608–620. https://doi.org/10.1007/S00248-016-0826-5
Walters KE, Martiny JBH (2020) Alpha-, beta-, and gamma-diversity of bacteria varies across habitats. PLoS ONE 15:1–17. https://doi.org/10.1371/journal.pone.0233872
Wang R, Lin JQ, Liu XM et al (2019) Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front Microbiol 10:1–20. https://doi.org/10.3389/fmicb.2018.03290
Weisse T, Moser M, Scheffel U et al (2013) Systematics and species-specific response to pH of Oxytricha acidotolerans sp. nov. and Urosomoida sp. (Ciliophora, Hypotricha) from acid mining lakes. Eur J Protistol 49:255–271. https://doi.org/10.1016/J.EJOP.2012.08.001
Wu B, Liu F, Fang W et al (2021) Microbial sulfur metabolism and environmental implications. Sci Total Environ J 778:146085. https://doi.org/10.1016/j.scitotenv.2021.146085
Xu R, Li B, Xiao E et al (2020) Uncovering microbial responses to sharp geochemical gradients in a terrace contaminated by acid mine drainage. Environ Pollut 261:114226. https://doi.org/10.1016/J.ENVPOL.2020.114226
Acknowledgements
This study was partially funded by Fundação de Amparo à Pesquisa e Inovação do Estado de Santa Catarina (FAPESC) through a grant from the Universal Research Support Program (FAPESC-2021TR000671) and by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior–Brazil (CAPES)–Finance Code 001 (Grant no. 88882.438331/2019-01). We also acknowledge the financial resources provided by Biome4All.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Oren.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Odisi, E.J., de Freitas, R.C., do Amaral, D.S. et al. Metataxonomy of acid mine drainage microbiomes from the Santa Catarina Carboniferous Basin (Southern Brazil). Extremophiles 28, 8 (2024). https://doi.org/10.1007/s00792-023-01324-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00792-023-01324-0