Abstract
Microscopic eukaryotes play a key role in ecosystem functioning, but their diversity remains largely unexplored in most environments. To advance our knowledge of eukaryotic microorganisms and the factors that structure their communities, high-throughput sequencing was used to characterize their diversity and spatial distribution along the pollution gradient of the acid mine drainage at Carnoulès (France). A total of 16,510 reads were retrieved leading to the identification of 323 OTUs after normalization. Phylogenetic analysis revealed a quite diverse eukaryotic community characterized by a total of eight high-level lineages including 37 classes. The majority of sequences were clustered in four main groups: Fungi, Stramenopiles, Alveolata and Viridiplantae. The Reigous sediments formed a succession of distinct ecosystems hosting contrasted eukaryotic communities whose structure appeared to be at least partially correlated with sediment mineralogy. The concentration of arsenic in the sediment was shown to be a significant factor driving the eukaryotic community structure along this continuum.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Acid mine drainage (AMD) refers to extreme aquatic environments resulting from the dissolution of pyrite-rich mining wastes. Because of their acid pH and their high concentrations of toxic metals and metalloids, they are an environmental and public health threat [1]. In these ecosystems, adapted microorganisms are involved in biogeochemical mechanisms that control the behavior of metals and metalloids at the water/sediment interface. Indeed, the microorganisms that colonize AMD are responsible for the release of metals and metalloids and for the acidification resulting from oxidation of pyrite-rich waste. Microbial metabolism can also mitigate pollution, for example, by co-precipitation of arsenic with iron [2]. The prokaryotic communities that colonize AMD, particularly bacteria, have been extensively studied and generally include organisms involved in the iron, sulfur or arsenic cycles [3–5]. On the other hand, studies of the eukaryotic members of the microbial communities are rare and the involvement of eukaryotes in the biogeochemical cycles of these extreme environments is poorly understood [6–9]. Culture-independent gene surveys have revealed relatively high eukaryotic richness in the Río Tinto River in Spain including members of the Bacillariophyta, Chlorophyta, Rhodophyta, and Euglenophyta phyla as well as Ciliates, Cercomonads, Amoebae, Stramenopiles, Fungi, Heliozoans, and Rotifers [6, 8, 10, 11]. In contrast, studies at the Richmond mine, an extremely acid (pH < 1) subsurface AMD site at Iron Mountain in California, evidenced low eukaryotic diversity, mainly dominated by Fungi associated with the presence of a small proportion of Heterolobosea and protists affiliated with the deeply branching lineage named the acidophilic protist clade (APC) [9].
The ecological role of eukaryotic microorganisms is of great importance in mining environments. Some protists isolated from AMD graze on specific acidophilic bacteria [12, 13], suggesting they impact the abundance and the community composition of prokaryotes involved in key biogeochemical processes such as acid mine generation or mitigation via oxidation or reduction of iron and sulfur compounds. A strain of the algae Cyanidioschyzon sp. isolated from the Yellowstone National Park was shown to be directly implicated in the As cycle, oxidizing As(III), reducing As(V) and methylating As(III) to form trimethylarsine oxide and dimethylarsenate [14]. Some photosynthetic organisms like Euglena mutabilis or the filamentous algae Klebsormidium not only provide oxygen but can also be directly involved in the development of stromatolite-like and organo-sedimentary structures that are rich in iron and play an important role in remediation [15]. Because of their ecological importance and their potential for bioremediation, the full diversity and the precise role of eukaryotes in AMD along with the environmental parameters that structure their communities require further investigation.
The level of pollution at Carnoulès (France) is particularly high, with arsenic concentrations ranging from 80 to 350 mg L−1 in the Reigous Creek spring, which corresponds to the AMD draining the site [16]. Microbial activity is involved in the natural attenuation process that occurs at this site and results in a pollution gradient along Reigous Creek. In particular, microbial iron and arsenic oxidation leads to the removal of dissolved arsenic by co-precipitation with Fe(III) and formation of As(III)-Fe(III) and As(V)-Fe(III) phases associated with sulfate in the Reigous riverbed [2, 16, 17]). The concentration of arsenic subsequently decreases by nearly 95 % between the source of Reigous Creek and its confluence with the Amous River, 1500 m downstream. Bacterial and archaeal communities have been extensively studied at this site [5, 18–21]. However, to date, the eukaryotic community thriving in this ecosystem remains largely unexplored although cultivation-based studies revealed the presence of Fungi along the creek [22] and the presence of the acidophilic protozoan Euglena mutabilis [23, 24]. This photosynthetic organism may play an indirect but important role in natural AMD attenuation through primary production [24]. Indeed, an increase in the concentration of dissolved oxygen in these oligotrophic waters probably increases metal precipitation rates [25]. Furthermore, Euglena mutabilis is able to sequester and store iron and other metals both intracellularly and extracellularly [23, 25].
Determining eukaryotic diversity is the first step in assessing the potential influence of eukaryotes on metal and metalloid cycling and on ecosystem functioning in the Carnoulès As-rich AMD. To this end, eukaryotic diversity in Reigous riverbed sediments was investigated using a barcoded pyrosequencing approach targeting the hypervariable V2-V3 regions of the eukaryotic small-subunit 18S ribosomal RNA (rRNA) gene [26, 27]. To help understand how environmental stressors determine the structure of the microbial community, the eukaryotic spatial pattern was characterized in relation with the physicochemical and mineralogical conditions observed in situ along the pollution gradient.
Methods
Site Description
The Pb-Zn Carnoulès mine is located in southern France in the drainage basin of the River Rhône. Mining activity stopped in 1962 but left about 1.2 Mt of solid wastes containing 0.7 % Pb, 10 % Fe, and 0.2 % As stored behind a dam. Water percolating through the wastes emerges at the base of the tailing dam and forms the source of Reigous Creek. This water is acidic (pH around 3), sub-oxic (dissolved oxygen concentration below 1 mg L−1) and rich in dissolved sulfate, iron and arsenic (2000–7700, 500–1000, and 50–350 mg L−1, respectively). Iron and arsenic are mainly present in their reduced forms Fe(II) and As(III) [16]. Mean annual rainfall is 1320 mm with a distribution typical of the Mediterranean climate characterized by long periods of drought and intense rainfall events mainly in spring and fall.
Sampling Procedure and Physicochemical Analysis
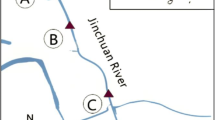
A sampling campaign was carried out in November 2009 at five sampling sites along Reigous Creek. Samples were collected at the spring (S1), 30 m downstream (COWG), 150 m downstream at two sampling sites characterized by different sediment textures (stromatolite-like structure with laminated hard concretions called GAL and soft and creamy sediment called GALm), and 1500 m downstream (CONF) just before the confluence of the Reigous and the Amous River (Fig. 1). Around 5-cm deep pale yellow loosely packed sediments were collected in triplicate at the bottom of the creek using a sterile spatula. The three samples were pooled and homogenized. Solid phases were harvested by centrifugation and dried under vacuum prior to mineralogical and spectroscopic analyses. The main physicochemical parameters (pH, temperature, redox potential and dissolved oxygen concentration) and concentrations of SO4 2-, Fe, and As in the running water covering the sediments were measured as previously described [21].
Map of the Carnoulès mining site and location of the sampling sites [62]
Physicochemical Characterization of Sediments
The total concentrations of arsenic, iron, sulfate, lead, and zinc were measured in sediments after bulk mineralization. About 100 mg of sediment was digested in closed Teflon reactors on hot plates at 95 °C for 24 h successively with (1) H2O2 35 % (2) a 4:3:0.13 mL concentrated HNO3-HF-HClO4 mixture (3) a 1:3 mL concentrated HNO3-HCl mixture. Samples were cooled and evaporated to dryness between each step and at the end of the procedure. Samples were brought to 30 mL using 3 mL of HNO3 and double deionized water (Milli-Q®). Finally, samples were filtered to remove possible residues. For each set of samples, method blanks and international certified reference materials digestion (Stream sediments LGC6189 from the UK Accreditation Service and NCS DC70317 from LGC Standards) were performed. Chemical analysis was then carried out as previously described [21].
The mineralogical composition of the collected sediment samples was determined qualitatively using powder X-ray diffraction analysis (XRD). Data were collected with Co K-alpha radiation on an X’Pert PRO P analytical diffractometer equipped with an X’Celerator detector, in continuous mode and a 4 h counting time per sample. X-ray absorption spectroscopy data were collected on the five sediment phases. X-ray absorption near edge structure (XANES) and extended X-ray absorption fine structure (EXAFS) spectra were recorded at a temperature of 10–15 K in fluorescence mode on the FAME BM30B bending magnet beamline at ESRF (Grenoble, France). Data for the COWG sample were previously collected and analyzed in Volant et al. [5]. Experimental details and data reduction procedures are reported in previous studies [2, 28, 29]. XANES and EXAFS data were interpreted by linear combination fitting using a set of model compound spectra. This set includes As(V)- and As(III)–Fe(III) oxyhydroxides and oxyhydroxysulfates synthesized via biotic and abiotic pathways [2, 30].
DNA Isolation
Genomic DNA was extracted in triplicate from sediments using the UltraClean Soil DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA, USA) according to the manufacturer’s recommendations. The three extractions were pooled before PCR amplification. All extracted genomic DNA samples were stored at −20 °C until further analysis.
Pyrosequencing of Eukaryotic 18S rRNA Genes
The set of primers, 18S-82F (5′-AxxxGAAACTGCGAATGGCTC-3′) and Ek-516r (5′-BxxxACCAGACTTGCCCTCC-3′) was used to amplify a 480 bp region targeting the complete V2 and V3 domains of all eukaryote SSU 18S rRNA genes [26, 27, 31]. A and B represent the two FLX Titanium adapters (A adapter sequence: 5′-CGTATCGCCTCCCTCGCGCC ATCAG-3′; B adapter sequence: 5′-CTATGCGCCTTGCCAGCCCGCTCAG-3′) and xxx represent the sample-specific barcode sequence. Triplicate PCR amplifications were performed with the same barcode on each sample. The reaction mixture contained 1 μl of DNA template, 1 μl of both primers (10 μM), and 12.5 μl of PCR Master Mix Ampli Taq Gold 360 (Applied Biosystems, Foster City, CA, USA). Sterile distilled water was added to obtain a final volume of 25 μl. PCR conditions were as follows: denaturation at 95 °C for 10 min, 35 cycles at 95 °C for 45 s, 56 °C for 45 s, and 72 °C for 45 s, followed by 10 min at 72 °C. The PCR products with a proximal length of 480 bp were excised from a 1 % agarose gel and purified with the QIAquick Gel Extraction Kit (QIAGEN Inc., Valencia, CA, USA). DNA concentrations of the purified PCR products were measured using a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA). To minimize random PCR bias, triplicates were pooled in equimolar ratios to obtain a total of 500 ng of DNA per sample. Pyrosequencing of the five amplicon libraries was performed on a GS-FLX-Titanium sequencer (Roche 454 Life Sciences) at GATC Biotech (Konstanz, Germany).
18S rRNA Gene Sequence Analysis and Taxonomic Classification
Preliminary quality checks, sorting, and trimming of the 454-reads were performed by GATC Biotech according to the manufacturer’s recommendations. The sequences were then analyzed with Mothur software, version 1.31 [32]. Preprocessing of unaligned sequences included removing sequences < 200 pb, all sequences containing ambiguous characters, and sequences with more than eight homopolymers. In addition, we removed sequences that did not align over the same span of nucleotide positions. Identical sequences were grouped and representative sequences were aligned against the SILVA eukaryotic reference database using the Needleman–Wunsch algorithm [33]. Chimeric sequences were detected and removed using the implementation of Chimera Uchime [34]. A further screening step (pre-cluster) was applied to reduce sequencing noise by clustering reads that differed by only one base every 100 bases [35]. The remaining high-quality reads were used to generate a distance matrix and clustered into operational taxonomic units (OTUs) defined at 97 % cutoff using the average neighbor algorithm. The 97 % similarity level has been shown to be the most suitable to reproduce original eukaryotic diversity [36]. Next, taxonomic affiliation of the OTUs was determined using the Naïve Bayesian Classifier (confidence threshold 80 %) trained on the RDP’s taxonomic outline implemented in Mothur with the PR2 database [37]. Note that OTUs classified as Metazoa and Embryophyta were excluded from further analysis to reduce the number of non-microbial eukaryotes. Sequences were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the accession number SRP075900.
Statistical Analysis
Non-parametric Chao1 and Shannon alpha diversity estimates, as well as coverage and rarefaction curves were calculated with Mothur v.1.31. To obtain comparable data, we performed random sampling so that the number of sequences per sample was the same (541 sequences). To estimate community similarity among samples, a distance matrix was calculated based on the Bray–Curtis dissimilarities of the OTU composition of all sampling sites. The normalized abundances of OTUs were square root transformed before analysis. A second dissimilarity matrix was computed based on the Euclidean distance of log(N + 1) sediments chemical characteristics (Table 1). Variations in the structure of eukaryotic communities among the sites were visualized using principal coordinate analysis (PCoA) based on the distance matrix of normalized OTU abundances. Environmental variables were vector fitted to the PCoA ordination to assess possible explanatory variables. Pearson correlations were used to identify the relationships between physicochemical parameters and the relative abundance of dominant taxa (>5 % of total abundance per sample). In addition to pairwise comparison, Venn diagrams were constructed for graphical descriptions of unshared and shared OTUs between two, three, four, and five samples. All statistical analyses were performed with R 3.0.1 [38] using the packages vegan and stats and custom R scripts.
Results
Water and Sediment Characteristics
The main physicochemical characteristics of the water collected from the different sites are listed in ESM 1: Table S1. Reigous creek was characterized by a low pH (around 3) that remained relatively constant along the creek. Redox potential (Eh) showed an increasing trend with values ranging from 476 to 715 mV. Extremely high concentrations of As (222 mg L−1), iron (1904 mg L−1), and sulfate (2226 mg L−1) were found at the almost anoxic upstream site S1. However, natural remediation occurred along the 1500 m of the creek characterized by a clear decreasing gradient of arsenic (222 to 0.32 mg L−1), iron (1904 to 23 mg L−1), and sulfate (2226 to 853 mg L−1) associated with a substantial increase in the concentration of dissolved oxygen (0.5 to 9.4 mg L−1). The reduced forms Fe(II) and As(III) were dominant at all sites except at the downstream site CONF, which was the most oxygenated and had the lowest concentrations of iron, sulfate and arsenic.
Unlike the clear pollution gradient evidenced in waters, metals and metalloid concentrations in the sediments were unevenly distributed along the Reigous (Table 1). The concentration of As was particularly high in samples GAL, S1 and GALm (191, 197, and 230 mg g−1, respectively) and lower at COWG (23 mg g−1) and CONF (21 mg g−1). No clear trend was observed along the Reigous for Fe and S, whose concentrations remained stable at all the sampling sites. Pb concentrations ranged between 1.43 and 3.98 mg g−1 with the highest concentration measured at COWG. The concentration of Zn was one order of magnitude higher at CONF than in the other sediments (Table 1).
The nature and structure of the sediment samples were also investigated using mineralogical and spectroscopic methods. XRD analyses (ESM 2: Figure S1) showed that the mineralogical composition evolved from mainly tooeleite (Fe6(AsO3)4SO4(OH)4 · 4H2O) upstream (sample S1: spring), to amorphous ferric arsenate hydroxysulfate (samples GAL and GALm), and to schwertmannite downstream (Fe8O8(OH)6SO4) (sample CONF). Sample COWG was sandy with quartz and micas. XANES analyses for COWG, GAL, GALm and CONF at the arsenic K-edge, showed that despite the presence of an As(III) component accounting for 12–34 ± 2 % of total arsenic, the oxidized arsenic form As(V) predominated in these samples (ESM 3: Figure S2). In contrast, sample S1 was characterized by a larger proportion of As(III) reaching 61 ± 2 % of total arsenic, which was expected since this sample essentially consisted of tooeleite, a rare iron-arsenite mineral found in large amounts in the Carnoulès sediments [2]. EXAFS data confirmed that As(III) dominated over As(V) in sample S1, with the presence of tooeleite and As(III)-sorbed schwertmannite (ESM 4: Figure S3). Downstream in Reigous Creek, As(V) was mainly present as amorphous Fe(III)–As(V) hydroxysulfate phase, As(III) likely being sorbed to poorly ordered schwertmannite. These findings are in good agreement with previously observed As speciation in Reigous Creek in winter [2].
Eukaryotic Community Composition
A total of 16,510 sequence reads were generated in a single run of 454 pyrosequencing from the five independent 18S rRNA libraries. After trimming and processing with Mothur, 9046 reads remained including 14.4 % classified as Metazoa and Embryophyta (Table 2). As this study focuses on microorganisms, Metazoa and Embryophyta sequences were removed from the dataset. However, they accounted for highly variable percentages of eukaryotic sequences at the different sites (50.6, 3.8, 25.9, 19.0, and 4.5 %, respectively, for S1, COWG, GALm, GAL, and CONF) leading to a significant decrease in the number of sequences in some samples. To compare the five samples, we randomly resampled each dataset to make the number of sequences the same for each sample (500 sequences). Clustering of the 2500 remaining sequences led to the identification of 323 OTUs (including 179 singletons) defined at 97 % identity.
The relative abundance of each taxon in each sample was determined at a confidence threshold of 80 % (Table 3). Across all the samples, eight high-level eukaryotic lineages were identified including a total of 37 classes. The majority of the sequences clustered into four high-level taxonomic groups: Fungi (76.6 %), Stramenopiles (8.4 %), Alveolata (8.2 %), and Viridiplantae (3.6 %). Four minor taxa represented less than 1 % of the sequences (Amoebozoa, Choanoflagellida, Rhizaria, and Apusozoa). Fungi formed the dominant group. They were highly diverse and distributed among 17 classes mainly associated with two dominant phyla, Basidiomycota and Ascomycota (40.8 % and 33.7 % of the total sequences) (Table 3). These phyla were dominated by Microbotryomycetes and Leotiomycetes, respectively (Fig. 2a). The Stramenopiles were related to six taxonomic groups characterized by the dominance (91 %) of the photosynthetic lineages Chrysophyceae-Synurophyceae (Fig. 2b). Alveolata were composed of four distinct groups (Apicomplexa, Ciliophora, Dinoflagellida and Perkinsea) (Fig. 2c). Ciliophora was dominant (87.4 %) and mainly characterized by Spirotrichea and Colpodea. Most of the Viridiplantae (84.4 %) were affiliated with the Chlorophyta phylum, mainly represented by Trebouxiophyceae. The Streptophyta phylum was composed of Klebsormidiophyceae and mainly poorly characterized sequences (Fig. 2d).
Spatial Variation Along the Reigous Creek
Diversity and species richness of the eukaryotic community varied considerably in the streambed sediments (Table 2). The Shannon diversity (H) and Chao1 richness indices ranged from 1.86 to 4.68 and 41 to 457, respectively. While the arsenic-rich sediments S1, GALm, and GAL displayed similar levels of species richness and diversity, COWG located only 30 m downstream from Reigous spring, exhibited the highest values (Chao1 = 457; H = 4.68). This is consistent with the rarefaction curves that tended to reach an asymptote for all the samples except COWG (Fig. 3). The CONF site represented an intermediate situation (Chao1 = 118; H = 3.28). Good’s coverage ranged from 73 to 98 % with an average value of 92 %, suggesting that the majority of eukaryotic phylotypes were recovered in the majority of samples (Table 2).
To investigate the spatial distribution of the eukaryotic community along the Reigous Creek, we compared eukaryotic communities based on community membership. A Venn diagram showing the unique and the shared OTUs at the five sites revealed that a high proportion of OTUs in each sediment sample was site-specific (57, 85, 54, 42, and 77 %, respectively, for S1, COWG, GALm, GAL and CONF) (Table 2; Fig. 4). Only one OTU corresponding to 54 sequences was common to the five sediments and showed 99 % of similarity with Escovopsis sp. Esc19. The samples GALm and GAL, retrieved from distinct sediments located at the same site in the creek, shared only 13 OTUs, corresponding to 42 and 33 % of their respective taxa. More generally, the proportion of shared OTUs between at least two sites was very low (12 %) but corresponded to a high proportion of the total sequences (68 %) (Fig. 4). To analyze these shifts in community composition along the pollution gradient in more detail, we examined the dynamics of the four dominant taxa. The distribution of Fungi varied along the creek with the dominance of Basidiomycota in the As-rich sediments S1, GAL and GALm, whereas a major proportion of Ascomycota was found at both COWG and CONF (Fig. 2a; Table 3). Within the Stramenopiles, the Clade C of the Chrysophyceae-Synurophyceae was dominant at all sites in varying proportions along the creek (Fig. 2b). Alveolata were characterized by the dominance of Ciliophora at all sites, except at GAL and GALm (Fig. 2c). These two sites were characterized by significantly lower abundances (<2 %) of Alveolata than the other sites. Among Viridiplantae, Chlorophyta prevailed at all sites, again except at GAL and GALm where no Viridiplantae were retrieved (Fig. 2d; Table 3).
Analyses at the genus level also highlighted variations in community composition (Table 4). Rhodotorula were particularly abundant at the As rich sediment site S1, GAL and GALm representing respectively around 47, 63, and 47 % of the sequences. While at the upstream site S1, Rhodotorula, Pezizomycotina, Arthrinium, unc. Oxytrichidae, and Escovopsis were the main genera detected, the downstream site CONF was mainly characterized by distinct genera (Calyptrozyma, Monascostroma, Ochromonas, Microscypha, Calyptrozyma, and Escovopsis).
Environmental Factors as Drivers of Eukaryotic Community Structure
To disentangle the environmental factors shaping the eukaryotic community, a principal coordinate analysis (PCoA) based on normalized abundance of OTUs was performed to explore the relationships between the microbial diversity patterns and the sediment physicochemical characteristics. PCoA ordination demonstrated that the eukaryotic community composition varied significantly along Reigous Creek (Fig. 5). The first ordination axis accounted for 39 % of the community variability, separated the samples into two clusters opposing the most diverse communities (COWG and CONF) and the least diverse communities (S1, GALm and GAL). COWG and CONF were characterized by higher concentrations of Zn and Pb, whereas the others were particularly rich in As. The second ordination axis separated the samples into three clusters (COWG, S1 + GALm + GAL, and CONF) according to Fe and S concentrations and accounted for an additional 28 % of community variability. Vector fitting of possible explanatory environmental parameters revealed that the concentration of As was the main factor shaping the eukaryotic communities along Reigous Creek (r 2 = 0.99, p = 0.041) (Table 1; Fig. 5). Thus, the eukaryotic community structure at COWG and CONF was mainly correlated with low concentrations of As, whereas high concentrations explained the composition of the community at S1, GAL, and GALm.
Principal coordinate analysis (PCoA) of the eukaryotic community matrix for normalized OTU abundances. Arrows are projections of possible explanation variables gained by vector fitting. The red and blue arrows represent significant and non-significant fittings, respectively (Bonferroni corrected, P < 0.05). The corresponding r 2 correlation coefficients and p values are listed in Table 1
To go further in the identification of physicochemical parameters potentially controlling the abundance of eukaryotic taxa, we also performed Pearson correlation tests on dominant OTUs (>5 % of total abundance per sample) as displayed in Table 4. Seven of the 13 dominant OTUs exhibited significant correlations with environmental variables. The relative abundances of four OTUs (3, 7, 8, and 15, respectively Calyptrozyma, Monascostroma, Ochromonas, and Microscypha) were positively correlated with the concentration of Zn. Moreover, As concentrations were significantly correlated with Rhodotorula mucilaginosa (OTU 1, positive correlation) and Monascostroma sphagnophilum (OTU 7, negative correlation). Unclassified Oxytrichidae (OTU 12) were positively correlated with the concentration of S and Penidiella columbiana (OTU 14) was positively correlated with the concentration of Pb.
Discussion
Diversity Patterns and Methodological Considerations
In agreement with a previous study conducted at Richmond Mine [39], our results suggest that despite the extreme characteristics of these systems, diverse organisms can grow in AMD although they are dominated by a few taxa.
Most of the sequences retrieved correspond to lineages previously found in other AMDs, including members of the Chlorophyta, Streptophyta, and Bacillariophyta phyla as well as Rhizaria, Alveolata, Amoebozoa, Stramenopiles and Fungi [8, 11, 40]. However, some taxa detected in the present study have not been previously identified in any AMD system, such as Choanoflagellida. Fungi were the dominant group in Carnoulès sediments and exhibited high diversity with 17 different classes identified. This phylogenetic group was previously evidenced in this ecosystem using culture-based approaches that led to the detection of 16 yeast species, mainly belonging to the phylum Basidiomycota [22]. Moreover, the eukaryotic community appears to be more diverse than previously described in other AMD ecosystems [9, 15, 41]. However, substantial eukaryotic diversity was encountered in the Río Tinto River in Spain where the eukaryotic community was mainly composed of photosynthetic Algae, using cloning sequencing techniques [6, 42, 43] or pyrosequencing [40].
In the present study, Euglena mutabilis was not detected. This photosynthetic acidophilic protist isolated from the Carnoulès ecosystem, forming continuous, green, macroscopic biofilms, has been shown to be particularly abundant at the COWG sampling site [23, 24]. We probably only captured a portion of the entire eukaryotic diversity present in the AMD, because a single PCR primer set does not amplify the 18S rRNA from all Eukaryotes [44, 45]. Furthermore, different targeted SSU rRNA gene sequences might not be PCR-amplified with equal efficiency by the different primers, and variability within the amplified regions may have a significant effect on the apparent species richness [46]. In addition, richness results should be interpreted with caution, given the technological limits (e.g., differential cell lysis, pyrosequencing errors, etc. [35]) and the unknown variability of the multiple copies of rRNA operons from the same organism [27].
Ecological Role of Microbial Eukaryotes
The eukaryotic populations found in Reigous Creek can be grouped according to their potential ecological role as primary producers, decomposers, and consumers inferred from the metabolic capabilities of related microorganisms.
Previous studies showed that archaea and bacteria in Carnoulès AMD sediment mainly derive energy from sulfur/iron oxidation or reduction [5, 21]. The chemolithoautrotrophic prokaryotes together with the photosynthetic eukaryotes evidenced in this study act as primary producers of biomass in this ecosystem. The photosynthetic eukaryotes detected in Carnoulès mainly belonged to the Chlorophyta (green algae), Streptophyta and autotrophic classes of the Stramenopiles such as the Bacillariophyceae (diatoms), Chrysophyceae (golden algae), Synurophyceae or Euglena. Photoautrotrophic eukaryotes may sustain the development of diverse heterotrophic lineages such as Acidiphilium detected in this ecosystem. Halter et al. [24] showed that the acidophilic protist Euglena mutabilis isolated at the water/sediment interface of Carnoulès AMD promotes the growth of heterotrophic bacteria by providing them with organic substrates like amino acids, polyamine compounds, urea or some sugars. Moreover, as suggested by Gonzalez-Toril et al. [47], the aerobic oxidation of iron, sulfur, or arsenic by prokaryotes (Acidithiobacillus ferrooxidans, Thiomonas spp., Thermoplasmatales) could be enhanced by the oxygen produced by photosynthesis, thereby highlighting an indirect role for eukaryotes in the bio-attenuation processes that occur at Carnoulès.
Our study also revealed the predominance and diversity of Fungi in all the sampling sites. Fungi are decomposers and the main producers of organic substances. The concentration of easily degradable organic carbon in Carnoulès sediment is probably very low (since in acid water, the degradation of organic carbon, such as leaves, is limited by the absence of invertebrates [48]). Thus, Fungi may play an important role as primary degraders of complex organic matter [49]. Moreover, by maintaining low organic carbon levels and by producing dissolved carbonate ions, they could promote the growth of chemolithoautotrophic acidophilic prokaryotes [12]. They may also allow the development of less metal-tolerant species by removing toxic metals from their environment through several mechanisms such as biosorption, sequestration or extracellular precipitation [43, 49, 50]. Indeed, the most abundant OTU, affiliated with Rhodotorula mucilaginosa, retrieved from S1, GAL and GALm may accumulate Zn and Pb and produce siderophores involved in metal resistance and complexation [51–53]. Members of Rhodotorula genus are also known to enhance ferrous iron oxidation carried out by Acidithiobacillus ferrooxidans (formely Thiobacillus ferrooxidans), which are active in this ecosystem [54]. They also can convert arsenate to arsenite, dimethylarsinic acid and methylarsonic acid [55]. Furthermore, the filamentous fungi Eurotiomycetes and Dothideomycetes detected in the present study may improve the anchorage of biofilms to pyrite-rich sediment in AMD by increasing their rigidity and organization [12].
The Carnoulès microbial consumer community consists of heterotrophic protozoa characterized by representatives of the Rhizaria, Alveolata, Amebozoa, Apusozoa, and Choanoflagellida. Protozoa such as Oxytrichidae sp. present in relatively high proportions in S1 can impact microbial populations and biomass by grazing on heterotrophic and acidophilic bacteria including iron/sulfur-oxidizers and archaea [41, 56]. Top-down control of the bacterial community by the mixotrophic flagellate Ochromonas sp., commonly encountered in AMD [8, 47] and, in this study, was also evidenced by Schmidtke et al. [57]. Moreover, acidophilic protozoa have been reported to graze preferentially on Acidithiobacillus ferrooxidans over Leptospirillum ferrooxidans, resulting in the dominance of L. ferrooxidans in mixed cultures of iron-oxidizers and revealing the impact of protozoa on prokaryotic community structure [56].
Our results confirm that diverse eukaryotes colonize extreme environments like AMD, where they probably play a substantial role particularly in carbon and metal cycling through direct or indirect mechanisms, making them good candidates for remediation strategies [9, 22, 49].
Spatial Distribution of the Eukaryotic Community and Influence of Environmental Drivers
The sediment samples were retrieved from different locations along the AMD reflecting distinct environmental conditions. The water chemistry varied significantly, with a clear decreasing gradient of As, Fe and sulfates associated with an increasing trend of oxygen content and pH, between the spring (S1, the most extreme site) and the confluence with the Amous River (CONF, the least extreme site). The composition of the sediments also differed according to the mineralogical analyses, including the two samples collected in the same area (GALm and GAL). However, no upstream to downstream gradient in sediment composition was identified.
These variations in geochemical and mineralogical conditions along Reigous Creek were associated with important variations in the structure and composition of the eukaryotic community. Only one OTU corresponding to 54 sequences was common to the five sediments, and a small proportion of OTUs was shared between at least two sites. The marked differences in the sediment eukaryotic community along Reigous Creek show that distinct communities occupy specific niches. However, the shared OTUs correspond to a high proportion of the total sequences. The distribution of the eukaryotic community is thus characterized by a low proportion of dominant OTUs omnipresent along the creek and a high proportion of minor taxa specific to each site. Moreover, the samples GALm and GAL, retrieved from distinct sediments geographically close in the creek, shared only 13 OTUs, highlighting the probable selective pressure of local sediment characteristics on eukaryotic membership patterns.
Contrary to what was observed in the Río Tinto by Amaral-Zettler et al. [7], eukaryotic richness was not highest at the most extreme sites of the AMD. Nevertheless, diversity was linked to the composition of the sediment. Indeed, the sediments sampled at the most diverse site (COWG) had the lowest Fe and As concentrations while the least diverse site (GAL) had the highest concentration of As. The eukaryotic diversity encountered at COWG could be indicative of mature and well-established communities able to sustain relatively complex trophic interactions. Sediments present along Reigous Creek are flushed away during the intensive rainfall events that characterize the Mediterranean climate of this region, generally in spring and fall. However, the COWG sampling site is located in a small depression and is protected by a low wall, allowing the sediments to remain in place over the years, and could thus sustain the development of more complex communities.
Based on PCoA analysis, the Reigous riverbed sediments formed a succession of distinct ecosystems hosting contrasted eukaryotic communities whose structure was linked to sediment composition. The concentration of arsenic in the sediment was identified as the main factor controlling the composition of the eukaryotic community; a high level of As being associated with low eukaryotic diversity. In addition to arsenic, Pearson’s correlation revealed that S, Zn, and Pb were also significantly correlated with the composition of the eukaryotic community. In accordance with our results, the presence of other heavy metals (Zn, Cu, Ni) has been previously suggested to shape eukaryotic community diversity and biomass in the Río Tinto more than the influence of pH [10]. A recent study suggested that seasonality, associating light intensity and water temperature, significantly influences eukaryotic distribution in AMD biofilms [15].
Distinct mineralogical conditions were evidenced in this study. They impacted the microbial assemblages in Carnoulès sediments and may have selected specific communities adapted to the local environmental properties. These selected communities probably influence biogeochemical processes and ecosystem functioning. As an illustration, the most abundant OTU retrieved from the As-rich sediments S1, GAL and GALm belonged to the genus Rhodotorula, which is known to be involved in arsenic cycling [55]. This genus was not detected at other sites characterized by a lower concentration of As, revealing a possible selective effect of arsenic.
The present study showed that eukaryotic community structure and composition both responds to, and impacts, ecosystem characteristics. To gain insight into the specific contribution of Eukaryotes in Carnoulès ecosystem functioning, further characterizations of the species, their mode of nutrition (autotrophy, heterotrophy, mixotrophy), their interactions (food web, saprotrophism, parasitism, commensalism, etc.) are now required. The capacity of eukaryotic microorganisms to interact with arsenic thereby impacting the fate of arsenic in the aquatic ecosystem also requires future investigation. The potential for arsenic methylation was recently evidenced in Reigous Creek and the River Amous [58, 59]. Since Eukaryotes (Fungi, Algae, etc.) are known for their biomethylation capacity [59, 60], targeting this microbial function to assess its contribution to arsenic cycling in AMD impacted water will be of particular interest.
References
Motsi T, Rowson NA, Simmons MJH (2009) Adsorption of heavy metals from acid mine drainage by natural zeolite. Int J Miner Process 92(1-2):42–48
Morin G, Juillot F, Casiot C et al (2003) Bacterial formation of tooeleite and mixed arsenic(III) or arsenic(V)-iron(III) gels in the Carnoulès acid mine drainage, France. A XANES, XRD, and SEM study. Environ Sci Technol 37(9):1705–1712
Johnson DB, Hallberg KB (2003) The microbiology of acidic mine waters. Res Microbiol 154(7):466–473
Johnson DB (2012) Geomicrobiology of extremely acidic subsurface environments. FEMS Microbiol Ecol 81(1):2–12
Volant A, Desoeuvre A, Casiot C et al (2012) Archaeal diversity: temporal variation in the arsenic-rich creek sediments of Carnoulès Mine, France. Extremophiles 16(4):645–657
Aguilera A (2013) Eukaryotic organisms in extreme acidic environments, the Río Tinto case. Life 3(3):363–374
Amaral-Zettler LA, Zettler ER, Theroux SM, Palacios C, Aguilera A, Amils R (2011) Microbial community structure across the tree of life in the extreme Río Tinto. ISME J 5(1):42–50
Amils R, González-Toril E, Fernández-Remolar D et al (2007) Extreme environments as Mars terrestrial analogs: the Río Tinto case. Planet Space Sci 55(3):370–381
Baker BJ, Tyson GW, Goosherst L, Banfield JF (2009) Insights into the diversity of eukaryotes in acid mine drainage biofilm communities. Appl Environ Microbiol 75(7):2192–2199
Aguilera A, Manrubia SC, Gómez F, Rodríguez N, Amils R (2006) Eukaryotic community distribution and its relationship to water physicochemical parameters in an extreme acidic environment, Río Tinto (Southwestern Spain). Appl Environ Microbiol 72(8):5325–5330. doi:10.1128/aem.00513-06
Aguilera A, Zettler E, Gómez F, Amaral-Zettler L, Rodríguez N, Amils R (2007) Distribution and seasonal variability in the benthic eukaryotic community of Río Tinto (SW, Spain), an acidic, high metal extreme environment. Syst Appl Microbiol 30(7):531–546
Baker BJ, Lutz MA, Dawson SC, Bond PL, Banfield JF (2004) Metabolically active eukaryotic communities in extremely acidic mine drainage. Appl Environ Microbiol 70(10):6264–6271
McGinness S, Johnson DB (1992) Grazing of acidophilic bacteria by a flagellated protozoan. Microb Ecol 23(1):75–86
Qin J, Lehr CR, Yuan C, Le XC, McDermott TR, Rosen BP (2009) Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci U S A 106(13):5213–5217
Brake SS, Arango I, Hasiotis ST, Burch KR (2014) Spatial and temporal distribution and characteristics of eukaryote-dominated microbial biofilms in an acid mine drainage environment: implications for development of iron-rich stromatolites. Environ Earth Sci 72(8):2779–2796
Casiot C, Morin G, Juillot F et al (2003) Bacterial immobilization and oxidation of arsenic in acid mine drainage (Carnoulès creek, France). Water Res 37(12):2929–2936
Egal M, Casiot C, Morin G, Elbaz P, Françoise, Cordier MA, Bruneel O (2010) An updated insight into the natural attenuation of As concentrations in Reigous Creek (southern France). Anglais 25(12):1949–1957
Bertin PN, Heinrich-Salmeron A, Pelletier E et al (2011) Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta- and proteo-genomics. ISME J 5(11):1735–1747
Bruneel O, Duran R, Casiot C, Elbaz-Poulichet F, Personné JC (2006) Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoulès, France. Appl Environ Microbiol 72(1):551–556. doi:10.1128/aem.72.1.551-556.2006
Bruneel O, Pascault N, Egal M et al (2008) Archaeal diversity in a Fe-As rich acid mine drainage at Carnoulès (France). Extremophiles 12(4):563–571
Bruneel O, Volant A, Gallien S et al (2011) Characterization of the active bacterial community involved in natural attenuation processes in arsenic-rich Creek sediments. Microb Ecol 61(4):793–810
Delavat F, Lett M-C, Lièvremont D (2013) Yeast and bacterial diversity along a transect in an acidic, As-Fe rich environment revealed by cultural approaches. Sci Total Environ 463–464:823–828
Casiot C, Bruneel O, Personne JC, Leblanc M, Elbaz-Poulichet F (2004) Arsenic oxidation and bioaccumulation by the acidophilic protozoan, Euglena mutabilis, in acid mine drainage (Carnoulès, France). Sci Total Environ 320(2-3):259–267
Halter D, Goulhen-Chollet F, Gallien S et al (2012) In situ proteo-metabolomics reveals metabolite secretion by the acid mine drainage bio-indicator, Euglena mutabilis. ISME J 6(7):1391–1402
Brake SS, Dannelly HK, Connors KA (2001) Controls on the nature and distribution of an alga in coal mine-waste environments and its potential impact on water quality. Environ Geol 40(4):458–469
López-García P, Philippe H, Gail F, Moreira D (2003) Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc Natl Acad Sci U S A 100(2):697–702
Monchy S, Sanciu G, Jobard M et al (2011) Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environ Microbiol 13(6):1433–1453
Hohmann C, Morin G, Ona-Nguema G, Guigner J-M, Brown GE Jr, Kappler A (2011) Molecular-level modes of As binding to Fe(III) (oxyhydr)oxides precipitated by the anaerobic nitrate-reducing Fe(II)-oxidizing Acidovorax sp. strain BoFeN1. Geochim Cosmochim Acta 75(17):4699–4712
Ona-Nguema G, Morin G, Juillot F, Calas G, Brown GE Jr (2005) EXAFS analysis of arsenite adsorption onto two-line ferrihydrite, hematite, goethite, and lepidocrocite. Environ Sci Technol 39(23):9147–9155
Maillot F, Morin G, Juillot F et al (2013) Structure and reactivity of As(III)- and As(V)-rich schwertmannites and amorphous ferric arsenate sulfate from the Carnoulès acid mine drainage, France: comparison with biotic and abiotic model compounds and implications for As remediation. Geochim Cosmochim Acta 104:310–329
Díez B, Pedrós-Alió C, Marsh TL, Massana R (2001) Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl Environ Microbiol 67(7):2942–2951
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541. doi:10.1128/aem.01541-09
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48(3):443–453
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12(7):1889–1898
Behnke A, Engel M, Christen R, Nebel M, Klein RR, Stoeck T (2011) Depicting more accurate pictures of protistan community complexity using pyrosequencing of hypervariable SSU rRNA gene regions. Environ Microbiol 13(2):340–349
Guillou L, Bachar D, Audic S et al (2013) The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res 41(D1):D597–D604
R Development Core Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org. edn., Vienna, Austria
Aliaga Goltsman DS, Comolli LR, Thomas BC, Banfield JF (2015) Community transcriptomics reveals unexpected high microbial diversity in acidophilic biofilm communities. ISME J 9(4):1014–1023
Amaral-Zettler LA (2013) Eukaryotic diversity at pH extremes. Front Microbiol 3:441
Zirnstein I, Arnold T, Krawczyk-Barsch E, Jenk U, Bernhard G, Roske I (2012) Eukaryotic life in biofilms formed in a uranium mine. Microbiologyopen 1(2):83–94
Amaral-Zettler LA, Gomez F, Zettler E, Keenan BG, Amils R, Sogin ML (2002) Microbiology: eukaryotic diversity in Spain’s river of fire. Nature 417(6885):137–137
Amaral-Zettler LA, Messerli MA, Laatsch AD, Smith PJ, Sogin ML (2003) From genes to genomes: beyond biodiversity in Spain’s Río Tinto. Biol Bull 204(2):205–209
Caron DA, Countway PD, Brown MV (2004) The growing contributions of molecular biology and immunology to protistan ecology: molecular signatures as ecological tools. J Eukaryot Microbiol 51(1):38–48
Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS (2006) A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157(1):31–43
Casamayor EO, Massana R, Benlloch S et al (2002) Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol 4(6):338–348
González-Toril E, Aguilera A, Souza-Egipsy V, Lopez Pamo E, Sanchez Espana J, Amils R (2011) Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain). Appl Environ Microbiol 77(8):2685–2694
Siefert J, Mutz M (2001) Processing of leaf litter in acid waters of the post-mining landscape in Lusatia, Germany. Ecol Eng 17(2-3):297–306
Das BK, Roy A, Koschorreck M, Mandal SM, Wendt-Potthoff K, Bhattacharya J (2009) Occurrence and role of algae and fungi in acid mine drainage environment with special reference to metals and sulfate immobilization. Water Res 43(4):883–894
Zafar S, Aqil F, Ahmad I (2007) Metal tolerance and biosorption potential of filamentous fungi isolated from metal contaminated agricultural soil. Bioresour Technol 98(13):2557–2561
Ji LY, Zhang WW, Yu D, Cao YR, Xu H (2012) Effect of heavy metal-solubilizing microorganisms on zinc and cadmium extractions from heavy metal contaminated soil with Tricholoma lobynsis. World J Microbiol Biotechnol 28(1):293–301
Purchase D, Scholes LN, Revitt DM, Shutes RB (2009) Effects of temperature on metal tolerance and the accumulation of Zn and Pb by metal-tolerant fungi isolated from urban runoff treatment wetlands. J Appl Microbiol 106(4):1163–1174
Rajpert L, Sklodowska A, Matlakowska R (2013) Biotransformation of copper from Kupferschiefer black shale (Fore-Sudetic Monocline, Poland) by yeast Rhodotorula mucilaginosa LM9. Chemosphere 91(9):1257–1265
Fournier D, Lemieux R, Couillard D (1998) Essential interactions between Thiobacillus ferrooxidans and heterotrophic microorganisms during a wastewater sludge bioleaching process. Environ Pollut 101(2):303–309
Vidal FV, Vidal VMV (1980) Arsenic metabolism in marine bacteria and yeast. Mar Biol 60(1):1–7. doi:10.1007/bf00395600
Johnson DB, Rang L (1993) Effects of acidophilic protozoa on populations of metal-mobilizing bacteria during the leaching of pyritic coal. J Gen Microbiol 139(7):1417–1423. doi:10.1099/00221287-139-7-1417
Schmidtke A, Bell EM, Weithoff G (2006) Potential grazing impact of the mixotrophic flagellate Ochromonas sp. (Chrysophyceae) on bacteria in an extremely acidic lake. J Plankton Res 28(11):991–1001. doi:10.1093/plankt/fbl034
Héry M, Casiot C, Resongles E et al (2014) Release of arsenite, arsenate and methyl-arsenic species from streambed sediment affected by acid mine drainage: a microcosm study. Environ Chem 11(5):514–524. doi:10.1071/EN13225
Desoeuvre A, Casiot C, Hery M (2015) Diversity and distribution of arsenic-related genes along a pollution gradient in a river affected by acid mine drainage. Microb Ecol 71(3):672–685
Bentley R, Chasteen TG (2002) Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol Mol Biol Rev 66(2):250–271. doi:10.1128/mmbr.66.2.250-271.2002
Egal M, Casiot C, Morin G et al (2009) Kinetic control on the formation of tooeleite, schwertmannite and jarosite by Acidithiobacillus ferrooxidans strains in an As(III)-rich acid mine water. Chem Geol 265(3-4):432–441
Bruneel O, Personne JC, Casiot C et al (2003) Mediation of arsenic oxidation by Thiomonas sp. in acid-mine drainage (Carnoulès, France). J Appl Microbiol 95(3):492–499
Acknowledgments
The study was financed by the “Observatoire de Recherche Méditerranéen en Environnement” (OSU-OREME). Aurélie Volant was supported by a grant from the French Ministry of Education and Research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1: Table S1
Physical-ochemical characteristics of the water body (mg L−1) measured directly above the sediment at the sampling sites of Reigous Creek. (DOCX 14 kb)
ESM 2: Figure S1
X-ray powder diffraction patterns of selected sediment samples collected along Reigous Creek (To: tooeleite; Schw: schwertmannite; Qz: quartz; Mi: micas). (TIF 382 kb)
ESM 3: Figure S2
XANES spectra of selected sediment samples. Arsenic oxidation states were determined using the linear combination fit of As(III) and As(V) ferric hydroxides spectra. Precision is ±2 %. (PDF 726 kb)
ESM 4: Figure S3
Extended X-ray absorption fine structure (EXAFS) data at the As K-edge of selected sediment samples. Experimental spectra were interpreted using linear composition fitting (LCF) using 3 model compound spectra: biogenic amorphous ferric arsenate hydroxysulfate from Thiomonas sp. strain B2 (Tm As(V)-am); arsenic(V) sorbed onto synthetic schwertmannite (As(V)/schw); and arsenic(III) sorbed onto biogenic schwertmannite from Acidithiobacillus ferrooxidans strain CC1(Af As(III)/schw). See [2, 30, 61] for details on these biogenic and abiotic mineral model compounds. As(III) proportions are underestimated with respect to XANES fit results because of the lower sensitivity of EXAFS to redox composition. (PDF 826 kb)
Rights and permissions
About this article
Cite this article
Volant, A., Héry, M., Desoeuvre, A. et al. Spatial Distribution of Eukaryotic Communities Using High-Throughput Sequencing Along a Pollution Gradient in the Arsenic-Rich Creek Sediments of Carnoulès Mine, France. Microb Ecol 72, 608–620 (2016). https://doi.org/10.1007/s00248-016-0826-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0826-5