Abstract
Herein, we report a rapid and sensitive colorimetric detection of Hg2+ by designing a specific DNA probe with phosphorothioate RNA modification (PS-probe) for Hg2+ recognition and utilizing DNA-modified gold nanoparticles (DNA-AuNPs) as the transducer. The distance between two DNA-AuNPs is controlled by a linker DNA, providing the linker DNA-regulated aggregation or dispersion status of AuNPs in solution. Exonuclease III (Exo III) can trigger the recycled digestion of linker DNA strands, inhibiting the reformation of aggregated nanoparticles and hence leading to a color shift from purple to red. However, the Hg2+-induced cleavage of the PS-probe can efficiently prevent the digestion of linker DNA strands by Exo III and hence reassemble the modified AuNPs to form aggregates in purple color. Thus, a positive correlation between the linker DNA strands left and the addition of Hg2+ provides a quantitative basis for Hg2+ sensing. A linear range of A520/A700 versus Hg2+ concentration is achieved in the range 2–100 nM associated with a detection limit as low as 1.30 ± 0.04 nM. Moreover, the biosensor exhibits excellent selectivity for Hg2+. The strong selectivity behavior was confirmed by recoveries ranging from 96 to 114% in real water samples.

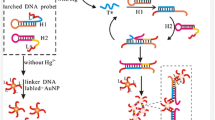
Schematic representation of sensing mechanism of Hg2+ using a DNA probe with phosphorothioate RNA modification (PS-probe) and Exo III-assisted signal amplification
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mercury is generally considered to be one of the most toxic metals found in nature, which is mainly released from mercury-related industries [1]. Considering its bioaccumulation characteristics, the severe adverse health effect of mercury on human beings cannot be overlooked even at trace concentration level [2]. While the traditional instrumental techniques, for example, inductively coupled plasma mass spectrometry (ICP-MS), are accurate and widely used for mercury detection, their applications are still limited to the laboratory due to the expensive and large-scale equipment and complicated detection procedure. Therefore, developing facile and sensitive detection methods of mercury in the environments is still of great importance. Considering the divalent mercury (Hg2+ ions) are water-soluble and one of the most common and stable existing form in water environments among the mercury pollution [3], the development of novel Hg2+ detection has been focused and received a lot of attentions.
To meet the goals of facile and sensitive analysis, the biosensor technique emerges as an effective alternative to the traditional analytical techniques. By virtue of their high sensitivity, specificity, and facile synthesis, functional nucleic acid (FNA)-based biosensors have received numerous attentions and witnessed their broad applications ranging from environmental monitoring [4], diagnostics [5], and food safety to basic research [6]. Since Hg2+ was reported to be specifically bound between two thymine bases, thus promoting the T-T mismatch [7], various T-Hg2+-T interaction-based biosensors for Hg2+ detection were reported with different transduction modes used, including electrochemistry [8], colorimetry [3], surface plasmon resonance [9], fluorescence [10], and surface-enhanced Raman scattering [11]. Although these biosensors need less professional personnel and simpler sensing instrument than traditional methods, the Hg2+-driven T-T mismatch was reported to be strongly affected by reaction buffer conditions, including pH, temperature, ionic strength, and ion species [12,13,14]. DNAzyme is used as another DNA-based strategy for sensing Hg2+ [15, 16]. While Hg2+ can activate some catalysis of DNAzymes, the high detection limits and low catalytic efficiency limited their downstream applications [10]. Although some DNAzymes have been reported with high sensitivity, the T-Hg2+-T interaction has to be incorporated in the design of such DNAzyme [17], which makes the sensing system very complicated. Recently, a new strategy for specific Hg2+ sensing was reported by using a specifically designed DNA probe, which incorporated phosphorothioate (PS)-modified RNA linkages [18]. This DNA probe inserted with PS-modified RNA can efficiently and specifically cleaved by Hg2+ ions due to their strong thiophilicity. Considering that the cleavage procedure is a chemical reaction, this sensing strategy was reported to be less affected by the reaction conditions and show excellent selectivity [12, 18].
Colorimetric biosensors, especially by using the gold nanoparticles (AuNPs) as indicators, have been extensively utilized due to its simplicity of operation and visibility of observation [19]. The peak of the absorption spectra of AuNPs is dependent on their size and shape, providing a quantitative basis for colorimetric sensing. Compared with the unmodified AuNPs (in the general sense, citrate-stablized AuNPs), DNA-modified AuNPs (DNA-AuNPs) by using thiol-gold chemistry can provide harsher environment resistance and higher reliability, which have been developed and utilized in detecting different targets, such as DNA sequence [20], metal ions [21], and bacteria [22].
Stated thus, by merging the virtues of the DNA probe inserted with PS-modified RNA for Hg2+-specific recognition, Exo III-assisted signal amplification, and DNA-AuNPs as stable indicators of colorimetry, we herein develop a visible, cost-effective, easy-to-handle yet sensitive biosensor for the colorimetric detection of Hg2+ in aqueous environments. As a result, the specificity provided by the DNA probe inserted with PS-modified RNA towards Hg2+ is exclusive; moreover, the limit of detection can reach 1.30 nM via the color variance of AuNPs, which allows the turnaround time after sample collection from suspicious sites to be limited to 40 min.
Methods
Preparation of DNA probes
The list of chemicals used is described in Electronic Supplementary Material (ESM). The stock solution of different DNA probes was prepared by using 10-mM HEPES buffer (pH 7.5) to dissolve the oligonucleotides to 10 μM strand concentration. The solution was heated at 95 °C for 5 min and then gradually cooled to room temperature in 2 h. All stock solutions were maintained at 4 °C before the use. If not specified, all buffers used in this work were prepared either in biomolecule grade deionized water or DEPC-treated deionized water (in general referring to DI water).
Preparation and modification of AuNPs
AuNPs in diameter of 13 nm were prepared by reducing HAuCl4 using sodium citrate [23]. Briefly speaking, 100 mL of 1 mM HAuCl4 was heated to reflux under the condition of stirring, and subsequently, 10 mL of 38.8 mM sodium citrate was added. The color of mixture changed from light yellow to heavy red within a few minutes. After that, the mixture was left to reflux for another half hour, completing the reduction of HAuCl4 to gold nanoparticles. The morphology of AuNPs was characterized using TEM (Fig. S1), and their average size was determined to be approximately 12 ± 2 nm by counting more than 100 particles from TEM pictures.
The functionalization of AuNPs with 3′- or 5′-thiol-modified DNA strands followed the previously reported procedures [24]. Briefly speaking, 90 μL of 100 μM DNA1 or DNA2 was activated by tris(2-carboxyethyl)phosphine hydrochloride (TCEP) for 1 h. Subsequently, 3 mL AuNPs was added into the mixture and incubated for 16 h. After that, 31 μL of 500 mM sodium acetate was then added drop by drop and incubated for another 24 h. After centrifugation, the supernatant was discarded, and 1.2 mL buffer containing 25 mM Tris-acetate and 100 mM NaCl (pH 8.2) was then added. After centrifugation again, the supernatant was removed, and the DNA-modified AuNPs were dispersed in 3 mL buffer (25 mM Tris-Acetate and 300 mM NaCl, pH 8.2).
Nondenaturing PAGE
For a typical nondenaturing PAGE, the DNA samples were diluted by using 6 x loading buffer with a volume ratio of 5:1. The 15% polyacrylamide (19:1 acrylamide/bisacrylamide) gels were freshly prepared. The nondenaturing PAGE were performed in 1 x TBE under the conditions of 80 V, 50 min, and 20 °C. Gels were stained with GelRed Acid Gel Stain and imaged under UV exposure by FLS-5100 film (Fuji, Japan). The measurements were performed in 10 mM HEPES buffer (pH 7.5).
Absorbance measurement
Under the optimized conditions (see Fig. S2), the absorbance measurement methods towards Hg2+ were developed. A series of Hg2+ standard solutions (0.3, 1.5, 3.0, 4.5, 7.5, 11.25, 15, and 30 μM) was prepared by diluting the stock solution of Hg2+ with DI water. 10 μM strand concentration of probes and linker were prepared by diluting the DNA stock solutions, respectively, with 10 mM HEPES buffer (pH 7.5). The optimal steps for Hg2+ detection were listed as follows: 1 μL Hg2+ standard solution under various concentrations was successively added into 4 μL of 150 nM PS-probe for incubation of 15 min. Subsequently, 1 μL 150 mM Mg2+ solution, 2 μL purified water, 1 μL 10 μM linker strand, and 5 U Exo III were added to the solution described above to catalyze the hydrolysis reaction for another incubation of 30 min at 37 °C. The reaction mixture was then heated to 65 °C for 10 min to stop the digestion reaction by inactivating the Exo III activities. Finally, 20 μL DNA1-Au, 20 μL DNA2-Au, and 100 μL of 15 mM HEPES buffer (pH 7.5) were added for final incubation of 10 min before the absorbance measurements. The Hg2+ concentrations in the samples were 2, 10, 20, 30, 50, 75, 100, and 200 nM, respectively. The optical chamber (1-cm path length, 0.35-mL volume) was used for absorbance measurements in the wavelength range of 400 to 750 nm at room temperature (approximately 25 °C).
Selectivity and recovery
The selectivity of this method was tested using Mn2+, Pb2+, Ca2+, Cd2+, Cu2+, Ni2+, Fe2+, Cr3+, and Co2+ (each at 100 nM) in the presence or absence of 10 nM Hg2+, respectively. The absorption ratios of A520/A700 for competing species were recorded. Furthermore, the metal ions described above were incubated with the PS-probe (1 μM) for 10 min. The product after incubation was separated on 15% nondenaturing PAGE gels and analyzed by using a FLS-5100 film imaging system.
Environmental water samples were taken from Guanting reservoir (Beijing, China) to test the reliability of this method. Sample solutions were taken and then passed through a 0.2-μm pore-size membrane filter before test. Subsequently, the water samples were spiked with various Hg2+ to final concentrations of three levels (0, 5, and 10 nM). Detection procedures were performed based on the optimal conditions for Hg2+ detection described above.
Results and discussion
Working principle
Figure 1 illustrates the principle scheme of colorimetric biosensing of Hg2+ using a DNA probe inserted with phosphorothioate-modified RNA and Exo III-assisted signal amplification. The colorimetric determination system comprises a PS-probe, a linker strand of DNA, Exo III, and two sets of different DNA-AuNPs (called DNA1-Au and DNA2-Au, respectively) with sequences complementary to both ends of the linker strand of DNA, respectively. It should be noted that the linker DNA strand is rationally designed in such a way that, if Hg2+ is absent, the linker will hybridize with the PS-probe to form a specific duplex with a recessed 3′ terminal end, which can be cleaved by Exo III. Considering that Exo III can only digest the 3′ recessed single-stranded DNA of linker strand, the PS-probe will hence be recycled to hybridize more linker strands to undergo new digestion reactions. Such an Exo III-assisted cyclic hybridization-hydrolysis process may trigger the digestion of numerous linker DNA strands, resulting in a significantly amplified signal transduction. By contrast, the presence of Hg2+ ions can efficiently cleave the phosphorothioate RNA linkage incorporated in the PS-probe due to their extremely strong thiophilicity [18]. This will prevent the digestion of linker strands by the enzyme of Exo III. Upon completion of the digestion/cleavage cycle, the mixtures of DNA1-Au and DNA2-Au are added into the solution. As a result of the complementary hybridization of DNA1-Au and DNA2-Au to both ends of the linker DNA strand, a color change of AuNPs from red to purple would be obtained by shortening their distance between DNA1-Au and DNA2-Au and used to quantify the amount of the remaining linker DNA strands. Considering a positive correlation between the linker strand left and the addition of Hg2+, Hg2+ concentration can be quantified accordingly.
The UV–vis spectra of colorimetric modified-AuNP system in the presence of different Hg2+ concentrations are shown in Fig. 2a. Initially, the linker strand complementary to the 12-mer DNAs on AuNPs assembled the nanoparticles closer to form purple-colored aggregates, exhibiting the plasmon resonance absorption at about 600 nm (curve a). Meanwhile, the linker hybridized with PS-probe strand to form a 3′ terminus end recessed duplex that was digestible by Exo III. Due to the function of Exo III, the 3′ recessed linker strand was cleaved into single bases, which was not able to assemble the modified AuNPs of DNA1-Au and DNA2-Au. The dispersed AuNPs were accompanied with a plasmon resonance absorption peak at about 520 nm (curve b). However, following the addition of 200 nM Hg2+, the absorbance of AuNPs decreased significantly at 520 nm and increased in a longer wavelength range with a peak value at approximately 600 nm, resulting into the red-to-purple color shift (curve c). It was attributed to the Hg2+-triggered cleavage of PS-probe, which ended the cyclic Exo III enzymatic amplification. By contrast, the DNA sequence without phosphorothioate-modified RNA linkage (WPS-probe) was designed, and its responses towards Hg2+ were investigated for comparison. As shown in curve d, the absorbance spectra had a slight change when 200 nM Hg2+ were incubated with WPS-probe compared to the sensing system without Hg2+ (curve a). Inset of Fig. 2a shows the photos under four conditions, accordingly. The colorimetric visualization with the naked eye was achieved without using the complicated instrumentation. These results concurred with our speculation that Hg2+ trigger the cleavage of the phosphorothioate-modified RNA linkage rather than unmodified one and hence change the aggregation status of AuNPs. Meanwhile, the phenomena described above accorded well with numerous previous studies [22].
a Absorption spectra of the modified AuNP system in the presence of (a) linker strand; (b) linker strand, PS-probe, and Exo III; (c) linker strand, PS-probe, and Exo III after incubation with Hg2+; and (d) linker strand, WPS-probe, and Exo III after incubation with Hg2+. DNA1-Au, 3.2 nM; DNA2-Au, 3.4 nM; linker, 67 nM; PS-probe, 4 nM; WPS-probe, 4 nM; Exo III 5 units (U); Hg2+, 200 nM; and pH 7.5. Inset shows the photos under four conditions, accordingly. b TEM images of AuNPs containing PS-probe. DNA1-Au, 3.2 nM; DNA2-Au, 3.4 nM; linker, 67 nM; PS-probe, 4 nM; Exo III 5 U; pH 7.5; and Hg2+, 0 (Left) and 200 nM (Right). c Nondenaturing 15% PAGE image (lane 1 to lane 5 from left to right) exhibiting the cleavage products of PS-probe incubated with 0, 1, 5, 10, and 50 μM Hg2+, respectively, for 10 min
The changes in the aggregation status of DNA1-Au and DNA2-Au nanoparticles in the Exo III amplification system were verified by the TEM technique. As illustrated in Fig. 2b, DNA-AuNPs were well dispersed with no addition of Hg2+ due to the effective Exo III-assisted digestion of the linker strand in a duplex form with PS-probe (Left). However, upon adding Hg2+, two sets of DNA-AuNPs were assembled by the linker strands considering that their digestion was apparently inhibited due to the significant cleavage ability of Hg2+ on the PS-probe (Right). In addition, the nondenaturing PAGE was used to characterize the cleavage process of PS-probe induced by Hg2+ ions. Figure 2c demonstrates the images of nondenaturing PAGE for PS-probe in 10 mM HEPES buffer (pH 7.5) at room temperature. We observed three distinct bands with unequal mobilities in the PS-probe system, implying the DNA sequences with different lengths formed. The faster two bands exhibited that the PS-probe underwent cleavage, while an increase in the cleaved fraction was found when more Hg2+ existed. This result confirmed that the existence of Hg2+ can induce the cleavage of PS-probe. The DNA sequence at the slowest band was explained to its desulfurization to the normal phosphate backbone and was not cleaved by Hg2+again [12]. The cleavage yield that has yet to be optimized may also contribute to this phenomenon [25].
Optimization of bioassay conditions
Optimization processes were conducted to identify the optimal bioassay conditions of this colorimetric method for Hg2+ detection. Three key factors for the modified AuNPs-based colorimetric bioassay as suggested by previous studies [3, 26, 27], including the linker concentration, the magnesium ion concentration in buffer, and the Exo III concentration, were investigated. The linker concentration should have a great impact on the colorimetric biosensing because of it being responsible for the assembly of DNA1-Au and DNA2-Au nanoparticles. Unveiled by Fig. S2A, the higher linker concentrations resulted in greater color changes represented in the form of ration of absorption intensity A520/A700, however reached relatively steady when the linker concentration was higher than 20 nM. To increase the detection sensitivity, 20 nM linker strand was chosen for the subsequent experiments. The magnesium ion salt was added to enhance the linker DNA-induced nanoparticle assemblies, and therefore, the concentration of Mg2+ on the performance was exploited. Unveiled by Fig. S2B, no significantly observable change in A520/A700 was obtained until 1 mM Mg2+ ions; however, the value decreased sharply when the Mg2+ concentrations further increased. To obtain a better sensitivity in the color change, 1 mM Mg2+ was used in the following experiments. In the end, the influence of the Exo III concentration was investigated as illustrated in Fig. S2C. Under the condition of Exo III concentration below 5 U, A520/A700 changes with the enzyme concentration showed opposite trends in the absence (R1) and presence (R2) of target. However, R1 and R2 values reached steady with increased Exo III concentration under both conditions. The highest ratio R1 to R2 was 2.1, at which the concentration of Exo III was 5 U, so we selected 5 U Exo III, 20 nM linker strand, and 1 mM Mg2+ as the optimal detection conditions.
Moreover, the number of PS-modified RNA linkage has a great impact on the cleavage reaction yield [28]. The DNA sequence inserted with two, three, and four PS-modified RNA linkages (referring to PS2-probe, PS3-probe, and PS4-probe) were tested by incubating in the absence or presence of 100 nM Hg2+, respectively, under optimal conditions. The A520/A700 values are demonstrated in Fig. S2D. As we expected, the increase in the number of PS-modified RNA linkages resulted in more significant signal changes, indicating the greater cleavage reaction yield. Three and four PS-modified RNA linkages exhibited a slight difference to increase the cleavage performance. Compromising the cleavage capability and the price of the probe synthesis, the PS-probe with three PS-modified RNA linkage was chosen for the experiments.
Performance for Hg2+ determination
Under the optimal bioassay conditions, the sensitivity of this colorimetric sensing system for Hg2+ determination was evaluated. The visible spectra of modified AuNPs under different Hg2+ concentrations are demonstrated in Fig. 3a. Initially, this modified AuNPs (DNA1-Au, DNA2-Au) with modification of non-complimentary DNA sequences as the stabilizer [29] exhibited red in color, accompanied by a strong plasmon resonance absorption at about 520 nm (curve 1) due to the Exo III-assisted recycling of digesting the linker DNA strand via its hybridization with PS-probe. However, the existence of Hg2+ would cleave PS-probe into short fragments, which inhibited its hybridization with the linker DNA and hence avoided its digestion by Exo III. As a result, the remaining linkers would assemble the DNA1-Au and DNA2-Au nanoparticles closer, enabling AuNPs aggregated. Theoretically, the AuNPs aggregated quicker with the increased Hg2+ concentration, therefore leading to more aggregated AuNPs in a certain period of time. Following the addition of Hg2+, curves 2–9 displayed an observable decrease in the peak of absorption at 520 nm, accompanied by an increase in the longer wavelength range with a peak value at 600 nm. This result corresponded to a red-to-purple color change (Inset of Fig. 3a).
a Visible spectra of the colorimetric biosensor under various amounts of Hg2+ in the order of curves top to bottom (curve 1–9): 0, 2, 10, 20, 30, 50, 75, 100, and 200 nM, Inset of the color display from left to right corresponds to the concentration of Hg2+ (0, 2, 10, 20, 30, 50, 75, and 100 nM) and b absorption ratio of A520/A700 vs different Hg2+ concentration with linear fitting in the range of 2–100 nM
We compared the sensitivity by means of the signal represented by the ratios of absorbance intensities, A520/A700 and A520/A600, respectively. The linear relationship between A520/A700 values and different Hg2+ concentration is represented in Fig. 3b. The linear regression range exhibited 2–100 nM Hg2+ (R2 = 0.99) and a limit of determination (LOD) of 1.30 nM using the three-sigma method [30]. It is indicated that A520/A700 is more sensitive than A520/A600 to the change of Hg2+ concentration (Fig. S3) although their linear fitting results with Hg2+ concentration shared a same linear range (see Table S2).
Nanomaterials especially Au and Ag in the form of nanoparticles and nanorods are widely used in designing colorimetric strategies for Hg2+ determination; however, these colorimetric assays often suffered from low sensitivity [26]. A comparison of the analytical performance of this work with those obtained by several other approaches based on for the determination of Hg2+ is shown in Table 1. Obviously, the LOD of our method was comparable with most of the reported Hg2+ biosensing techniques. In our system, such an attractive LOD was mostly given to the recycling of PS-probe and hence induced the Exo III-assisted signal amplification. Meanwhile, this technique was simple, easy-to-handle, and obtained the test results within 40 min. Furthermore, sensitive and rapid visualization determination can facilitate in-field and on-site applications without the assistance of instrumentation.
Selectivity
To confirm the practical applicability of this colorimetric biosensor, the competition experiments and selectivity trials were conducted to evaluate the potential interferences from potentially coexisting metal ions in water environments. Frequently encountered metal ions were chosen for the interference experiments under the optimal conditions same as for Hg2+ measurement. As illustrated in Fig. 4, only the presence of Hg2+ ions can result in a significant signal variation even though the interfering ions, including Mn2+, Pb2+, Ca2+, Cd2+, Cu2+, Ni2+, Fe2+, Cr3+, and Co2+, existed at a high concentration in the samples.
Change of absorption ratio (A520/A700) for the colorimetric biosensor responding to different metal ions, e.g., Mn2+, Pb2+, Ca2+, Cd2+, Cu2+, Ni2+, Fe2+, Cr3+, Co2+, and Hg2+, and coexisting species as mentioned above in the absence and presence of Hg2+. Inset: Nondenaturing 15% PAGE images exhibiting the cleavage of PS-probe incubated with 10 μM different metal ions for 10 min
Moreover, the colorimetric biosensor was further challenged with Hg2+ determination in the coexistence of interfering metal ions. Results showed that the coexistence of interfering metal ions posed a negligible impact on the performance of our protocol towards Hg2+. These results further demonstrated the excellent selectivity of this sensing platform for Hg2+ determination even in the complicated matrixes. The selectivity was further confirmed by investigating the cleavage of the PS-probe incubated with 10 μM metal ions for 10 min. As unveiled by nondenaturing 15% PAGE image (Inset of Fig. 4), distinct bands with clearly different mobilities appeared only in the lanes contained Hg2+ ions, indicating the selective cleavage of the PS-probe caused by Hg2+ ions. This result accorded well with the absorption ratio described above.
Application in real water sample analysis
To test the applicability of this biosensor for Hg2+ determination in real samples, two environmental water samples were taken from Guanting reservoir located in the north of Beijing, China, and tap water in lab. The colorimetric biosensor demonstrated a negligible signal change compared to the blank samples towards the raw water, suggesting no presence of Hg2+ or below the LOD of this method. Because the Hg2+ concentration of environment water was nondetectable, therefore the concentrations of 0, 5.0, and 10.0 nM of Hg2+ were spiked and analyzed, using this bioassay system and inductively coupled plasma mass spectrometry (ICP-MS), respectively. Each analysis was conducted in triplicate to minimize any possibilities of error. Therefore, a spike-recovery test was carried out by spiking Hg2+ ions at three concentration levels. The testing results of the biosensor towards the Hg2+-spiked samples are demonstrated in Table S3. The quantitative recoveries in the range of 96–114% were achieved by using this method. The good accordance of measured values with the spiked Hg2+ concentrations suggested the prospective applicability of this method for Hg2+ determination in real water environments.
Conclusions
By incorporating a PS-probe and a recycled enzymatic cleavage to amplify the signal, we described a colorimetric biosensor for the rapid and sensitive determination of Hg2+ in water environments. This colorimetric biosensor obtained a LOD of 1.30 nM for Hg2+ determination. It exhibited an excellent selectivity for Hg2+ ions rather than other interfering metal ions even existed at high concentration. The applicability of this method was confirmed by results showing good recoveries (96–114%) in environmental water samples. In view of its simplicity, easy-to-handle, rapid, and sensitive visual determination, this biosensing method paves great potential for applications in on-site determination of Hg2+ without the aid of instrumentation for water pollution alarming and control. However, the cleavage yield of PS-probe that be further improved requires more research to in the future.
References
Jarup L (2003) Hazards of heavy metal contamination. Br Med Bull 68:167–182. https://doi.org/10.1093/bmb/ldg032
Zahir F, Rizwi SJ, Haq SK, Khan RH (2005) Low dose mercury toxicity and human health. Environ Toxicol Pharmacol 20(2):351–360. https://doi.org/10.1016/j.etap.2005.03.007
Li T, Dong S, Wang E (2009) Label-free colorimetric detection of aqueous mercury ion (Hg2+) using Hg2+-modulated G-quadruplex-based DNAzymes. Anal Chem 81(6):2144–2149. https://doi.org/10.1021/ac900188y
Nguyen VT, Kwon YS, Gu MB (2017) Aptamer-based environmental biosensors for small molecule contaminants. Curr Opin Biotechnol 45:15–23. https://doi.org/10.1016/j.copbio.2016.11.020
Kaur H, Bruno JG, Kumar A, Sharma TK (2018) Aptamers in the therapeutics and diagnostics pipelines. Theranostics 8(15):4016–4032. https://doi.org/10.7150/thno.25958
Zhang Y, Deng Y, Wang C, Li L, Xu L, Yu Y, Su X (2019) Probing and regulating the activity of cellular enzymes by using DNA tetrahedron nanostructures. Chem Sci 10(23):5959–5966. https://doi.org/10.1039/C9SC01912J
Ono A, Togashi H (2004) Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew Chem Int Ed Engl 43(33):4300–4302. https://doi.org/10.1002/anie.200454172
Zhuang J, Fu L, Tang D, Xu M, Chen G, Yang H (2013) Target-induced structure-switching DNA hairpins for sensitive electrochemical monitoring of mercury (II). Biosens Bioelectron 39(1):315–319. https://doi.org/10.1016/j.bios.2012.07.015
Ling B, Ma Y, Chen H, Wang L (2017) A SPR aptamer sensor for mercury based on AuNPs@NaYF4:Yb,Tm,Gd upconversion luminescent nanoparticles. Anal Methods 9(42):6032–6037. https://doi.org/10.1039/C7AY01810J
Qi L, Zhao Y, Yuan H, Bai K, Zhao Y, Chen F, Dong Y, Wu Y (2012) Amplified fluorescence detection of mercury(II) ions (Hg2+) using target-induced DNAzyme cascade with catalytic and molecular beacons. Analyst 137(12):2799–2805. https://doi.org/10.1039/C2AN35437C
Song X, Wang Y, Liu S, Zhang X, Wang H, Wang J, Huang J (2018) Ultrasensitive electrochemical detection of Hg2+ based on an Hg2+-triggered exonuclease III-assisted target recycling strategy. Analyst 143(23):5771–5778. https://doi.org/10.1039/C8AN01409D
Huang PJ, van Ballegooie C, Liu J (2016) Hg2+ detection using a phosphorothioate RNA probe adsorbed on graphene oxide and a comparison with thymine-rich DNA. Analyst 141(12):3788–3793. https://doi.org/10.1039/C5AN02031J
Kiy MM, Jacobi ZE, Liu J (2012) Metal-induced specific and nonspecific oligonucleotide folding studied by FRET and related biophysical and bioanalytical implications. Chemistry 18(4):1202–1208. https://doi.org/10.1002/chem.201102515
Kiy MM, Zaki A, Menhaj AB, Samadi A, Liu J (2012) Dissecting the effect of anions on Hg2+ detection using a FRET based DNA probe. Analyst 137(15):3535–3540. https://doi.org/10.1039/C2AN35314H
Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM (2008) A highly selective DNAzyme sensor for mercuric ions. Angew Chem Int Ed Engl 47(23):4346–4350. https://doi.org/10.1002/anie.200800960
Zhang XB, Kong RM, Lu Y (2011) Metal ion sensors based on DNAzymes and related DNA molecules. Annu Rev Anal Chem (Palo Alto, Calif) 4:105–128. https://doi.org/10.1146/annurev.anchem.111808.073617
Liu J, Lu Y (2007) Rational design of "turn-on" allosteric DNAzyme catalytic beacons for aqueous mercury ions with ultrahigh sensitivity and selectivity. Angew Chem Int Ed Engl 46(40):7587–7590. https://doi.org/10.1002/anie.200702006
Huang PJ, Wang F, Liu J (2015) Cleavable molecular beacon for Hg2+ detection based on phosphorothioate RNA modifications. Anal Chem 87(13):6890–6895. https://doi.org/10.1021/acs.analchem.5b01362
Rosi NL, Mirkin CA (2005) Nanostructures in biodiagnostics. Chem Rev 105(4):1547–1562. https://doi.org/10.1021/cr030067f
Borghei YS, Hosseini M, Dadmehr M, Hosseinkhani S, Ganjali MR, Sheikhnejad R (2016) Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal Chim Acta 904:92–97. https://doi.org/10.1016/j.aca.2015.11.026
Yang X, Xu J, Tang X, Liu H, Tian D (2010) A novel electrochemical DNAzyme sensor for the amplified detection of Pb2+ ions. Chem Commun (Camb) 46(18):3107–3109. https://doi.org/10.1016/j.aca.2015.11.026
Zhu D, Yan Y, Lei P, Shen B, Cheng W, Ju H, Ding S (2014) A novel electrochemical sensing strategy for rapid and ultrasensitive detection of Salmonella by rolling circle amplification and DNA-AuNPs probe. Anal Chim Acta 846:44–50. https://doi.org/10.1016/j.aca.2014.07.024
Turkevich J, Stevenson PC, Hillier J (1951) A study of the nucleation and growth processes in the synthesis of colloidal gold. Discussions of the Faraday Society 11:55–75. https://doi.org/10.1039/DF9511100055
Liu J, Lu Y (2004) Accelerated color change of gold nanoparticles assembled by DNAzymes for simple and fast colorimetric Pb2+ detection. J Am Chem Soc 126(39):12298–12305. https://doi.org/10.1021/ja046628h
Wang H, Liu Y, Liu G (2018) Electrochemical biosensor using DNA embedded phosphorothioate modified RNA for mercury ion determination. ACS Sens 3(3):624–631. https://doi.org/10.1021/acssensors.7b00892
Ye BC, Yin BC (2008) Highly sensitive detection of mercury (II) ions by fluorescence polarization enhanced by gold nanoparticles. Angew Chem Int Ed 47(44):8386–8389. https://doi.org/10.1002/anie.200803069
Liu S, Leng XQ, Wang X, Pei QQ, Cui XJ, Wang Y, Huang JD (2017) Enzyme-free colorimetric assay for mercury (II) using DNA conjugated to gold nanoparticles and strand displacement amplification. Microchim Acta 7(184):1969–1976. https://doi.org/10.1007/s00604-017-2182-7
Geng Z, Zhang H, Xiong Q, Zhang Y, Zhao H, Wang G (2015) A fluorescent chitosan hydrogel detection platform for the sensitive and selective determination of trace mercury (II) in water. J Mater Chem A 3(38):19455–19460. https://doi.org/10.1039/C5TA05610A
Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA (1997) Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277(5329):1078–1081. https://doi.org/10.1126/science.277.5329.1078
Li L, Li B, Qi Y, Jin Y (2009) Label-free aptamer-based colorimetric detection of mercury ions in aqueous media using unmodified gold nanoparticles as colorimetric probe. Anal Bioanal Chem 393(8):2051–2057. https://doi.org/10.1007/s00216-009-2640-0
Guo YM, Wang Z, Qu WS, Shao HW, Jiang XY (2011) Colorimetric detection of mercury, lead and copper ions simultaneously using protein-functionalized gold nanoparticles. Biosens Bioelectron 26(10):4064–4069. https://doi.org/10.1016/j.bios.2011.03.033
Chen YJ, Yao L, Deng Y, Pan DD, Ogabiela E, Cao JX, Adeloju SB, Chen W (2015) Rapid and ultrasensitive colorimetric detection of mercury (II) by chemically initiated aggregation of gold nanoparticles. Microchim Acta 182(13–14):2147–2154. https://doi.org/10.1007/s00604-015-1538-0
Liang GX, Wang L, Zhang HQ, Han ZX, Wu XX (2012) A colorimetric probe for the rapid and selective determination of mercury (II) based on the disassembly of gold nanorods. Microchim Acta 179:345–350. https://doi.org/10.1007/s00604-012-0882-6
Li K, Liang AH, Jiang CN, Li F, Liu QG, Jiang ZL (2012) A stable and reproducible nanosilver-aggregation-4-mercaptopyridine surface-enhanced Raman scattering probe for rapid determination of trace Hg2+. Talanta 99:890–896. https://doi.org/10.1016/j.talanta.2012.07.052
Funding
This research is supported by the Beijing Municipal Natural Science Foundation-Haidian Primitive Innovation Joint Fund Project (L182045).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 451 kb)
Rights and permissions
About this article
Cite this article
Xing, Y., Zhu, Q., Zhou, X. et al. A gold nanoparticle-based colorimetric mercury(II) biosensor using a DNA probe with phosphorothioate RNA modification and exonuclease III-assisted signal amplification. Microchim Acta 187, 214 (2020). https://doi.org/10.1007/s00604-020-4184-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-4184-0