Abstract
This study surveyed physicochemical properties and bacterial community structure of water and sediments from an acid mine drainage (AMD) dam in South Africa. High-throughput sequence analysis revealed low diversity bacterial communities affiliated within 8 dominant phyla; Acidobacteria, Actinobacteria, Chloroflexi, Firmicutes, Nitrospirae, Proteobacteria, Saccharibacteria, and ca. TM6_(Dependentiae). Acidiphilium spp. which are common AMD inhabitants but rarely occur as dominant taxa, were the most abundant in both AMD water and sediments. Other groups making up the community are less common AMD inhabitants; Acidibacillus, Acidibacter, Acidobacterium, Acidothermus, Legionella, Metallibacterium, Mycobacterium, as well as elusive taxa (Saccharibacteria, ca. TM6_(Dependentiae) and ca. JG37-AG-4). Although most of the taxa are shared between sediment and water communities, alpha diversity indices indicate a higher species richness in the sediments. From canonical correspondence analysis, DOC, Mn, Cu, Cr, Al, Fe, Ca were identified as important determinants of community structure in water, compared to DOC, Ca, Cu, Fe, Zn, Mg, K, Mn, Al, sulfates, and nitrates in sediments. Predictive functional profiling recovered genes associated with bacterial growth and those related to survival and adaptation to the harsh environmental conditions. Overall, the study reports on a distinct AMD bacterial community and highlights sediments as microhabitats with higher species richness than water.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Acid mine drainage (AMD) sites are globally distributed highly acidic extreme environments associated with mining activities (Akcil and Koldas 2006). AMD is generated primarily through microbially catalyzed oxidation of metal sulfides, such as iron sulfides, in the presence of air and water (Kuyucak 2002). Although the process involves several steps, it is often modelled through the oxidation of pyrite (FeS2), as given by Eq. 1:
Overall, the process generates high amounts of sulfuric acid which reduces the solution pH and promotes the dissolution of metals (Kuyucak 2002). Typically, AMD is characterized by excessive amounts of total dissolved solids, pronounced mineral acidity, and low pH (Akcil and Koldas 2006). These physical and chemical characteristics represent an extreme environment which maybe intolerable to most life forms. Nonetheless, it has been established that AMD is populated by distinct microorganisms (mostly prokaryotes) endowed with novel cellular mechanisms adaptive of the harsh conditions (Johnson and Hallberg 2003). AMD environments have been a subject of intensive microbial ecology studies (Chen et al. 2016). Owing to the low diversity of AMD microbial communities and geochemical simplicity, AMD environments are perceived as ideal model systems for discerning the microbial ecology, processes, and interaction networks in natural microbial assemblages (Baker and Banfield 2003; Denef et al. 2010; Kuang et al. 2016). In light of the role of some AMD inhabitants in its natural attenuation, these microbes are perceived to be perfect candidates for limiting AMD production as well as promoting the bioremediation of existing sites (Johnson and Hallberg 2005). However, a deeper understanding of the role of individual AMD microbes in driving nutrient cycles is necessary for successful biotechnological exploitation of AMD microbes. This can be achieved in part through describing the diversity and structure of AMD microbial communities as well as their variation in space at time and across environmental gradients (Hallberg 2010; Volant et al. 2014).
In the last decade, molecular techniques using Next-Generation Sequencing (NGS) platforms have taken center stage in microbial ecology (Shendure and Ji 2008). These are high-throughput techniques that allow a robust sampling of microbial diversity, enabling a comprehensive scrutiny of broad trends of microbial distribution (Liang et al. 2017). Accordingly, the applications of NGS platforms have greatly improved our perspectives on the structure and diversity of environmental microbial communities (Caporaso et al. 2011). Particularly, high-throughput targeted sequencing of phylogenetic marker genes such 16S rRNA has emerged as an efficient, fast, and low-cost technique, for probing microbial diversity (Logares et al. 2012). In this present work, the diversity and taxonomic composition of planktonic and benthic bacterial communities from an AMD containment dam were characterized using amplicon gene sequencing targeting the 16S rRNA gene.
Materials and methods
Sample collection
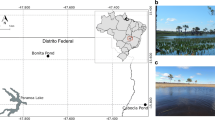
Water and sediment samples were collected from the Lancaster Dam (S 26° 07 39.9′, E 27° 46 46.2′) located in Randfontein in the Westrand of the Gauteng Province of the Republic of South Africa (RSA). The Randfontein area is well known for its long history of mining activities. The Lancaster Dam is a natural dam that was initially a source of pristine water. However, over the years, it has been infiltrated by acidic mine water emanating from nearby mine tailings and mine wastes generated by several gold mines in the area. To date, the dam is filled with acidic water.
Sampling in the Lancaster Dam was done over a period of 5 months between July and November 2017. Three sample collection points were identified around the dam based on ease of accessibility. From each sampling site, 5 L of water was collected into sterile polypropylene bottles and later pooled into one composite sample representative of each sampling occasion from which DNA was extracted. An additional 1 L of water was collected from each site for chemical analysis. At each sampling site sediments were collected from a 10 cm depth into 15 mL centrifuge tubes. These were later pooled and homogenized to a composite sample and used for DNA extraction. Sediments for geochemical analysis were collected into separate centrifuge tubes. All samples were kept on ice (4 °C) and transported to the laboratory for further processing within 48 h.
Analysis of water physicochemical parameters
Physicochemical parameters, including pH, temperature (T), total dissolved solids (TDS), salinity (SAL), and electrical conductivity (EC), were measured in situ using a multi-probe field meter (YSI ™ 6 series, Sonde Marion, Germany). In the laboratory, water samples for chemical analysis were prefiltered through Porafil 0.45 µm, 47 mm cellulose acetate membrane filters under negative pressure. Filtered samples for metal analysis were acidified to pH 2 using 70% nitric acid and stored at 4 °C until analysis. Elemental analysis was performed in an Inductively Coupled Optical Emission Spectrometer (Agilent Technologies 700 series ICP-OES). Anions (sulfates, nitrates) and chemical oxygen demand (COD) concentrations were measured spectrophotometrically from non-acidified pre filtered water using a Spectroquant Pharo 300 (Merck, RSA) as per the manufacturer’s instructions. Dissolve organic carbon (DOC) was measured in a torch combustion carbon analyzer (Teledyne Tekmar, USA).
Geochemical analysis of sediments
Sediment samples were dried in a benchtop vacuum freeze-dryer (Labconco, USA). Dried samples were homogenized by grinding in a pestle and mortar before passing through a 200-mesh sieve. Prior to metal analysis powdered sediment samples were digested using a microwave (SINEO MDS-6G, SepSci, RSA) following a protocol provided by the manufacturer. Samples were weighed (0.5 g), placed in microwave vessels and mixed with 9 mL nitric acid and 3 mL hydrochloric acid (37%). Digestion was performed at 175 °C for 60 min at 6 Watts power. After digestion, samples were allowed to cool and centrifuged at 5000×g for 10 min. The supernatant was collected and quantitatively made up to 50 mL with de ionized water and the total metals were quantified from this solution as described for water samples. For physicochemical analysis, 2 g of dry sediment was suspended in 10 mL deionized water and shaken at 150 rpm for 4 h. The suspension was allowed to settle overnight at room temperature. The liquid part was carefully decanted, and used to measure pH, sulfates, nitrates, and DOC as described for water samples (Sun et al. 2016).
Genomic DNA extraction, amplification, and sequencing
Pooled water samples for DNA extraction were pre filtered through Macherey–Nagel 1.2 µm, 47 mm glass fiber membrane filters to remove suspended particles. Microbial biomass was harvested through filtration in Pall Supor 0.22 µm, 47 mm polyethersulfone membrane filters under vacuum. The membranes were stored in sterile Petri dishes and kept at − 20 °C until DNA extraction. The filters were used as templates for DNA extraction using a D’Neasy PowerWater kit (Qiagen, Germany) following the manufacturer’s instructions. DNA from sediments was extracted from 9 g of pooled sediment sample using the D’Neasy PowerMax Soil kit (Qiagen, Germany) following the manufacturer’s instructions. Extracted DNA was quantified on a Qubit 3.0 Fluorometer (Life Technologies, RSA) and the purity was determined by measuring the A260/280 and A260/230 ratios in a Biodrop µLite spectrophotometer (Biochrom, USA). Polymerase chain reactions (PCR) were performed in triplicate to amplify the bacterial 16S rRNA gene using the universal primer pair 27F (5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG AGA GTT TGA TCM TGG CTC AG-3′) and 518R (5′- GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GAT TAC CGC GGC TGC TGG–3′) with adapters. This pair of primers amplifies the bacterial V1–V3 region of the 16S rRNA gene sequence and produces 500 bp amplicons (Caporaso et al. 2011). PCR reactions were performed with a 50 µL reaction mixture containing 25 µL Qiagen Top Taq Master Mix (2.5 units Taq DNA polymerase, dNTPs (200 µM each), 1.5 Mm MgCl2), 1 µL each of forward primer and reverse primer, metagenomic DNA template (50–100 ng/µL), and sterile nuclease-free water added to make up the final reaction volume of 50 µL. PCR reactions were performed in a BioRad T100 thermal cycler using the following cycling conditions: initial denaturation step at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min 30 s, with a final extension at 72 °C for 10 min. To monitor contamination, a template-free control was included in every reaction. The PCR products were loaded onto a horizontal gel of 1% agarose in 1× TAE buffer stained with ethidium bromide. Gels were run at 80 V for 90 min in 1× TAE buffer. After electrophoresis, gels were viewed under a Biorad UV illuminator. Band size and authenticity was confirmed by comparison to a CSL-MDNA-100BP DNA ladder. Authentic PCR products were pooled together for each respective sample. The PCR products were sent for sequencing at the Agricultural Research Council Biotech Platform (Pretoria, South Africa). Sequencing was done on the Illumina Miseq using the paired-end sequencing chemistry.
Sequence data analysis
Prior to analysis, the raw sequence data sets in .fastq format were trimmed to remove the Illumina tags, PCR artefacts, and poor quality reads. Following the initial quality filtering, all data sets were uploaded onto the Mothur pipeline v.1.40.0 for further analysis (Schloss et al. 2009). During the course of analysis, sequence reads with < 50 nucleotides, > 2% of ambiguities or 7% of homopolymers and those with mitochondrial or chloroplast origins were discarded. Similarly, chimeric sequences were removed using default parameters in UCHIME (Edgar et al. 2011). Non-chimeric sequence reads (490 nt) were subsequently aligned against the SILVA 16S rRNA gene reference database (version 128) at 80% confidence threshold (Quast et al. 2013). The aligned data sets were clustered into Operational Taxonomic Units (OTUs) at 97% sequence similarity using a pairwise distance matrix. Dominant OTUs were further subjected to a BLAST search to compare their identities with sequences in the NCBI-BLAST tool (Johnson et al. 2008). Alpha diversity among the 7 sequence data sets was inferred from the number of observed OTUs per sample, as well as alpha non-parametric indices including Chao_1, Dominance, Shannon_H, Simpson_D, and Species evenness (Zakrzewski et al. 2017). All non-parametric diversity indices were calculated at a genetic distance of 0.03. The relative abundance of individual taxa was expressed as a percentage of the number of sequences affiliated with the specific taxon against the total number of sequences obtained for that sample. In addition, similarities and differences in sediment and water bacterial communities were inferred from principal coordinate analysis (PCoA) of weighted UniFrac distances obtained through QIIME (Lozupone and Knight 2005). A canonical correspondence analysis (CCA) was also performed in the Paleontological Statistics Software (PAST v 2.17) to identify possible correlations between bacterial community structure and diversity with physicochemical parameters (Hammer et al. 2001). CCA was done on abundant bacterial genera and selected environmental parameters (pH, DOC, sulfates, nitrates, and metal ions).
Predictive functional profiling
The functional profiles of the bacterial communities were predicted from the 16S rRNA gene sequence data set using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) (Langille et al. 2013). The OTU relative abundance data were mapped against genes available in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The resultant data were then normalized by dividing each OTU by the known 16S copy-number abundance. The normalized data were used as input for the metagenome predictions and subsequent collapse into functional pathways (Langille et al. 2013).
Nucleotide Sequence Accession numbers
Sequences generated in this study have been deposited with the NCBI under project number PRJNA550971.
Results
Physicochemical profiles of water and sediment samples
The water samples collected from Lancaster Dam are characterized by remarkably low pH (2.33–2.93), elevated electrical conductivity (5743–6400 µS cm−1), chemical oxygen demand (142.74–167.50 mg L−1), and sulfate concentrations (571–1573 mg L−1). The sediment samples were also acidic, although slightly less acidic than the water samples (pH 3.07–3.35). These also depicted high levels of sulfates with concentrations ranging between 1267 and 2775 mg kg−1. Other physicochemical parameters of water and sediment samples are presented in Tables 1 and 2, respectively. Dissolved metals in water samples were quantified in the ICP-OES and the concentrations are presented in Fig. 1. Aluminium was by far the most prevalent dissolved metal quantified in the water samples with concentrations above 500 mg L−1. This was followed by iron, magnesium, sodium, and manganese all with concentrations above 100 mg L−1. Other metal ions detected in substantial amounts are zinc, calcium, potassium, and copper. Chromium and silver were measured in trace amounts (≤ 1 mg L−1). In the sediment samples, iron was detected as the most abundant metal with concentrations ranging between 35,619 and 65,314 mg kg−1. Aluminium, calcium, magnesium, potassium, and manganese were also detected in substantial amounts (above 500 mg kg−1), whilst zinc, chromium, copper, and silver were detected in trace amounts.
Bacterial community diversity
After quality filtering and trimming, a total of 66,914 sequence reads were obtained from the 7 sequencing libraries. A higher number of sequences were generated from water samples (7212–15,396 reads) compared to sediment samples (6612–10,851 reads). Rarefaction curves (Fig. S1) drawn from the raw sequence data indicate that the samples were sequenced at different depths, but at a genetic distance of 0.03, bacterial diversity was adequately covered for all of them. The sequence read tags were assigned to different OTUs at a 97% similarity threshold and a total of 1836 OTUs were recovered from the 7 sequence libraries. Despite the high number of sequence reads observed in water samples, they recorded the lowest number of OTUs (201–286) compared to (218–356) from sediment samples. This points towards a higher species richness in the sediment compared to the water bacterial community. The observed trend is further corroborated by the non-parametric alpha diversity indices (Dominance_D, Chao_1, Shannon_H, Simpson_D, and Species evenness) calculated (Table 3). These all indicate a significant shift (p < 0.05) in the species richness and evenness between water and sediment bacterial communities. For sediment samples, the Chao-1 index values ranged between 225.1 and 365.1 compared to 215.8 and 291.2 in water samples. Similarly, the Dominance values were almost close to zero (0.028–0.043) in sediment samples indicating a high degree of species evenness in the community. For water samples on the other hand, Dominance values are significantly higher (0.347–0.431) indicative of the predominance of a few OTUs. In accordance, the Shannon, Simpson, and Species evenness indices were higher in sediment than water samples.
Compositional similarities and dissimilarities between the bacterial communities in the two Lancaster Dam microhabitats were further determined from a PCoA of weighted UniFrac distances constructed from OTU count data. The PCoA plot (Fig. 2) shows a clear segregation of the water and sediment bacterial communities. Bacterial communities from water samples collected at different sampling dates clustered along the second axis, whilst bacterial communities from the sediment samples are distributed over the first and second quadrants of the plot in line with the heterogeneity observed in their physicochemical profiles. Furthermore, an Unweighted Pair Group Method with Arithmetic mean (UPGMA) tree generated from weighted UniFrac distance matrix with 100% support at all nodes (Fig. S2) shows a similar pattern of clustering for the two microbial communities.
Phylogenetic structure of bacterial communities
Of the total sequence, reads recovered over 99% were classified under Kingdom bacteria and approximately 1% remained unclassified. The recovered sequences fall into 22 phyla, 45 classes, 73 orders, 122 families, and 132 genera. At phylum level, the overall sequence reads were classified into 8 dominant phyla (relative abundance ≥ 1% in at least 1 sequence library) namely; Acidobacteria, Actinobacteria, Chloroflexi, Firmicutes, Nitrospirae, Proteobacteria, Saccharibacteria, and ca. TM6_(Dependentiae). The list of recovered phyla is presented in Table S1. In both the water and sediment sequence libraries, Proteobacteria was the most abundant phylum with higher abundance in water (72.74–83.43%) compared to (30.31–52.16%) in sediments. In agreement with the trend noted with the non-parametric indices, there are clear differences in the diversity of water and sediment bacterial communities (Fig. 3). In the water samples, 4 additional phyla, Actinobacteria (4.0–11.74%), Firmicutes (5.7–8.9%), ca. TM6_(Dependentiae) (1.49–4.28%), and Saccharibacteria (0.1–2.02%) had relative abundances greater than 1%. On the contrary, in sediment bacterial communities, 6 additional phyla were dominant; Saccharibacteria (7.80–21.27%), Actinobacteria (14.20–17.57%), Acidobacteria (10.10–18.83%), Chloroflexi (2.57–8.67), Nitrospirae (1.56–7.62%), and Firmicutes (0.45–3.28%).
At class level, a majority of the bacterial community was distributed across 12 major classes: three classes of Proteobacteria (Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria), Acidimicrobiia, Acidobacteria, Actinobacteria, Bacilli, Clostridia, ca. JG37-AG-4, Nitrospira, Saccharibacteria, and ca. TM6_(Dependentiae). The relative abundance of each class is variable across sediment and water samples, as shown in Fig. S3. Both the water and sediment bacterial communities are largely composed of Alphaproteobacteria with a higher abundance in water (69.63–78.07%) than sediments (24.04–39.36%). Other notable differences between sediment and water samples are observed in the relative abundance of Bacilli, Clostridia, Betaproteobacteria, and ca. TM6_(Dependentiae) which are all highly enriched in water samples and almost non-existent in sediments. Furthermore, Acidimicrobiaa, Actinobacteria, Saccharibacteria, Acidobacteria, Nitrospira, Gammaproteobacteria, and ca. JG37-AG-4 are significantly enriched in sediment than in water bacterial communities.
More than 90% of the bacterial sequence reads were confidently classified to genus level and were distributed in about 15 major genera. A heatmap showing the variation in the abundance of the top genera amongst the 7 sequence libraries is shown in Fig. 4. The most prominent genera are; Acidiphilium, Acidobacterium, Acidothermus, Acidibacter, Bacillus, unclassified ca. JG37-AG-4, Metallibacterium, Mycobacterium, Legionella, Leptospirillum, Propionibacterium, Saccharibacteria, and unclassified ca. TM6_(Dependentiae). In both the water and sediment communities, Acidiphilium spp. is dominant with the relative abundance ranging between 68 and 76% in water and 7–19% in sediments. Again, there are remarkable dissimilarities in the water and sediment bacterial community structure. Water bacterial communities are particularly enriched with sequences related to Mycobacterium (2.44–5.24%), ca. TM6_(Dependentiae) (1.48–4.48%), Legionella (1.21–2.03%), Propionibacterium (1.26–1.23%), and Acidibacillus (1.28–1.35%) which all occur in lower proportions in the sediments. On the other hand, sediment bacterial communities depict high abundances of sequences related to Saccharibacteria (7.8–21.27%), Metallibacterium (2.07–11.06%), Acidobacterium (3.73–8.99%), ca. JG37-AG-4 (2.19–8.67%), Acidibacter (1.01–4.60%), Leptospirillum (1.56–7.62%), and Acidothermus (1.95–2.46%). In addition, a substantial amount of sequences with high similarity to unclassified Acidobacteriaceae (2.74–9.91%), Acetobacteraceae (2.02–2.86%), Acidimicrobiaceae (1.55–5.56%), as well as uncultured bacteria (1.45–7.98%) are notably enriched in the sediments.
Effects of physicochemical parameters on the bacterial community structure
Canonical correspondence analysis (CCA) was performed to ascertain possible correlations between physicochemical variables and the community structure as well as individual dominant taxa (genera). For the water bacterial communities’ CCA plot (Fig. 5a), the first axis explained 77.73% of the bacterial community–environment relationship. This axis showed a positive correlation with DOC, Mn, Mg, Cu, Cr, Na, Al, and Fe and negative correlations with K, Zn, Ag, Ca, pH, sulfates, and nitrates. The second axis explained 22.27% of the variability and correlated positively with K, Zn, Ag, Ca, pH, sulfates, and nitrates, and negatively with DOC, Mn, Mg, Cu, Cr, Na, Al, and Fe. In accordance with the PCoA analysis, the three water samples clustered together at the centroid of the triplot, implying that they are all equally affected by the environmental parameters. With reference to correlation networks, the abundant genera Acidiphilium, Legionella, and Acidibacter depict a co-occurrence, and all show a positive correlation with DOC, sulfates, Mn, Mg, Cu, Cr, Na, Fe, Al, and Ca. Mycobacterium and ca. TM6_(Dependentiae) as well co-occur and show positive correlations with nitrates, pH, Ag, Zn, and K. Notably, those taxa that occur in low abundance; Saccharibacteria, Leptospirillum, ca. JG37_AG_4, Metallibacterium, Acidobacterium, and Acidothermus are scattered on the margin of the plot showing no co-occurrence with other taxa or correlations with any of the physical parameters.
CCA biplot showing correlations between a water and b sediment bacterial community structure and physicochemical parameters. The black circles indicate the microbial communities. Red dots indicate individual genera. Green lines represent physicochemical parameters associated with bacterial community structures. The position of each genus in relation to that of a physicochemical parameter is a measure of the correlation between the two. Sac, Saccharibacteria; Lept, Leptospirillum; Acidib, Acidibacterium; Leg, Legionella; Acido, Acidithermus; Acidip, Acidiphilium; Myco, Mycobacterium; Meta, Metallibacterium; Acidib, Acidibacter
For the sediment bacterial communities’ CCA plot (Fig. 5b), the first axis explained 59.83% of observed dissimilarity depicting positive correlations with Al, Ca, Cu, Fe, Zn, Mg, Mn, DOC, sulfate, and nitrates. The second axis explained 22.08% of the variability and is positively correlated with Ca, Cu, Zn, Mg, K, Al, Cr, Na, DOC, sulfates, and nitrates. In the sediments, Ca appears to be the strongest determinant of microbial community structure. Nonetheless, the four sediment samples were grouped at the center of the biplot, implying that there is no dissimilarity on how each of them is shaped by the environmental parameters. The genus Acidimicrobium and ca. TM6_(Dependentiae) co-occur and are positively correlated with most of the environmental parameters: DOC, nitrates, sulfates, and the metals Ca, Cu, Zn, Fe, Mg, K, Mn, and Al. The second cluster of Acidothermus, Saccharibacteria, Mycobacterium, ca. JG37_AG_4, Leptospirillum, and Metallibacterium depicts a co-occurrence and positive correlations with pH, Ag, and Cr. Notably, the two genera Acidiphilium and Acidibacter show some co-occurrence and together with Legionella do not depict any clear relationships with any of the physicochemical parameters tested.
Predictive functional profiling of bacterial communities
The metabolic function of bacterial communities in water and sediments was predicted using PICRUSt and a heatmap showing the variation in the expression of the predominant functions was generated (Fig. 6). Across all 7 metagenomes, the recovered functions can be classified broadly into two groups: those that are associated with the normal general functioning of prokaryotic cells and those that are associated with mechanisms related to survival and resistance to the harsh environmental conditions in AMD. Putative functions associated with normal functioning include: general function predictive only, metabolism (amino acid, butanoate, pyruvate, pyrimidine, chlorophyll, purine, and methane), biosynthesis (chromosome, ribosomes, peptidases, RNA, and bacterial motility proteins), carbon fixation, and glycolysis.
Recovered functions that may be deemed crucial for survival in the harsh environmental conditions in AMD include DNA recombination, replication and repair, transporters, chaperons, and folding catalysts as well as the bacterial secretion system. Among the recovered functions, the top three most abundant are transporters, DNA recombination, replication, and repair as well as ABC transporters. All three functions are related to mechanisms of survival in the harsh conditions. Indeed, there is mounting evidence to the effect that AMD systems are populated by specialist microorganisms endowed with novel cellular mechanisms adaptive of the harsh conditions (Johnson and Hallberg 2003). Accordingly, there are reports on the overexpression of genes related to adaptation compared to those related to normal general metabolism in AMD systems (Ram et al. 2005).
Discussion
The physicochemical profile of water samples collected from Lancaster Dam is characteristic of AMD with high acidity, high electrical conductivity, chemical oxygen demand, and sulfate concentrations. Sediment samples as well showed high acidity and sulfate concentrations, suggesting that the prolonged exposure to AMD results in the deposition of large amounts of sulfur in sediments. Nonetheless, pH values in sediments were slightly higher than in the water. The direct effect of low pH in mine waters is the increase in the solubility and subsequently bioavailability of metals (Akcil and Koldas 2006). Indeed, metals species such as Al, Ag, Ca, Cd, Cr, Cu, Fe, K, Mg, Mn, and Zn were detected in both the water and sediments at levels comparable to those previously reported in other AMD systems (Kamika et al. 2016). In this particular site, the water and sediments showed different profiles of metal concentrations. For instance in water, aluminium was detected as the most abundant metal ion compared to iron in sediments. Furthermore, sodium was detected only in the water and not in sediment samples. Often times, sediments act as reservoirs for metal deposition, and may be characterized by a high metal content. However, transition of metals between the water column and sediments is inevitable with changes in several physicochemical variables such as pH, salinity, and redox potential (Balintova et al. 2012).
The low pH, coupled with high metal concentrations and limited availability of organic carbon in mine wastes, presents an extreme environment that is hostile for microbial growth (Dopson et al. 2003; Slonczewski et al. 2009). However, through culture based and culture independent studies, it has been established that AMD is populated by a specialist microbial community (primarily unicellular prokaryotes) despite the high content of pollutants (Johnson et al. 2005; Kuang et al. 2013; Falagán et al. 2014). In this study, the bacterial community structure of AMD was measured through high-throughput sequencing of 16S rRNA gene amplicons obtained from total genomic DNA extracted from water and sediments. The results are indicative of a low diversity sediment and water bacterial community which is common in these extreme environments. The recovered bacterial OTUs that are shared in the two microhabitats correspond to lineages previously found in other highly acidic and metal-rich environments (Kuang et al. 2013; Yang et al. 2014; Méndez-García et al. 2015; Gupta et al. 2017; Sajjad et al. 2018). These were affiliated within 8 major phyla; Acidobacteria, Actinobacteria, Chloroflexi, Firmicutes, Nitrospirae, Proteobacteria, Saccharibacteria, and ca. TM6_(Dependentiae). The phylum Proteobacteria was detected as the most abundant in both water and sediment bacterial communities. Members of this phylum are ubiquitous and have frequently been observed in extreme environments including those with extreme acidity such as mine water and tailings (Edwards et al. 2000; Bruneel et al. 2006; He et al. 2007; Kuang et al. 2013; Kamika and Momba 2014; Keshri et al. 2015; Kamika et al. 2016). Members of this phylum are endowed with physiological properties that enable them to adapt to extreme environmental conditions including low pH, high metal content, and low nutrient availability (Yergeau et al. 2012).
At genus level, the recovered taxon includes those that are renowned as AMD inhabitants and directly or indirectly linked to the carbon, iron, and sulfur cycles. Nonetheless, other genera that are rarely reported in AMD systems were also recovered suggestive of a tentatively distinctive or novel AMD bacterial community. Acidithiobacillus and Leptospirillum have been identified as key players in the generation of AMD (Chen et al. 2016) and they have been detected globally in AMD sites (Auld et al. 2013, 2017). However, in this study, Acidithiobacillus was not detected at all, whereas Leptospirillum was detected only in the sediments as one of the dominant genera. Both water and sediment bacterial communities showed a high prevalence of sequences related to Acidiphilium spp. Members of this genus are primarily heterotrophs that can also reduce iron (Baker and Banfield 2003). They are widely distributed in acidic metal-rich environments and are important in the maintenance of AMD by enhancing the growth of chemoautotrophs through their metabolism of low-molecular-weight organic compounds that are toxic to chemoautotrophs (Chen et al. 2016). Acidiphilium spp. show enhanced growth under conditions of elevated aluminium concentrations (Wakao et al. 2002). However, heterotrophic acidophiles are rarely reported as the dominant members in AMD bacterial communities (Johnson and Hallberg 2007). The occurrence of Acidiphilium as dominant members of a bacterial community was first reported in an AMD dam in China (Hao et al. 2010). The authors attributed the dominance of Acidiphilium and limited abundance of chemoautotrophs to low ferrous iron concentrations and high DOC levels. Under conditions of low ferrous iron concentration, the growth of iron-oxidizing chemolithotrophs may be restricted (González-Toril et al. 2003; Druschel et al. 2004; Rzhepishevska et al. 2005). Furthermore, surface AMD sites often receive excess DOC from soils and plant material washed off from the surroundings. This may promote the growth of heterotrophic bacteria. However, these may undergo photolysis or acid hydrolysis to form small molecular weight organic acids which are toxic to chemoautotrophs (Tittel et al. 2005). Although ferrous iron concentrations were not measured in this study, the paucity of chemoautotrophic acidophiles could be attributed to the high DOC concentrations as the Lancaster Dam does receive external carbon loads.
Other heterotrophic genera that were recovered include Acidibacter (Gammaproteobacteria), Acidobacterium (Acidobacteria), Metallibacterium (Gammaproteobacteria), Acidothermus (Actinobacteria) and the newly described genera Acidibacillus (Firmicutes). Acidibacter, and Acidobacterium spp. are well distributed in acidic environments as minor members of bacterial communities. The recovery of sequences with close similarity to Metallibacterium spp. is in line with previous studies in which members of this genus were detected as dominant members of the community in several acidic metal-rich environments (acid streamers, sediments, water, biofilms) (Kay et al. 2013, 2014; Brantner and Senko 2014; Brantner et al. 2014; González-Toril et al. 2014; Sun et al. 2015, 2016; Falteisek et al. 2016; Gupta et al. 2017). This genus is represented by a sole species, M. scheffleri first isolated from acidic biofilms and characterized as being able to oxidize Fe(II) as well as reduce Fe(III) (Ziegler et al. 2013). The enrichment of this genus in the sediments is not surprising as it has previously been reported that surface attached growth appears to select for the preferential growth of M. scheffleri and related organisms, which is reflected by their abundance in biofilms. The occurrence of Acidothermus in the sediments is puzzling. The only cultured organism from this genus A. cellulolyticus first isolated from Yellowstone acidic hot springs displayed a thermophilic growth, yet water temperatures at Lancaster Dam were around 20 °C throughout the sampling period (Barabote et al. 2009). This could suggest the existence of mesophilic members within this genus or the sequences may have arised from sequencing errors (Huse et al. 2010).
Although AMD microbes are known to remain recalcitrant to in vitro cultivation (Aguilera and Johnson 2016), techniques using high-throughput NGS platforms have the added advantage of being able to capture both the cultivable and non-cultivable members within a community (De Mandal et al. 2015). In this study, sequence reads affiliated with three uncultivated phyla often referred to as the “dark matter”; Saccharibacteria, ca. TM6_(Dependentiae), and ca. JG37-AG-4 were detected in high abundance. Sequence reads with high similarity to the phylum Saccharibacteria (previously candidate division TM7) were recovered in high abundance in sediment samples. The phylum is currently described from 16S rRNA gene sequences and genome data only. It represents a highly ubiquitous group of bacteria distributed in soils, seawater, sludge, animal and human sources, as well as clinical environments (Ferrari et al. 2014). Most importantly, some members of this phylum have been recovered in acidic metal-rich environments such as acidic soils (Ferrari et al. 2014; Winsley et al. 2014) acid water (García-Moyano et al. 2015) as well as oxic and suboxic biofilms (Méndez-García et al. 2014). Due to a lack of cultured isolates and the small number of 16S rRNA gene sequences in reference databases, the metabolic traits, general biology of Saccharibacteria, and hence functions in AMD remain elusive (Méndez-García et al. 2014; García-Moyano et al. 2015; Mesa et al. 2017). Moreover, sequences related to the uncultured phylum ca.TM6_(Dependentiae) were detected (more enriched in water samples than sediments). 16S rRNA sequence signatures from this phylum have been found in diverse habitats including sulfur hot springs, arsenic-rich sediments, and AMD-contaminated creek sediments (Youssef et al. 2012; Escudero et al. 2013; Sun et al. 2016). Initially, sequenced genomes of members within this phylum were suggestive of a facultative anaerobic metabolism characteristic of free living organisms (McLean et al. 2013). However, recently constructed genomes are characterized by a small genome size, AT bias, and lack of biosynthetic pathways, pointing towards a symbiotic lifestyle (Yeoh et al. 2016). Another yet-uncultured taxa that was identified to be dominant in the bacterial community is ca. JG37-AG-4 from Chloroflexi which has previously been detected in uranium waste piles and the Río Tinto sediments (Selenska-Pobell et al. 2001; García-Moyano et al. 2012).
The results in this study also portray quite a different microbial community compared to other AMD sites, as sequences related to Mycobacterium and Legionella spp. were recovered with their relative abundance greater than 1%. Both genera occur in higher abundance in the water. The occurrence of Mycobacterium has been reported in aquatic and soil environments often as minor members (Falkinham 2002). Reports of the occurrence of Mycobacterium spp. in acidic environments remain limited thus far. Their distribution has been reported in moderately acidic forest soils (pH 4) (Iivanainen et al. 1997) and recently in an endolithic bacterial community at Yellowstone Geothermal sites as the dominant taxa (Walker et al. 2005). Their role in acidic environments remains poorly understood, although they are postulated to be heterotrophs (Walker et al. 2005). Sequence reads classified as Legionella spp. were recovered from water samples albeit in low abundance (≤ 2%). Members of the genus Legionella are neutrophilic organisms responsible for the disease Legionnaires Pneumonia in humans. Although they have recently been detected in AMD systems, their occurrence still remains unusual (Hao et al. 2012; Auld et al. 2013, 2017). Previous workers have opined that Legionella occurs in AMD as endosymbionts of AMD amoeboid protists (Auld et al. 2017). The same can be assumed in this study, although the eukaryotic community was not profiled.
Variation in the bacterial community in water and sediment samples
At all taxonomic levels, the sediment and water-column bacterial communities shared most of the recovered OTUs. This implies that most of the taxa inhabiting the water column can also be tracked in the sediments. This observation is in agreement with a report in which intimate associations among bacterial populations residing in different microhabitats (water, sediments, and biofilms,) at Río Tinto were noted (García-Moyano et al. 2012; Falteisek et al. 2016). However, the relative abundance of the shared bacterial populations varies between the sediment and water communities. Sediment bacterial communities show a significantly higher species richness than water bacterial communities. Higher bacterial diversity in sediments might be attributed to the higher pH and hence less extreme environmental conditions (Sánchez-Andrea et al. 2011; García-Moyano et al. 2012; Méndez-García et al. 2014). A substantial proportion (up to 7%) of sequences from sediments were affiliated with yet uncultivated bacterial clades suggesting possible untapped bacterial diversity in the sediments. Studies on the characterization and profiling of microbial communities from sediments in AMD environments remain this far limited as compared to the water column and biofilms (Sánchez-Andrea et al. 2011; Sanz et al. 2011). However, it is clear that these microhabitats may serve as niches for enormous bacterial diversity and hence the need to be considered in trying to understand the integrated microbial ecology of these peculiar extreme systems.
Physicochemical parameters affect bacterial diversity
Free living organism often shows non-random distribution patterns across diverse habitats at various spatial and temporal scales (Liu et al. 2014). However, environmental parameters have also been shown to drive microbial community structure and diversity. Whereas, some studies have often identified individual parameters such as pH, TOC, or metal concentrations as key parameters structuring microbial diversity in AMD (Hao et al. 2010, 2017; Streten-Joyce et al. 2013; Yang et al. 2014; Sun et al. 2016; Auld et al. 2017) in this study, the tested physicochemical parameters showed an almost even effect on both the water and sediment bacterial communities. Notably, correlations of physicochemical parameters with individual genus were evident. In water bacterial communities, DOC, Mn, Cu, Cr, Al, Fe, and Ca appear to have correlations with most members of the community compared DOC, Ca, Cu, Fe, Zn, Mg, K, Mn, Al, sulfates, and nitrates in sediment bacterial communities. A clear demarcation between taxa that are positively correlated to low pH, and high metal concentrations (strict acidophiles) as well as those that prefer growth under moderate acidity and metal concentrations (moderate acidophiles) was evident. The different taxa within the sediment and water bacterial communities show some correlation networks that may be indicative of biotic or species–species interactions within the microbial community.
Predictive functional profile
Metagenomic sequencing of acidophilic microbial communities has revealed the predominance of gene clades involved in iron and sulfur oxidation, iron and sulfur reduction, carbon fixation, nitrogen metabolism, and resistance to the low pH and high metal concentrations (Huang et al. 2016). Similarly, in this study, PICRUSt analysis recovered functions related to mechanisms essential to the normal function in prokaryotic cells. Furthermore, functions associated with adaptation and survival in the harsh environment such as maintenance of a neutral intracellular pH, metal resistivity, and cellular protection were recovered in high abundance.
An abundance of putative functions associated with transport (general transport, ABC transporters, and ion-coupled transporters) was evident in both the sediment and water metagenomes. In acidophiles, the transport system has been identified as crucial for the maintenance of a neutral intracellular pH (Dopson et al. 2003). Particularly, transporters are used to pump out excess protons in the cytoplasm to maintain a neutral pH that is conducive for enzyme activity (Michels and Bakker 1985). Accordingly, genes encoding putative proton efflux systems have been recovered from the sequenced genomes of acidophiles such as Ferroplasma type II and Leptospirillum group II (Tyson et al. 2004). In addition, the transport system plays a role in pumping out excess metal ions within cells thus minimizing their toxicity (Dopson et al. 2014). For example, Acidithiobacillus ferroxidans survives high copper concentrations by forming copper-phosphate complexes intracellularly and pumping them out of the cell using the efflux pump systems (Alvarez and Jerez 2004). Furthermore, the water and sediment metagenomes depict an abundance of putative functions associated with bacterial secretion systems. The bacterial secretion system is perceived to be necessary for the secretion of protective molecules that enable bacteria to withstand the harsh conditions in AMD. For example, Leptospirillum group II UBA type strains secrete a novel lyso phosphatidylethanolamine (PE) lipid (Fischer et al. 2012). This compound has a high affinity for iron and calcium ions and apparently minimizes excessive cellular uptake of metal ions (Fischer et al. 2012). Similarly, taurine and hydroxyectoine detected through a metabolomics study of microbial biofilm communities from the Richmond Mine are linked to cellular osmotic stress protection (Mosier et al. 2013). Accordingly, the recovery of functions associated with bacterial secretion systems in the water and sediment metagenomes may be linked to survival strategies in the harsh environment. DNA repair recombination as well as chaperons and folding catalysts were other putative functions recovered in high abundance in both the sediments and water metagenomes. These as well are associated with mechanisms for cellular protection. Since the extreme conditions in AMD systems also cause a lot of damage to biomolecules, AMD microorganisms have evolved an efficient repair system and often depict an overexpression of genes involved in DNA repair and recombination as well as chaperons and folding catalysts for protein repair. Accordingly, genomes of several extreme acidophiles such as Picrophilus torridus show a high prevalence of DNA and protein repair genes (Crossman et al. 2004). Furthermore, chaperones involved in protein refolding were recovered in high prevalence in the metaproteome of Richmond Mine biofilms (Ram et al. 2005).
Other studies have reported on the recovery of functions with significant bioremediation and biotechnological applications such as dioxin, tolune, and polyaromatics degradation (Sibanda et al. 2019). However, such functions were not recovered in this study. This could either mean that these functions are lacking or they may have been missed, highlighting the inadequacy of PICRUSt in environmental studies. PICRUSt has been shown to be more effective in predicting the metagenomes of environments for which extensive gene collections are available in reference gene databases (Langille et al. 2013). For peculiar environments such as extreme environments, gene predictions may be sparse and less accurate (Mesa et al. 2017).
Conclusions
The present study reports on the physicochemical properties of two microhabitats within a highly acidic, metal-rich, and oligotrophic environment. Furthermore, the bacterial community structure was profiled using a high-throughput Illumina sequencing approach. The bacterial community structure in sediments and the water column are characterized by low diversity which is often encountered in extremely acidic environments. The phylum Proteobacteria occurred in high abundance in both the water and sediment bacterial communities. Although the bacterial communities were characterized by common AMD inhabitants such as Acidiphilium, other taxa that are less frequently encountered in AMD sites were detected. The estimated community diversity and species richness indicate that the sediments harbour higher bacterial diversity and species richness compared to the water column. Judging from the abundance of sequence reads associated with uncultured bacterial clades, this often overlooked microhabitat may be a source of novel yet untapped bacterial diversity. A suite of physicochemical parameters play a role in structuring the bacterial community and taxon co-occurrence networks in both microhabitats.
References
Johnson DB, Aguilera A (2016) The microbiology of extremely acidic environments. In: Yates M, Nakatsu C, Miller R, Pillai S (eds) Manual of environmental microbiology, 4th edn. ASM Press, Washington, DC, pp 4.3.1-1-4.3.1-24. https://doi.org/10.1128/9781555818821.ch4.3.1.
Akcil A, Koldas S (2006) Acid mine drainage (AMD): causes, treatment and case studies. J Clean Prod 14:1139–1145. https://doi.org/10.1016/j.jclepro.2004.09.006
Alvarez S, Jerez CA (2004) Copper ions stimulate polyphosphate degradation and phosphate efflux in Acidithiobacillus ferrooxidans. Appl Environ Microbiol 70:5177–5182. https://doi.org/10.1128/AEM.70.9.5177-5182.2004
Auld RR, Myre M, Mykytczuk NCS et al (2013) Characterization of the microbial acid mine drainage microbial community using culturing and direct sequencing techniques. J Microbiol Methods 93:108–115. https://doi.org/10.1016/j.mimet.2013.01.023
Auld R, Mykytczuk N, Leduc L, Merritt T (2017) Seasonal variation in an acid mine drainage microbial community. Can J Microbiol 63:137–152. https://doi.org/10.1139/cjm-2016-0215
Baker BJ, Banfield JF (2003) Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152. https://doi.org/10.1016/S0168-6496(03)00028-X
Balintova M, Petrilakova A, Singovszka E (2012) Study of metals distribution between water and sediment in the Smolnik Creek (Slovakia) contaminated by acid mine drainage. Chem Eng Trans 28:73–78. https://doi.org/10.3303/CET1228013
Barabote RD, Xie G, Leu DH et al (2009) Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus IIB provides insights into its ecophysiological and evolutionary adaptations. Genome Res 19:1033–1042. https://doi.org/10.1101/gr.084848.108
Brantner JS, Senko JM (2014) Response of soil-associated microbial communities to intrusion of coal mine-derived acid mine drainage. Environ Sci Technol 48:8556–8563. https://doi.org/10.1021/es502261u
Brantner JS, Haake ZJ, Burwick JE et al (2014) Depth-dependent geochemical and microbiological gradients in Fe(III) deposits resulting from coal mine-derived acid mine drainage. Front Microbiol 5:1–15. https://doi.org/10.3389/fmicb.2014.00215
Bruneel O, Duran R, Casiot C et al (2006) Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoulès, France. Appl Environ Microbiol 72:551–556. https://doi.org/10.1128/AEM.72.1.551-556.2006
Caporaso JG, Lauber CL, Walters WA et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 108:4516–4522. https://doi.org/10.1073/pnas.1000080107
Chen L, Huang L, Méndez-García C et al (2016) Microbial communities, processes and functions in acid mine drainage ecosystems. Curr Opin Biotechnol 38:150–158. https://doi.org/10.1016/j.copbio.2016.01.013
Crossman L, Holden M, Pain A, Parkhill J (2004) Genomes beyond compare. Nat Rev Microbiol 2:616–617. https://doi.org/10.1038/nrmicro961
De Mandal S, Panda AK, Bisht SS, Kumar NS (2015) Microbial ecology in the era of next generation sequencing. J Next Gener Seq Appl 01:1–6. https://doi.org/10.4172/2469-9853.S1-001
Denef VJ, Mueller RS, Banfield JF (2010) AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J 4:599–610. https://doi.org/10.1038/ismej.2009.158
Dopson M, Baker-Austin C, Koppineedi PR, Bond PL (2003) Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic micro-organisms. Microbiology 149:1959–1970. https://doi.org/10.1099/mic.0.26296-0
Dopson M, Ossandon FJ, Lövgren L, Holmes DS (2014) Metal resistance or tolerance? Acidophiles confront high metal loads via both abiotic and biotic mechanisms. Front Microbiol 5:10–13. https://doi.org/10.3389/fmicb.2014.00157
Druschel GK, Baker BJ, Gihring TM, Banfield JF (2004) Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem Trans 5:13–32. https://doi.org/10.1063/1.1769131
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Edwards KJ, Bond PL, Druschel GK et al (2000) Geochemical and biological aspects of sulfide mineral dissolution: lessons from Iron Mountain, California. Chem Geol 169:383–397. https://doi.org/10.1016/S0009-2541(00)00216-3
Escudero LV, Casamayor EO, Chong G et al (2013) Distribution of microbial arsenic reduction, oxidation and extrusion genes along a wide range of environmental arsenic concentrations. PLoS One 8:e78890. https://doi.org/10.1371/journal.pone.0078890
Falagán C, Sánchez-España J, Johnson DB (2014) New insights into the biogeochemistry of extremely acidic environments revealed by a combined cultivation-based and culture-independent study of two stratified pit lakes. FEMS Microbiol Ecol 87:231–243
Falkinham JO (2002) Nontuberculous Mycobacteria in the environment. Clin Chest Med 23(3):529–551. https://doi.org/10.1016/S0272-5231(02)00014-X
Falteisek L, Duchoslav V, Čepička I (2016) Substantial variability of multiple microbial communities collected at similar acidic mine water outlets. Microb Ecol 72:163–174. https://doi.org/10.1007/s00248-016-0760-6
Ferrari B, Winsley T, Ji M, Neilan B (2014) Insights into the distribution and abundance of the ubiquitous candidatus Saccharibacteria phylum following tag pyrosequencing. Sci Rep 4:1–9. https://doi.org/10.1038/srep03957
Fischer CR, Wilmes P, Bowen BP et al (2012) Deuterium-exchange metabolomics identifies N-methyl lyso phosphatidylethanolamines as abundant lipids in acidophilic mixed microbial communities. Metabolomics 8:566–578. https://doi.org/10.1007/s11306-011-0344-x
García-Moyano A, González-Toril E, Aguilera Á, Amils R (2012) Comparative microbial ecology study of the sediments and the water column of the Río Tinto, an extreme acidic environment. FEMS Microbiol Ecol 81:303–314. https://doi.org/10.1111/j.1574-6941.2012.01346.x
García-Moyano A, Austnes A, Lanzén A et al (2015) Novel and unexpected microbial diversity in acid mine drainage in Svalbard (78° N), revealed by culture-independent approaches. Microorganisms 3:667–694. https://doi.org/10.3390/microorganisms3040667
González-Toril E, Llobet-Brossa E, Casamayor EO et al (2003) Microbial ecology of an extreme acidic environment, the Tinto River. Appl Environ Microbiol 69:4853–4865. https://doi.org/10.1128/AEM.69.8.4853-4865.2003
González-Toril E, Santofimia E, López-Pamo E et al (2014) Comparative microbial ecology of the water column of an extreme acidic pit lake, Nuestra Señora del Carmen, and the Río Tinto basin (Iberian Pyrite Belt). Int Microbiol 17:225–233. https://doi.org/10.2436/20.1501.01.225
Gupta A, Dutta A, Sarkar J et al (2017) Metagenomic exploration of microbial community in mine tailings of Malanjkhand copper project, India. Genom Data 12:11–13. https://doi.org/10.1016/j.gdata.2017.02.004
Hallberg KB (2010) New perspectives in acid mine drainage microbiology. Hydrometallurgy 104:448–453. https://doi.org/10.1016/j.hydromet.2009.12.013
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package. Palaeontol Electron 4:1–9. https://doi.org/10.1016/j.bcp.2008.05.025
Hao C, Wang L, Gao Y et al (2010) Microbial diversity in acid mine drainage of Xiang Mountain sulfide mine, Anhui Province, China. Extremophiles 14:465–474. https://doi.org/10.1007/s00792-010-0324-5
Hao C, Zhang L, Wang L et al (2012) Microbial community composition in acid mine drainage lake of Xiang Mountain Sulfide Mine in Anhui Province, China. Geomicrobiol J 29:886–895. https://doi.org/10.1080/01490451.2011.635762
Hao C, Wei P, Pei L et al (2017) Significant seasonal variations of microbial community in an acid mine drainage lake in Anhui Province, China. Environ Pollut 223:507–516. https://doi.org/10.1016/j.envpol.2017.01.052
He Z, Xiao S, Xie X et al (2007) Molecular diversity of microbial community in acid mine drainages of Yunfu sulfide mine. Extremophiles 11:305–314. https://doi.org/10.1007/s00792-006-0044-z
Huang LN, Kuang JL, Shu WS (2016) Microbial ecology and evolution in the acid mine drainage model system. Trends Microbiol 24:581–593. https://doi.org/10.1016/j.tim.2016.03.004
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898. https://doi.org/10.1111/j.1462-2920.2010.02193.x
Iivanainen EK, Martikainen PJ, Räisänen ML, Katila ML (1997) Mycobacteria in boreal coniferous forest soils. FEMS Microbiol Ecol 23:325–332. https://doi.org/10.1016/S0168-6496(97)00040-8
Johnson DB, Hallberg KB (2003) The microbiology of acidic mine waters. Res Microbiol 154:466–473. https://doi.org/10.1016/S0923-2508(03)00114-1
Johnson DB, Hallberg KB (2005) Acid mine drainage remediation options: a review. Sci Total Environ 338:3–14. https://doi.org/10.1016/j.scitotenv.2004.09.002
Johnson DB, Hallberg KB (2007) Techniques for detecting and identifying acidophilic mineral-oxidizing microorganisms. Biomining. https://doi.org/10.1007/978-3-540-34911-2_12
Johnson DB, Okibe N, Hallberg KB (2005) Differentiation and identification of iron-oxidizing acidophilic bacteria using cultivation techniques and amplified ribosomal DNA restriction enzyme analysis. J Microbiol Methods 60:299–313. https://doi.org/10.1016/j.mimet.2004.10.002
Johnson M, Zaretskaya I, Raytselis Y et al (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:5–9. https://doi.org/10.1093/nar/gkn201
Kamika I, Momba MNB (2014) Microbial diversity of Emalahleni mine water in South Africa and tolerance ability of the predominant organism to vanadium and nickel. PLoS One 9:e86189. https://doi.org/10.1371/journal.pone.0086189
Kamika I, Azizi S, Tekere M (2016) Microbial profiling of South African acid mine water samples using next generation sequencing platform. Appl Microbiol Biotechnol 100:6069–6079. https://doi.org/10.1007/s00253-016-7428-5
Kay C, Rowe O, Rocchetti L et al (2013) Evolution of microbial “streamer” growths in an acidic, metal-contaminated stream draining an abandoned underground copper mine. Life 3:189–211. https://doi.org/10.3390/life3010189
Kay CM, Haanela A, Johnson DB (2014) Microorganisms in subterranean acidic waters within Europe’s deepest metal mine. Res Microbiol 165:705–712. https://doi.org/10.1016/j.resmic.2014.07.007
Keshri J, Mankazana BBJ, Momba MNB (2015) Profile of bacterial communities in South African mine-water samples using Illumina next-generation sequencing platform. Appl Microbiol Biotechnol 99:3233–3242. https://doi.org/10.1007/s00253-014-6213-6
Kuang JL, Huang LN, Chen LX et al (2013) Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J 7:1038–1050. https://doi.org/10.1038/ismej.2012.139
Kuang J, Huang L, He Z et al (2016) Predicting taxonomic and functional structure of microbial communities in acid mine drainage. ISME J 10:1527–1539. https://doi.org/10.1038/ismej.2015.201
Kuyucak N (2002) Role of microorganisms in mining: generation of acid rock drainage and its mitigation and treatment. Eur J Miner Process Environ Prot 2:179–196
Langille MGI, Zaneveld J, Caporaso JG et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. https://doi.org/10.1038/nbt.2676
Liang J-L, Li X-J, Shu H-Y et al (2017) Fine-scale spatial patterns in microbial community composition in an acid mine drainage. FEMS Microbiol Ecol 93:1–8. https://doi.org/10.1093/femsec/fix124
Liu J, Hua ZS, Chen LX et al (2014) Correlating microbial diversity patterns with geochemistry in an extreme and heterogeneous environment of mine tailings. Appl Environ Microbiol 80:3677–3686. https://doi.org/10.1128/AEM.00294-14
Logares R, Haverkamp THA, Kumar S et al (2012) Environmental microbiology through the lens of high-throughput DNA sequencing: synopsis of current platforms and bioinformatics approaches. J Microbiol Methods 91:106–113. https://doi.org/10.1016/j.mimet.2012.07.017
Lozupone CA, Knight R (2005) UniFrac: a new phylogenetic method for omparing microbial communities. Appl Environ Microbiol 71:8228–8235. https://doi.org/10.1128/AEM.71.12.8228
McLean JS, Lombardo M-J, Badger JH et al (2013) Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci 110:E2390–E2399. https://doi.org/10.1073/pnas.1219809110
Méndez-García C, Mesa V, Sprenger RR et al (2014) Microbial stratification in low pH oxic and suboxic macroscopic growths along an acid mine drainage. ISME J 8:1259–1274. https://doi.org/10.1038/ismej.2013.242
Méndez-García C, Peláez AI, Mesa V et al (2015) Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol 6:1–17. https://doi.org/10.3389/fmicb.2015.00475
Mesa V, Gallego JLR, González-Gil R et al (2017) Bacterial, archaeal, and eukaryotic diversity across distinct microhabitats in an acid mine drainage. Front Microbiol 8:1–17. https://doi.org/10.3389/fmicb.2017.01756
Michels M, Bakker EP (1985) Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J Bacteriol 161:231–237
Mosier AC, Justice NB, Bowen BP et al (2013) Metabolites associated with adaptation of microorganisms to an acidophilic, metal-rich environment identified by stable-isotope-enabled metabolomics. MBio 4:1–8. https://doi.org/10.1128/mBio.00484-12
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. https://doi.org/10.1093/nar/gks1219
Ram RJ, Verberkmoes NC, Thelen MP et al (2005) Community proteomics of a natural microbial biofilm. Science 308:1915–1920. https://doi.org/10.1126/science.1109070
Rzhepishevska OI, Lindström EB, Tuovinen OH, Dopson M (2005) Bioleaching of sulfidic tailing samples with a novel, vacuum-positive pressure driven bioreactor. Biotechnol Bioeng 92:559–567. https://doi.org/10.1002/bit.20609
Sajjad W, Zheng G, Zhang G et al (2018) Diversity of prokaryotic communities indigenous to acid mine drainage and related rocks from Baiyin Open-Pit Copper Mine Stope, China. Geomicrobiol J 35:580–600. https://doi.org/10.1080/01490451.2018.1430873
Sánchez-Andrea I, Rodríguez N, Amils R, Sanz JL (2011) Microbial diversity in anaerobic sediments at Río Tinto, a naturally acidic environment with a high heavy metal content. Appl Environ Microbiol 77:6085–6093. https://doi.org/10.1128/aem.00654-11
Sanz JL, Rodríguez N, Díaz EE, Amils R (2011) Methanogenesis in the sediments of Rio Tinto, an extreme acidic river. Environ Microbiol 13:2336–2341. https://doi.org/10.1111/j.1462-2920.2011.02504.x
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Selenska-Pobell S, Kampf G, Flemming K et al (2001) Bacterial diversity in soil samples from two uranium waste piles as determined by rep-APD, RISA and 16S rDNA retrieval. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol 79:149–161. https://doi.org/10.1023/A:1010237711077
Shendure J, Ji H (2008) Next-generation DNA sequencing. Nat Biotechnol 26:1135–1145. https://doi.org/10.1038/nbt1486
Sibanda T, Selvarajan R, Msagati T et al (2019) Defunct gold mine tailings are natural reservoir for unique bacterial communities revealed by high-throughput sequencing analysis. Sci Total Environ 650:2199–2209. https://doi.org/10.1016/j.scitotenv.2018.09.380
Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA (2009) Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol 55:1–317. https://doi.org/10.1016/S0065-2911(09)05501-5
Streten-Joyce C, Manning J, Gibb KS et al (2013) The chemical composition and bacteria communities in acid and metalliferous drainage from the wet-dry tropics are dependent on season. Sci Total Environ 443:65–79. https://doi.org/10.1016/j.scitotenv.2012.10.024
Sun W, Xiao T, Sun M et al (2015) Diversity of the sediment microbial community in the Aha watershed (Southwest China) in response to acid mine drainage pollution gradients. Appl Environ Microbiol 81:4874–4884. https://doi.org/10.1128/AEM.00935-15
Sun W, Xiao E, Krumins V et al (2016) Characterization of the microbial community composition and the distribution of Fe-metabolizing bacteria in a creek contaminated by acid mine drainage. Appl Microbiol Biotechnol 100:8523–8535. https://doi.org/10.1007/s00253-016-7653-y
Tittel J, Bissinger V, Gaedke U, Kamjunke N (2005) Inorganic carbon limitation and mixotrophic growth in Chlamydomonas from an acidic mining lake. Protist 156:63–75. https://doi.org/10.1016/j.protis.2004.09.001
Tyson GW, Chapman J, Hugenholtz P et al (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43. https://doi.org/10.1038/nature02340
Volant A, Bruneel O, Desoeuvre A et al (2014) Diversity and spatiotemporal dynamics of bacterial communities: physicochemical and other drivers along an acid mine drainage. FEMS Microbiol Ecol 90:247–263. https://doi.org/10.1111/1574-6941.12394
Wakao N, Yasuda T, Jojima Y et al (2002) Enhanced growth of Acidocella facilis and related acidophilic bacteria at high concentrations of aluminum. Microbes Environ 17:98–104. https://doi.org/10.1264/jsme2.2002.98
Walker JJ, Spear JR, Pace NR (2005) Geobiology of a microbiol endolithic community in the Yellowstone geothermal environment. Nature 434:1011–1014. https://doi.org/10.1038/nature03447
Winsley TJ, Snape I, McKinlay J et al (2014) The ecological controls on the prevalence of candidate division TM7 in polar regions. Front Microbiol 5:1–10. https://doi.org/10.3389/fmicb.2014.00345
Yang Y, Li Y, Sun QY (2014) Archaeal and bacterial communities in acid mine drainage from metal-rich abandoned tailing ponds, Tongling, China. Trans Nonferrous Met Soc China (English Ed) 24:3332–3342. https://doi.org/10.1016/s1003-6326(14)63474-9
Yeoh YK, Sekiguchi Y, Parks DH, Hugenholtz P (2016) Comparative genomics of candidate phylum TM6 suggests that parasitism is widespread and ancestral in this lineage. Mol Biol Evol 33:915–927. https://doi.org/10.1093/molbev/msv281
Yergeau E, Bokhorst S, Kang S et al (2012) Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J 6:692–702. https://doi.org/10.1038/ismej.2011.124
Youssef N, Steidley BL, Elshahed MS (2012) Novel high-rank phylogenetic lineages within a sulfur spring (Zodletone Spring, Oklahoma), revealed using a combined pyrosequencing-Sanger approach. Appl Environ Microbiol 78:2677–2688. https://doi.org/10.1128/aem.00002-12
Zakrzewski M, Proietti C, Ellis JJ et al (2017) Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics 33:782–783. https://doi.org/10.1093/bioinformatics/btw725
Ziegler S, Waidner B, Itoh T et al (2013) Metallibacterium scheffleri gen. nov., sp. nov., an alkalinizing gammaproteobacterium isolated from an acidic biofilm. Int J Syst Evol Microbiol 63:1499–1504. https://doi.org/10.1099/ijs.0.042986-0
Acknowledgements
The work was supported by the University of South Africa and the National Research Foundation of South Africa through a Grant (SFH170705248759), also authors would like to acknowledge Center for High Performance (CHPC), Pretoria for providing computational support for Metagenomic analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lukhele, T., Selvarajan, R., Nyoni, H. et al. Diversity and functional profile of bacterial communities at Lancaster acid mine drainage dam, South Africa as revealed by 16S rRNA gene high-throughput sequencing analysis. Extremophiles 23, 719–734 (2019). https://doi.org/10.1007/s00792-019-01130-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-019-01130-7