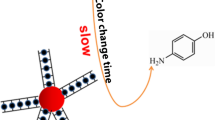
Abstract
It was found that dimension change of aptamer DNA significantly weakened the mimicking activity of gold nanozyme, which was contrary to previous research. Based on this, a rapid colorimetric method for the detection of low concentrations of mercury in environmental media was fabricated. It was observed that 40 nM Hg2+ causes color changes in solution. The detection limit of absorbance measurements was estimated to be 9.3 × 10–11 M. The assay was fast and could complete a single test in half an hour. The detection results for real environment samples confirmed the reliability of the colorimetric analysis in practical application. The proposed assay provides an alternative method for real-time monitoring of mercury in the environment. In particular, the charge effect on the affinity of nanozyme consummated the DNA regulation mechanism for the simulated enzyme activity.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Real-time and highly sensitive detection of low levels of mercury in environmental media is very important to prevent human health risks [1]. Various analytical methods, such as instrument-based analysis of chromatography [2], inductively coupled plasma-mass spectrometry (ICP-MS) [3, 4], atomic absorption spectroscopy (AAS) [5] and atomic fluorescence spectrometry (AFS) [6, 7], optical analysis of colorimetry [8,9,10,11,12,13,14,15,16], fluorescence [17,18,19], chemiluminescence [20] and surface-enhanced Raman spectroscopy (SERS) [21, 22], and electrochemical analysis [23,24,25] are used for the detection of mercury. Among them, colorimetric analysis is particularly suitable for monitoring of mercury in the environment due to outstanding simplicity.
At present, colorimetric analyses for mercury detection generally have two strategies. One is based on the change in color of gold nanoparticles (AuNPs) due to different dispersion states [8, 9, 12], and the other is based on the catalytic oxidation of enzyme substrate by nanozyme mimics [10, 11, 13,14,15,16]. Our group previously constructed the colorimetric analyses to detect mercury ions based on the change in the dispersed state of AuNPs induced by Hg (II) ions [9, 12]. Huang et al. developed the colorimetric assay for Hg (II) ions using DETL functionalized AuNPs based on AuNPs aggregation [8]. Many factors like pH value, ionic strength, and temperature can cause the dispersion state of AuNPs colloidal solution to change. The colorimetric analyses based on different dispersion states of AuNPs are susceptible. Nanozyme colorimetric analyses for mercury mainly rely on that mercury and other metals form amalgams to affect the catalytic activity of nanozyme. The colorimetric determination of Hg (II) was established by the stimulating oxidase-like ability of Ag3PO4 microcubes owing to the formation of Ag–Hg amalgam [16]. Lian et al. developed the colorimetric sensor for detection of mercuric ions by forming the CuO/Pt-Hg trimetallic amalgam [10]. Also, it was reported that the generation of the Pb-Cu/Hg-Cu alloy could induce the conversion of the catalase-like enzymatic activity of metallothionein-stabilized copper nanoclusters (MT-CuNCs) to peroxidase-like enzymatic activity, which was applied to establish the colorimetry for Pb2+/Hg2+ [11]. Since the metal that can form amalgams with mercury is not unique, the amalgam-based nanozyme colorimetric analyses for mercury have some interference. Aptamer recognition is a powerful means [20, 26]. However, only a few researches on nanozyme colorimetric analysis by aptamer recognition for mercury ions were reported. Wang’s group [15] and our group [13] constructed the colorimetric assays to detect mercury by using that mercury-induced conformational change of aptamer could restore the surface active sites of simulated enzyme and enhance the catalytic activity of nanozymes.
It is well known that AuNPs can easily interact with single-stranded DNA (ss-DNA), and DNAs with different conformations have different behaviors [27]. Previous research found that AuNPs had weak peroxidase mimics activity, and ss-DNA wrapped on AuNPs could significantly enhance peroxidase mimicking activity of AuNPs [28, 29]. Inspired by this, it is possible to construct a colorimetric assay to detect mercury by combining the different interactions of different DNAs and AuNPs with peroxidase mimicking activity of AuNPs. In this work, the catalytic oxidation of enzyme substrate 3,3,5,5-tetramethylbenzidine (TMB) by AuNPs before and after the interaction between mercury and aptamer was investigated. Interestingly, after the interaction of mercury with the aptamer, AuNPs catalyzed oxidation of TMB to produce very light blue product. The result was contrary to the previous colorimetric analysis based on aptamer with mercury enhancing the activity of nanozyme [13]. Further investigation showed that in addition to the surface active sites of the nanozyme, the affinity between nanozyme and substrate was also an important factor affecting the simulated enzyme activity. In this analysis, the change of the catalytic activity of the nanozyme was mainly due to the charge effect on the affinity between simulated enzyme and substrate. It was observed visually that 40 nM Hg could cause the solution to become pale in color. Using absorbance, the detection limit for mercury ions is 9.3 × 10–11, which is about one order of magnitude lower than previous aptamer-based nanozyme colorimetry [13, 15] and comparable to the colorimetry with AuNPs aggregation [9, 12]. The entire assay, including the preparation of AuNPs, can be completed in less than an hour. This analysis has prominent simplicity and can achieve rapid detection. In particular, both active site and affinity were important factors affecting the activity of the simulated enzyme, and there was a competitive effect between them. This can help to effectively regulate enzyme activity to construct sensing analysis.
Experimental section
Chemicals and instruments
Mercuric nitrate (Hg(NO3)2), chloroauric acid (HAuCl4·4H2O), 3,3,5,5-tetramethylbenzidine (TMB), and hydrogen peroxide (H2O2, 30%) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Mercury stock solutions were prepared by dissolving Hg(NO3)2 in dilute nitric acid, and the working mercury solution was obtained by serially diluting the stock solution with NaAc–HAc (0.10 M) buffered solution. The DNA aptamer for Hg2+ was chosen to be the oligonucleotides sequences of 5′-TTT TTT TTT T-3′ according to the reference [30]. The oligonucleotides sequences were purified and synthesized by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). The oligonucleotide stock solutions (100 μM) were obtained by dissolving with 20 mM PBS buffer (containing 10 mM KCl, 50 mM NaCl, pH = 7) and kept frozen until ready to use. The work oligonucleotide solutions were obtained by diluting stock solutions with the same PBS buffer. Before use, the oligonucleotide solutions were heated to 90 °C for 5 min, and then immersed in water (4 °C) immediately to unwind the single-stranded oligonucleotide [26, 31]. The nucleic acid base was exposed by this treatment, which facilitated the interaction of DNA aptamer with AuNPs or mercury. Other metal ion solutions were obtained by dissolving their respective nitrates with dilute nitric acid. All reagents and solvents in the experiments were purchased in their highest available purity and used without further purification. Ultrapure water (18 MΩ cm−1) was obtained from a Millipore Milli-Q water purification system (Billerica, MA) and used in all experiments.
A Hitachi U-3900H UV–Visible Spectrophotometer (Tokyo, Japan) was used to record UV–visible absorption spectra at room temperature in the wavelength range from 400 to 800 nm. All the photographs were obtained with Olympus C-370 digital camera. The transmission electron microscopy (TEM) image of AuNPs was obtained using a JEM-2100 TEM (Japan Electronics Co., Ltd) by placing a drop of colloidal solution sample on carbon-coated copper grid and being dried at room temperature. Zeta potentials were determined by using a Malvern Zetasizer 2000, and three rounds of operations were carried out to obtain the average data.
Rapid and convenient colorimetric assay for mercury using gold nanozyme
The nanozyme used in this analysis is AuNPs coated with citrate. AuNPs were prepared according to previous work [27, 32]. The detailed procedure is described in the Supporting Information. The prepared AuNPs were characterized by UV–Vis absorption spectroscopy and TEM image. The results (see Supporting Information, Fig. S1) showed that the prepared AuNPs had the obvious absorption peak at the wavelength of about 520 nm, which was the characteristic absorption of AuNPs [32]. The results of TEM (Insert in Fig. S1) showed that the AuNPs were spherical particles with the diameter of about 20 nm.
This nanozyme colorimetric assay was based on the fact that DNAs with different dimension structures could effectively regulate the mimicking peroxidase activity of AuNPs for the catalytic oxidation of enzyme substrates TMB. Typically, 100 μL AuNPs and 20 μL of 2.0-μM aptamers were incubated in PBS buffer (pH = 7.0) for 5 min to complete the coating of DNA on AuNPs. Then mercury ions solutions (50 μL) with different concentrations were added to AuNPs/DNA solutions to interact for 10 min for the affinity interaction of mercury and aptamer. Because mercury had strong affinity with the DNA aptamer and it was much stronger than DNA adsorption on the AuNPs, the DNAs coated on AuNPs were separated from the surface of AuNPs [9, 30]. After that, AuNPs/aptamer/Hg2+ solutions were added to the mixture of 15 μL of 5 mM TMB and 15 μL of 5 mM H2O2 containing 400 μL of 1 M acetate buffer (pH = 4.0) to react for 10 min at room temperature for catalytic oxidation of the enzyme substrate. Finally, the color change of the reaction solutions was observed visually by the naked eye. Or the resulting solutions were transferred into a slit quartz cuvette of 350-μL volume for scanning UV–Vis spectra. Standard curves were obtained using the absorbance values at 652 nm.
The genuine water sample needed to be filtered through the filtration membrane before testing. For the detection of genuine water sample, only the mercury ion solution in the above steps was replaced by the tested water sample after treatment, and other operations remained unchanged. The measured absorbance value at 652 nm was used to calculate the amount of mercury in the genuine sample by the standard curve.
Results and discussion
Colorimetric sensing mechanism for Hg2+
The colorimetric analysis is fabricated based on that different action of DNAs with different dimensions on AuNPs can change the surface condition of AuNPs and regulate the catalytic activity of gold nanozyme. The schematic diagram of sensing is shown in Scheme 1. AuNPs coated with citrate have weak peroxidase mimicking activity and catalyze the oxidation of enzyme substrates TMB to produce very pale blue product [28]. When there is no mercury ion in the system, the one-dimensional and linear ss-DNA is coated on the AuNPs. Since DNA is an anionic polymer with phosphoric acid skeleton, the coating of DNA on AuNPs will increase the electronegativity on the surface of AuNPs [27, 28]. TMB oxidation reaction is usually carried out in acidic conditions, under which TMB mainly exists in the form of cations [33, 34]. The gold nanozyme with increased surface electronegativity interacts closely with TMB cation, which enhances the affinity between the mimic enzyme and substrate. Thus, the DNA-coated AuNPs exhibit strong peroxidase mimic activity, catalyzing the oxidation of TMB to produce intense blue product. However, in the presence of mercury ions, the aptamer DNAs interact with mercury to form the rigid and three-dimensional spatial structure of T-Hg-T, which cannot be coated on the surface of AuNPs due to steric hindrance [20, 30]. The AuNPs remain in their original surface condition (no DNA coating) and catalyze the oxidation of TMB to produce very pale blue product. In this way, the interaction between mercury and aptamer induced the change of DNA dimension, which inhibited peroxidase mimetic activity of gold nanozyme.
In order to verify the above design, the oxidation reactions of enzyme substrate TMB with hydrogen peroxide under different cases were investigated. The results are shown in Fig. 1. In the absence of gold nanozyme, TMB hardly reacted with hydrogen peroxide and the solution was almost colorless, and the corresponding UV-Vis absorption spectrum had very weak absorption (Fig. 1a). AuNPs coated with citrate showed very weak peroxidase mimicking activity. AuNPs alone catalyzed the oxidation of TMB to produce very pale blue product (Fig. 1b). The results of UV-Vis absorption spectrum showed that the absorption at the wavelength of about 650 nm was weak (Fig. 1b). When AuNPs were coated with DNAs, the simulated enzyme activity of gold nanozyme was significantly enhanced, catalyzing the oxidation of TMB to produce intense blue product (Fig. 1e). The corresponding UV-Vis absorption spectrum showed that the absorption at the wavelength of 650 nm was also very strong (Fig. 1e). After the interaction of mercury with the aptamer DNA, the simulated enzyme activity of gold nanozyme was significantly weakened. The AuNPs-aptamer-Hg2+ catalyzed the oxidation of TMB to produce very pale blue product, and the corresponding UV-Vis absorption also obviously weakened (Fig. 1c). The UV-Vis absorption from AuNPs-aptamer-Hg2+ system (Fig. 1c) was slightly stronger than that of AuNPs alone (Fig. 1b), which could be due to the fact that there was still a small amount of DNAs wrapped on the AuNPs after the affinity interaction between mercury and the aptamer. In addition, it is considered that the formation of Au/Hg amalgam may affect the activity of gold nanozyme. The sensing response from the AuNPs-Hg2+ system was investigated. The results showed that the blue color of AuNPs-Hg2+ system solution (Fig. 1d) was slightly intenser than that of AuNPs alone, and the corresponding UV-Vis absorption was also slightly stronger. It agreed with the previous research [14]. These results further confirmed that mercury-induced dimension change of aptamer led to the weakening of gold nanozyme activity. Apparently, regulating the activity of gold nanozyme by DNA dimension can be used for visual or colorimetric detection of mercury ions.
UV–Visible absorption spectra for catalytic oxidation solution of peroxidase substrate TMB under different cases: a without AuNPs, b AuNPs, c AuNPs + aptamer + Hg2+, d AuNPs + Hg2+, and e AuNPs + aptamer. The insert shows the corresponding images. Experimental conditions: TMB, 5 mM; H2O2, 5 mM; AuNPs, 17 nM; aptamer, 2.0 μM; and Hg2+, 3.6 × 10–7 M
In order to ascertain the sensing mechanism for detecting mercury ions, the zeta potentials of AuNPs in different situations were measured. The results are shown in Table 1. The zeta potential of prepared AuNPs was measured to be − 39.27 ± 3.1 mV. The electronegativity on AuNPs increased obviously when DNAs were coated on AuNPs. The zeta potential of AuNPs/aptamer DNA was − 56.52 ± 2.9 mV, while after the interaction of mercury with the aptamer, the zeta potential of AuNPs+aptamer+Hg2+ decreased to − 42.54 ± 3.5 mV. The increase of electronegativity on AuNPs coated with ss-DNA supported the contact between AuNPs and TMB cation and enhanced the catalytic activity of gold nanozyme. However, after the interaction of mercury and aptamer DNA, ss-DNA left the surface of AuNPs to abate the electronegativity on AuNPs, which resulted in that the affinity of gold nanozyme and TMB was weakened and peroxidase mimicking activity of AuNPs was reduced. In order to further confirm the colorimetric sensing mechanism, the catalytic activity of AuNPs under different cases was studied by steady-state kinetics of enzymatic reaction. Typical Michaelis-Menten curves (Fig. 2a) were obtained respectively for prepared AuNPs, AuNPs/aptamer DNA, and AuNPs+aptamer+Hg2+ in the varied concentrations of TMB. The variation of absorbance value at 652 nm per unit time was calculated as the initial velocity. Lineweaver-Burk plots (1/V=(Km/Vmax) [S]+1/Vmax) (Fig. 2b) were obtained from the Michaelis-Menten equation (V=Vmax[S]/(Km+[S]). The two important parameters of Michaelis-Menten constant (Km) and maximum initial velocity (Vmax) were calculated. Km can indicate the affinity between enzyme and substrate, and the smaller Km value denotes the stronger affinity between enzyme and substrate [35]. The results are shown in Table 2. The Km value of AuNPs/aptamer DNA system was 0.187, which was less than that of AuNPs+aptamer+Hg2+ system (0.325). The results indicated that the AuNPs coated by ss-DNA had the stronger affinity with the substrate, leading to the very strong simulated enzyme activity of gold nanozyme. Moreover, the Vmax value of AuNPs/aptamer DNA system was obviously higher than that of AuNPs+aptamer+Hg2+ system, indicating that the catalytic rate of AuNPs/aptamer DNA was faster than that of AuNPs+aptamer+Hg2+. This also explained why the color response of TMB solution in the AuNPs/aptamer DNA system was very fast. The results further indicated that AuNPs coated by aptamer DNA had stronger peroxidase mimicking activity than AuNPs+aptamer+Hg2+ system. The results for steady-state kinetics of gold nanozyme were consistent with zeta potential results of AuNPs. In conclusion, the change of catalytic activity of gold nanozyme was due to the change of AuNPs surface electronegativity caused by spatial dimension of DNA aptamer, which resulted in the remarkable change of affinity between gold nanozyme and substrate. The proposed sensor is a colorimetric analysis of color fading based on affinity, which is different from the previous ones of enhancing nanozyme activity based on active site. This implies that there is more than one factor affecting the activity of the simulated enzyme, and the two factors of affinity and active site are competing.
Optimization of experimental conditions
The condition factors influencing the analysis sensitivity were investigated. The following parameters were optimized: (a) concentration of aptamer; (b) incubation time of aptamer and AuNPs; (c) reaction time of aptamer and Hg2+; and (d) concentration of TMB and H2O2. Respective data and figures are given in the Supporting Information (Fig. S2A and B). The optimized experimental conditions were (a) 2.0 μM; (b) 5 min; (c) 10 min; and (d) 5 mM and 5 mM.
Analytical performance of the colorimetric sensing for Hg2+
From the design principle of the colorimetric sensing, it can be seen that the color or absorbance of the sensing solution depends on the concentration of mercury in the system. Under the above optimized experimental conditions, the sensing responses of the systems with different concentrations of mercury were investigated. The results are shown in Fig. 3. As the mercury concentration increased, the blue color of the sensing solution became lighter (Fig. 3A). By naked eye, 40 nM Hg2+ can be observed to cause the color change of the solution. The result of UV–Vis absorption spectra (Fig. 3B) showed that the intensity of absorption peak at the wavelength of 652 nm gradually decreased with the increase of Hg2+ concentration. As shown in the inset of Fig. 3C, when the Hg2+ concentration increased from 2 nM to 0.8 μM, the absorbance at 652 nm decreased from 0.39 to 0.15. The concentration of Hg2+ in the range of 2.0 × 10–9 ~ 1.5 × 10–7 M was linearly correlated with the absorbance (A652). The linear curve is shown in Fig. 3C. Taken to be 3 times the standard deviation in the blank solution, the limit of detection (LOD) was estimated to be 9.3 × 10–11 M. It was about one order of magnitude lower than previous aptamer-based nanozyme colorimetry [13, 15] and comparable to the colorimetry based on AuNP aggregation [9, 12]. An overview on recently reported nanomaterial-based optical methods for the determination of mercury was listed in Supporting Information, Table S1. Most of the materials used for colorimetric analysis needed hours and even tens of hours to prepare, whereas the preparation of gold nanozyme in this method only required half an hour, which greatly reduced the analysis time. Although the present analysis was not the most sensitive in the existing optical analyses, the preparation of the used nanomaterials was much simpler. The proposed colorimetric analysis is especially suitable for real-time and rapid monitoring of heavy metal mercury.
A The photographs of the color change of the reaction solutions under different concentrations of Hg2+. B The corresponding absorption changes of UV–Vis absorption spectra. C The linear calibration plot for Hg2+ detection. Inset: the dependence of the absorbance at 652 nm on the concentration of target Hg2+. Experimental conditions: TMB, 5 mM; H2O2, 5 mM; AuNPs, 17 nM; and aptamer, 2.0 μM
The specificity of aptamer recognition lays the theoretical foundation for the selectivity of the proposed analysis. To verify the selectivity of this analysis for mercury detection, the systems from different metal ions and mercury aptamer were investigated respectively. The concentration of aptamer was 2.0 μM. The concentrations of mercury and other metal ions were 8.0 × 10–7 M and 8.0 × 10–6 M respectively and all other test conditions were the same. The result (Fig. 4A) was as expected. The difference of absorbance between only mercury ion system and the blank system was significantly larger than that of the other metal ions. It showed that the absorbance only from the mercury system decreased obviously. The developed analysis indeed exhibited high selectivity for mercury, which also confirmed the inherent specificity of aptamer recognition. The result provided a necessary precondition for the application of this analysis in real complex environment.
A Specificity of the colorimetric sensing for Hg2+ detection by binding Hg2+ aptamer with Hg2+ and other metal ions, where ΔA = A(blank, 652 nm) –A(with Hg2+ or other metal ions, 652 nm). Hg2+, 8.0 × 10–7 M and other metal ions, 8 μM. B Comparison of the colorimetric assay for the detection of Hg2+ solutions diluted with buffer and real water samples. Experimental conditions: TMB, 5 mM; H2O2, 5 mM; AuNPs, 17 nM; and aptamer, 2.0 μM. Error bars represent the standard deviations of three independent measurements
In addition, the sensing responses of the systems were investigated when other metal ions coexisted with mercury ions. The results found that 100 times of other metal ions (Na+, Ca2+, Cu2+, K+, Al3+, Ba2+, Ni2+, Pb2+, Mg2+, Fe3+, and Cd2+) had no influence on the system when the concentration of mercury ion was 1.0 × 10–6 M. Only 100 times of Cl− could interfere with the detection of mercury probably due to the formation of the complex of Hg2+ and Cl−. The results showed that this analysis was not suitable for the detection of mercury in environmental media with high chloride content.
Assay of mercury in real environmental samples
Tap water and lake water were used as real samples to investigate the potential application of the developed colorimetric analysis in actual environment. Tap water and lake water were taken from our laboratory and Qujiang Lake in Xi'an, respectively. Tap water sample needed being boiled to remove chlorine before testing. The lake water sample was filtered through a 0.22-mm membrane to remove suspended solids before detecting. The mercury stock solution was diluted by buffer solution, treated tap water and lake water separately, and then determined by this method. The measurement results are shown in Fig. 4B. The absorbance values of mercury solutions diluted by three different water samples were basically the same. It implied that the actual water samples like tap water and lake water did not interfere with the present analysis system. The results indicated that the colorimetric analysis had potential application to real environment water samples.
In order to further investigate the application of this analysis in real environmental media, the spiked recovery experiment was carried out for environmental water samples. The treated water samples of tap water and lake water were added with different concentrations of standard mercury and determined by this method and cold vapor atom adsorption spectrometry (CVAAS, the standard method for measuring mercury) respectively. The results are listed in Table 3. The recoveries of different spiked water samples were in the range of 95.2–104.3%, and the relative standard deviations (RSD) of three parallel measurements were all less than or equal to 4.2%. The results showed that this method had good precision and accuracy for detecting mercury in real water samples. And for the same water sample, the detection results by this method and the standard method (CVAAS) were basically consistent, which further confirmed the reliability of the proposed colorimetric analysis in practical application. In view of the simplicity, rapid test, and low cost, the developed colorimetric analysis is an ideal technique method for the rapid detection of heavy metal mercury in the environment and is especially applicable to economically backward areas.
Conclusions
The colorimetric sensor was developed for the detection of mercury in the environment by using that DNA dimension effectively regulated peroxidase-mimicking activity of AuNPs. The investigation for the sensing mechanism revealed that in addition to the surface active site, the affinity between nanozyme and substrate due to the charge effect was another important factor affecting the activity of simulated enzyme. And there was a competitive action between them. This colorimetric sensor was simple, rapid, and suitable for real-time monitoring of mercury in environmental media. The strategy of regulating peroxidase-mimicking activity of AuNPs by DNA dimension can be extended to other sensing systems. However, the poor stability of AuNPs colloidal solution also limits the practical application of the method. Nanozymes with high stability and simple preparation need to be further explored in the future work.
References
Yang LX, Zhang YY, Wang FF, Luo ZD, Guo SJ, Strähle U (2020) Toxicity of mercury: molecular evidence. Chemosphere 245:125586
Song YH, Ma QF, Cheng HY, Liu JH, Wang YC (2021) Simultaneous enrichment of inorganic and organic species of lead and mercury in pg L-1 levels by solid phase extraction online combined with high performance liquid chromatography and inductively coupled plasma mass spectrometry. Anal Chim Acta 1157:338388
Shih TT, Chen JY, Luo YT, Lin CH, Liu YH, Su YA, Chao PC, Sun YC (2019) Development of a titanium dioxide-assisted preconcentration/on-site vapor-generation chip hyphenated with inductively coupled plasma-mass spectrometry for online determination of mercuric ions in urine samples. Anal Chim Acta 1063(31):82–90
Sogame Y, Tsukagoshi A (2020) Development of a liquid chromatography-inductively coupled plasma mass spectrometry method for the simultaneous determination of methylmercury and inorganic mercury in human blood. J Chromatogr B 1136:121855
Volkov DS, Proskurnin MA, Korobov MV (2014) Survey study of mercury determination in detonation nanodiamonds by pyrolysis flameless atomic absorption spectroscopy. Diam Relat Mater 50:60–65
Liu YC, Zou J, Luo B, Yu HR, Zhao ZG, Xia H (2021) Ivy extract-assisted photochemical vapor generation for sensitive determination of mercury by atomic fluorescence spectrometry. Microchem J 169:106547
Zheng H, Hong JJ, Luo XL, Li S, Wang MX, Yang BY, Wang M (2019) Combination of sequential cloud point extraction and hydride generation atomic fluorescence spectrometry for preconcentration and determination of inorganic and methyl mercury in water samples. Microchem J 145:806–812
Huang DL, Liu XG, Lai C, Qin L, Zhang C, Yi H, Zeng GM, Li B, Deng R, Liu SY, Zhang YJ (2018) Colorimetric determination of mercury(II) using gold nanoparticles and double ligand exchange. Microchim Acta 186:31
Li L, Li BX, Qi YY, Jin Y (2009) Label-free aptamer-based colorimetric detection of mercury ions in aqueous media using unmodified gold nanoparticles as colorimetric probe. Anal Bioanal Chem 393(8):2051–2057
Lian Q, Liu H, Zheng XF, Li X, Zhang FJ, Gao J (2019) Enhanced peroxidase-like activity of CuO/Pt nanoflowers for colorimetric and ultrasensitive Hg2+ detection in water sample. Appl Surf Sci 483(31):551–561
Liu R, Zuo L, Huang XR, Liu SM, Yang GY, Li SY, Lv CY (2019) Colorimetric determination of lead(II) or mercury(II) based on target induced switching of the enzyme-like activity of metallothionein-stabilized copper nanoclusters. Microchim Acta 186:250
Qi YY, Ma JX, Chen XD, Xiu FR, Chen YT, Lu YW (2020) Practical aptamer-based assay of heavy metal mercury ion in contaminated environmental samples: convenience and sensitivity. Anal Bioanal Chem 412:439–448
Qi YY, Song DD, Chen YT (2021) Colorimetric oligonucleotide-based sensor for ultra-low Hg2+ in contaminated environmental medium: convenience, sensitivity and mechanism. Sci Total Environ 766:142579
Wang YW, Liu Q, Wang L, Tang S, Yang HH, Song H (2018) A colorimetric mercury(II) assay based on the Hg(II)-stimulated peroxidase mimicking activity of a nanocomposite prepared from graphitic carbon nitride and gold nanoparticles. Microchim Acta 186:7
Wang JJ, Zhou PL, Tao H, Wang XL, Wu YG (2020) Oligonucleotide-induced regulation of the oxidase-mimicking activity of octahedral Mn3O4 nanoparticles for colorimetric detection of heavy metals. Microchim Acta 187:99
Zhang Y, Ju P, Sun LP, Wang Z, Zhai XF, Jiang F, Sun CJ (2020) Colorimetric determination of Hg2+ based on the mercury-stimulated oxidase mimetic activity of Ag3PO4 microcubes. Microchim Acta 187:422
Guo XR, Huang JZ, Wei TB, Zeng Q, Wang LS (2020) Fast and selective detection of mercury ions in environmental water by paper-based fluorescent sensor using boronic acid functionalized MoS2 quantum dots. J Hazard Mater 381:120969
Li ZH, Sun HJ, Ma XY, Su RF, Sun R, Yang CY, Sun CY (2020) Label-free fluorescence “turn-on” strategy for mercury (II) detection based on the T-Hg2+-T configuration and the DNA-sensitized luminescence of terbium (III). Anal Chim Acta 1099(22):136–144
Maimaitiyiming X, Shi C (2021) Poly(1,4-diethynylphenylene-4,6-pyrimidine)s for fluorescence detection of mercury(II) ion. Mater Chem Phys 257:123783
Qi YY, Xiu FR, Yu GD, Huang LL, Li BX (2017) Simple and rapid chemiluminescence aptasensor for Hg2+ in contaminated samples: a new signal amplification mechanism. Biosens Bioelectron 87(15):439–446
Chen ZY, Gupta A, Chattopadhyay S (2021) Detection of mercury in spiked cosmetics by surface enhanced Raman spectroscopy using silver shelled iron oxide nanoparticles. Sensor Actuat B: Chem 337:129788
Zhao YB, Yamaguchi Y, Ni Y, Li MD, Dou XM (2020) A SERS-based capillary sensor for the detection of mercury ions in environmental water. Spectrochim Acta A 233:118193
Teodoro KB, Migliorini FL, Facure MH, Correa DS (2019) Conductive electrospun nanofibers containing cellulose nanowhiskers and reduced graphene oxide for the electrochemical detection of mercury(II). Carbohy Polym 207(1):747–754
Salandari N, Ensafi AA, Rezaei B (2021) Ultra-sensitive electrochemical aptasensor based on zeolitic imidazolate framework-8 derived Ag/Au core-shell nanoparticles for mercury detection in water samples. Sensor Actuat B: Chem 331:129426
Zou YS, Zhang YL, Xie ZZ, Luo SY, Zeng YM, Chen QZ, Liu GK, Tian ZQ (2019) Improved sensitivity and reproducibility in electrochemical detection of trace mercury (II) by bromide ion & electrochemical oxidation. Talanta 203(1):186–193
Qi YY, Li BX (2011) A sensitive, label-free, aptamer-based biosensor using a gold nanoparticle-initiated chemiluminescence system. Chem Eur J 17:1642–1648
Li HX, Rothberg L (2004) Colorimetric detection of DNA sequences based on electrostatic interactions with unmodifed gold nanoparticles. Proc Natl Acad Sci USA. 101(39):14036–14039
Hizir MS, Top M, Balcioglu M, Rana M, Robertson NM, Shen F, Sheng J, Yigit MV (2016) Multiplexed activity of perAuxidase: DNA-capped AuNPs act as adjustable peroxidase. Anal Chem 88(1):600–605
Yang Y, Yin YG, Li XL, Wang S, Dong YY (2020) Development of a chimeric aptamer and an AuNPs aptasensor for highly sensitive and specific identification of Aflatoxin B1. Sensor Actuat B: Chem 319:128250
Ono A, Togashi H (2004) Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew Chem Int Ed Engl 43(33):4300–4302
Song ZQ, Xiu F-R, Qi YY (2022) Degradation and partial oxidation of waste plastic express packaging bags in supercritical water: Resources transformation and pollutants removal. J Hazard Mater 423:127018
Zhang ZF, Cui H, Lai CZ, Liu LJ (2005) Gold nanoparticle-catalyzed luminol chemiluminescence and its analytical applications. Anal Chem 77:3324–3329
Huang K, Yang H, Zhou ZG, Yu MX, Li FY, Gao X, Yi T, Huang CH (2008) Multisignal chemosensor for Cr3+ and its application in bioimaging. Org Lett 10(12):2557–2560
Wu Y, Zheng JW, Li Z, Zhao YR, Zhang Y (2009) A novel reagentless amperometric immunosensor based on gold nanoparticles/TMB/Nafion-modified electrode. Biosens Bioelectron 24(3):1389–1393
Gao LZ, Zhuang J, Nie L, Zhang JB, Zhang Y, Gu N, Wang TH, Feng J, Yang DG, Perrett S, Yan XY (2007) Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat Nanotechnol 2:577–583
Funding
This work was supported financially by the National Natural Science Foundation of China (No. 21605018) and the Natural Science Basic Research Project of Shaanxi Province of China (No. 2020JM-528 and No. 2021JZ-52).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qi, Y., Wang, Y., Chen, Y. et al. Rapid color-fading colorimetric sensing of Hg in environmental samples: regulation mechanism from DNA dimension. Microchim Acta 189, 76 (2022). https://doi.org/10.1007/s00604-022-05177-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05177-w