Abstract
The objectives of this study were to analyze changes in gene expression and identify candidate genes and gene networks potentially inhibiting or triggering blossom-end rot (BER) in tomatoes treated with plant growth regulators. ‘Ace 55 (Vf)’ tomato plants were grown in a greenhouse and sprayed with Apogee (300 mg L−1), abscisic acid (ABA) (500 mg L−1), water (control), or gibberellins 4 + 7 (GA4 + 7) (300 mg L−1) weekly after pollination. The BER incidence rate was zero in Apogee- and ABA-, medium in water-, and high in GA4 + 7-treated plants from 26 to 40 days after pollination (DAP). At 26 DAP, healthy blossom-end fruit tissue still not showing visible BER symptoms was used for transcriptome analysis. Candidate genes potentially inhibiting or triggering BER were identified through a correlation analysis between gene expression levels at 26 DAP and BER incidence rate from 26 to 40 DAP. Genes inhibiting BER should be up-regulated in Apogee- and/or ABA-treated fruit and down-regulated in GA4 + 7-treated fruit. Genes triggering BER should be down-regulated in Apogee- and/or ABA-treated fruit and up-regulated in GA4 + 7-treated fruit. Most of the candidate genes inhibiting BER have functions leading to higher resistance to oxidative stress and toxic compounds, whereas most of the candidate genes triggering BER have functions leading to higher levels of oxidative stress and cell death. The results suggest that Apogee and ABA inhibited BER possibly by increasing fruit tissue resistance to reactive oxygen species (ROS) and other toxic compounds, whereas GA4 + 7 triggered BER possibly by increasing the levels of fruit oxidative stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although Ca2+ deficiency disorders have been known for a long time, studies have revealed that complex and conserved mechanisms are possibly regulating these disorders in different plant species (Saure 2005). Better understanding of the mechanisms triggering Ca2+ deficiency disorders in fruit will assist in selecting genotypes that are less susceptible to these disorders, as well as developing efficient control strategies for current cultivars.
In tomato fruit, blossom-end rot (BER) is believed to be triggered by a cell-localized Ca2+ deficiency that leads to plasma membrane damage, cell plasmolysis, and water-soaked tissue at the blossom-end region of the fruit that becomes dark-brown as cells die (Saure 2001; Suzuki and others 2003; Ho and White 2005; Freitas and others 2011b). Cell-localized Ca2+ deficiency may be triggered by mechanisms that reduce total fruit Ca2+ uptake and regulate cellular Ca2+ partitioning and distribution, which reduces Ca2+ binding to the plasma membrane, resulting in leaky membranes and cell death (Ho and White 2005; Freitas and others 2011b).
Plant growth regulators play important roles in Ca2+ deficiency disorder development in fruit tissue. Gibberellins (GAs) have been shown to trigger BER development in tomato fruit by reducing fruit Ca2+ uptake, decreasing apoplastic and water soluble Ca2+ concentration, and increasing plasma membrane leakage (Saure 2001, 2005; Freitas and others 2012a). Opposing results have been observed for abscisic acid (ABA) and a GA-biosynthesis inhibitor prohexadione, where these compounds prevented BER development by increasing fruit Ca2+ uptake, maintaining higher apoplastic and water soluble Ca2+ concentration, and decreasing membrane leakage (Freitas and others 2012a, 2014). Prohexadione inhibits active GAs biosynthesis by competing with the natural co-substrate, 2-oxoglutarate, at the active site of hydroxylases involved in the latter stages of the GA-biosynthesis pathway (Brown and others 1997). Although these studies suggest possible mechanisms by which GA and ABA control BER development in fruit tissue, the components of these mechanisms are largely unknown (Freitas and Mitcham 2012c). GA and ABA may control the expression of genes and gene networks that lead to independent and/or antagonistic responses that influence fruit susceptibility to BER.
In this study, a transcriptome approach was used to better understand the possible mechanisms triggering BER in response to GA and preventing BER development in response to ABA and the GA-biosynthesis inhibitor prohexadione. Our objectives were to analyze changes in gene expression and identify candidate genes and gene networks potentially inhibiting or triggering BER development in tomato fruit in response to growth regulators.
Materials and Methods
The fresh market tomato (Solanum lycopersicum. cultivar Ace 55 (Vf)) was grown from transplants in 9.5 L pots containing organic substrate (33.3% peat, 33.3% sand, 33.3% redwood compost with 2.6 kg dolomite lime m−3) in a greenhouse environment during the spring season at approximately 20 °C day and 18 °C night temperatures, without supplemental light. At full bloom, fully opened flowers were selected, tagged, and manually pollinated on each plant. Treatments started 1 day after manual pollination (DAP) by spraying the plants with solutions (200 mL plant−1) containing water (control), 300 mg L−1 GA4 + 7 (Typrus, Nufarm Americas, Burr Ridge, IL), 300 mg L−1 prohexadione-calcium (GA-biosynthesis inhibitor, Apogee®, BASF Corporation, Research Triangle Park, NC), or 500 mg L−1 ABA (Valent Biosciences, Libertyville, Illinois). Each solution contained 0.05% polysorbate 20 (Tween® 20) as a surfactant. There were four single-plant replications per treatment and evaluation time. Each replication contained 6–10 fruits. The treatments were applied every week to the same plants. Before initiating the treatments, the plants were irrigated once a day until the substrate in each plant was saturated with a nutrient solution containing N (7.2 mmol L−1), P (0.84 mmol L−1), K (3.1 mmol L−1), Ca (2.2 mmol L−1), Mg (1 mmol L−1), S (0.5 mmol L−1), Fe (0.03 mmol L−1), Mn (0.005 mmol L−1), Cu (0.003 mmol L−1), Zn (0.002 mmol L−1), B (0.024 mmol L−1), and Mo (0.00016 mmol L−1). On the day of manual pollination, 20 g of slow release fertilizer containing N (10.7 mol kg−1), P (2.9 mol kg−1), K (3 mol kg−1), Mg (0.41 mol kg−1), S (0.71 mol kg−1), Fe (0.08 mol kg−1), Mn (0.01 mol kg−1), Cu (0.008 mol kg−1), Zn (0.008 mol kg−1), B (0.018 mol kg−1), Mo (0.002 mol kg−1), but without Ca2+ (Osmocote Plus®, Scotts-Sierra Horticultural Products Co., Marysville, OH) was added to each plant pot. From this point on, the plants were irrigated once a day with deionized water only, as described previously (Freitas and others 2011a, 2012a, 2014). This approach was used to obtain fruit with low (Apogee and ABA), medium (water), and high (GA4 + 7) susceptibility to BER (Freitas and others 2011a, 2012a, 2014).
The relative BER incidence was calculated at 12, 26, and 40 DAP by multiplying the number of tagged fruit per plant with BER symptoms by 100, and dividing by the total number of tagged fruit per plant. The rate of BER incidence from 26 to 40 DAP was determined by subtracting the relative BER incidence at 40 DAP by the relative BER incidence at 26 DAP and dividing by 14, the number of days between 26 and 40 DAP. The results were expressed as the increase in percentage of BER incidence per day during this period (% day−1). The BER data were subjected to analysis of variance (ANOVA) following a completely randomized experimental design (SAS Institute 2002). The mean values were compared using Tukey’s test (p = 0.05) and are presented ± standard deviation (±SD).
Transcriptional analysis of gene expression was performed in four biological replications per treatment at 26 DAP, when the first symptoms of BER appeared in water- and GA4 + 7-treated fruit. Each biological replication included pericarp tissue sampled from the blossom-end region of healthy tomato fruit not showing visible BER symptoms. Total RNA was extracted following the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). The RNA concentration and purity were determined at 260 and 280 nm, respectively, using a UV spectrophotometer (NanoDrop 2000, Thermo Scientific, Wilmington, DE, USA). RNA quality was assessed by agarose gel electrophoresis. Total RNA (300 ng) was processed for the microarray hybridizations using the Affymetrix GeneChip 3′ IVT express kit. The resultant biotinylated cRNA was fragmented and hybridized to the GeneChip® Tomato Genome Array, which contains 10,209 tomato probe sets that interrogate more than 9200 Solanum lycopersicum genes. The arrays were washed, stained, and scanned at the Microarray Core Facility at the UC Davis Genome Center. The dChip software (http://www.dchip.org/dchip/) was used for background subtraction, normalization, and to compare samples. A t test was used to calculate p values, which were adjusted to cope with the problem of false discovery rates (Tusher and others 2001). Considering that BER incidence rate from 26 to 40 DAP was lowest in Apogee and ABA, intermediate in water (control), and highest in GA4 + 7-treated fruit, water treatment was used as the base level for gene expression analysis at 26 DAP. Expression values are presented as fold changes. Therefore, gene expression levels in Apogee-, ABA-, or GA4 + 7-treated fruit were presented as fold increase (+) or decrease (−) in relation to water (control)-treated fruit. Genes with adjusted p values ≤0.05 were considered to be differentially expressed between Apogee-, ABA-, or GA4 + 7-treated fruit and water (control)-treated fruit. The Affymetrix Tomato Genome Array functional annotations were updated according to the annotations in the Sol Genomics Network (SNG, http://www.solgenomics.net/), National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/), and UniProt (http://www.uniprot.org/) databases.
Candidate genes potentially regulating BER development in fruit tissue were identified according to the following criteria using water treatment as the base level for expression analysis: (1) gene must be statistically (p values ≤0.05) up- or down-regulated in Apogee- and/or ABA-, and GA4 + 7-treated fruit; (2) gene must have ≥ ± 0.10 fold change in expression in Apogee-, ABA-, or GA4 + 7-treated fruit; (3) gene up-regulated in treatment that inhibits BER (Apogee and ABA) must be down-regulated in treatment that triggers BER (GA4 + 7); and (4) gene down-regulated in treatment that inhibits BER (Apogee and ABA) must be up-regulated in treatment that triggers BER (GA4 + 7). After identifying the candidate genes, correlation analysis was performed between the expression levels at 26 DAP and the rate of increase in BER incidence from 26 to 40 DAP in fruit with low (Apogee and ABA), medium (water), and high (GA4 + 7) susceptibility to BER.
Candidate genes potentially inhibiting BER were identified according to the following criteria: (1) expression level at 26 DAP must be negatively correlated (−R2) to rate of increase in BER incidence from 26 to 40 DAP, meaning that expression levels at 26 DAP must be statistically up-regulated in Apogee and/or ABA (low susceptibility to BER) and down-regulated in GA4 + 7 (high susceptibility to BER)-treated tomato fruit, compared to water-treated fruit (medium susceptibility to BER). (2) Genes must have functional annotations potentially involved in mechanisms inhibiting BER symptom development.
Candidate genes potentially triggering BER were identified according to the following criteria: (1) expression level at 26 DAP must be positively correlated (+R2) to the rate of increase in BER incidence from 26 to 40 DAP, meaning that expression levels at 26 DAP must be statistically down-regulated in Apogee and/or ABA (low susceptibility to BER) and up-regulated in GA4 + 7 (high susceptibility to BER)-treated tomato fruit, compared to water-treated fruit (medium susceptibility to BER). (2). Genes must have functional annotations potentially involved in mechanisms leading to BER symptom development.
Quantitative Real-Time PCR (qRT-PCR) for genes potentially inhibiting or triggering BER was accomplished for each candidate gene to validate the GeneChip® Tomato Genome Array expression levels observed at 26 DAP. RNA extraction as well as concentration and quality analysis followed the same procedures described for the transcriptional analysis. Total RNA extracted was reverse transcribed with SuperScript III (Invitrogen, Carlsbad, CA, USA). qRT-PCR was then performed with the addition of 1x SYBR Green Mater Mix (Applied Biosystem, Foster City, CA, USA) to each sample containing about 100 ng of the synthesized cDNA. The data obtained were normalized based on the expression of the housekeeping tomato Clathrin adaptor complexes medium subunit (CAC) gene (González-Aguilera and others 2016). Target and housekeeping gene and primer information are presented in Supplemental Table 4. Primers were purified through standard desalting (Integrated DNA Technologies, Coralville, Iowa, USA). Expression level was determined as fold change for each gene, as described in the transcription analysis. Correlation analysis between GeneChip® Tomato Genome Array and qRT-PCR expression level for each candidate gene was accomplished to validate the GeneChip® Tomato Genome Array expression levels obtained at 26 DAP.
Results
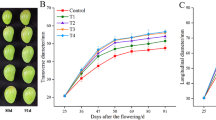
According to the results, BER incidence was higher in GA4 + 7-treated fruit followed by the water (control)-treated fruit during growth and development (Fig. 1). Apogee- and ABA-treated fruit showed no BER incidence during growth and development (Fig. 1). The rate of increase in BER incidence was higher in GA4 + 7 than in water-treated fruit from 26 DAP to 40 DAP (Fig. 2). Water-, GA4 + 7-, Apogee-, and ABA-treated fruit had 21.0 (±2.4), 19.9 (±2.4), 30.2 (±2.9), and 32.3 (±2.3) µg Ca2+ g−1 fresh weight at 40 DAP, respectively (data not shown). The correlation analysis between expression levels at 26 DAP and the rate of increase in BER incidence from 26 DAP to 40 DAP revealed 26 genes up-regulated in Apogee and down-regulated in GA4 + 7 (Fig. 3a, Supplemental Table 1), 23 genes down-regulated in Apogee and up-regulated in GA4 + 7 (Fig. 3b, Supplemental Table 1), 56 genes up-regulated in ABA and down-regulated in GA4 + 7 (Fig. 3c, Supplemental Table 2), 32 genes down-regulated in ABA and up-regulated in GA4 + 7 (Fig. 3d, Supplemental Table 2), 19 genes up-regulated in both Apogee and ABA and down-regulated in GA4 + 7 (Fig. 3e, Supplemental Table 3), and 39 genes down-regulated in both Apogee and ABA and up-regulated in GA4 + 7 (Fig. 3f, Supplemental Table 3).
Blossom-end rot incidence in ‘Ace 55 (Vf)’ tomato plants sprayed weekly with water (control), GA4 + 7, Apogee (GA-biosynthesis inhibitor prohexadione-Ca), or ABA. Mean values with different letters at each evaluation time are significantly different according to Tukey’s test (p = 0.05). Data shown, mean ± SD
Blossom-end rot incidence rate in ‘Ace 55 (Vf)’ tomato plants from 26 to 40 DAP. Plants were sprayed weekly with water (control), GA4 + 7, Apogee (GA-biosynthesis inhibitor prohexadione-Ca), or ABA. Mean values with different letters are significantly different according to Tukey’s test (p = 0.05). Data shown, mean ± SD
Gene expression changes in ‘Ace 55 (Vf)’ tomato fruit with low (Apogee and ABA), medium (water), or high (GA4 + 7) susceptibility to BER at 26 DAP. Genes presented are up-regulated (+) in Apogee and down-regulated (−) in GA4 + 7 (a), down-regulated (−) in Apogee and up-regulated (+) in GA4 + 7 treatments (b), up-regulated (+) in ABA and down-regulated (−) in GA4 + 7 (c), down-regulated (−) in ABA and up-regulated (+) in GA4 + 7 treatments (d), as well as up-regulated (+) in both Apogee and ABA and down-regulated (−) in GA4 + 7 (e), down-regulated (−) in both Apogee and ABA and up-regulated (+) in GA4 + 7 treatments (fb). Apogee and ABA (aa) expression level is represented by the average between Apogee and ABA treatments. Water treatment was used as the base level for gene expression analysis. Apogee-, ABA-, or GA4 + 7-regulated genes have expression levels ≥ ± 0.1 fold the expression observed in the water treatment at p value ≤0.05. Data show 26 Apogee up-regulated and GA4 + 7 down-regulated genes (a), 23 Apogee down-regulated and GA4 + 7 up-regulated genes (b), 56 ABA up-regulated and GA4 + 7 down-regulated genes (c), 32 ABA down-regulated and GA4 + 7 up-regulated genes (d), as well as 19 common Apogee and ABA (aa) up-regulated and GA4 + 7 down-regulated genes (e) and 39 common Apogee and ABA (aa) down-regulated and GA4 + 7 up-regulated genes (f)
Functional annotation showed that Apogee up- and GA4 + 7 down-regulated transcripts were represented by genes involved in protein metabolism and transport, cell wall metabolism, nutrient metabolism, carbohydrate metabolism, transcriptional regulation, hormone metabolism, and oxidation–reduction reactions (Table 1, Supplemental Table 1). Apogee down- and GA4 + 7 up-regulated transcripts were represented by genes involved in cell wall metabolism, transcription regulation, carbohydrate metabolism, cytoskeleton, protein metabolism, hormone metabolism, oxidation–reduction reactions, epidermal proteins, and aquaporins (Table 2, Supplemental Table 1). ABA up- and GA4 + 7 down-regulated transcripts were represented by genes involved in protein metabolism and transport, cell wall metabolism, nutrient metabolism, carbohydrate metabolism, transcription regulation, hormone metabolism, oxidation–reduction reactions, lipid metabolism, defense responses, epicuticular wax, aquaporins, cell cycle, and nucleoside diphosphate hydrolysis (Table 1, Supplemental Table 2). ABA down- and GA4 + 7 up-regulated transcripts were represented by genes involved in protein metabolism and transport, cell wall metabolism, nutrient metabolism, carbohydrate metabolism, transcription regulation, hormone metabolism, lipid metabolism, and storage proteins (Table 2, Supplemental Table 2). Common Apogee and ABA up- and GA4 + 7 down-regulated transcripts were represented by genes involved in protein metabolism and transport, cell wall metabolism, nutrient metabolism, carbohydrate metabolism, transcription regulation, hormone metabolism, lipid metabolism, defense responses, detoxification, and cell cycle (Table 1, Supplemental Table 3). Common Apogee and ABA down- and GA4 + 7 up-regulated transcripts were represented by genes involved in protein metabolism and transport, cell wall metabolism, nutrient metabolism, transcription regulation, hormone metabolism, oxidation–reduction reactions, lipid metabolism, membrane proteins, stress proteins, defense response, aquaporins, cell cycle, storage proteins, phosphatases, amino acid biosynthesis, and triterpenoid biosynthesis (Table 2, Supplemental Table 3).
The analysis of genes potentially regulating BER development revealed that candidate genes inhibiting BER were involved in transcription regulation (SRO2, DREB, MAF1, ERF109, ERF010, CBF1, NAC-NOR), hormone metabolism (MeJA- esterase, WAT1, AREB, PYL9, TAS14), oxidation–reduction reactions (TIC32, Class-III ADH), protein metabolism (FBXL2-like, 2 PIIF, SRP 7s, UBLCP1, 2 CIPK, RABA5a, DNAJC2, SDF2, PP2-A13, MSR-B5), lipid metabolism (2 GDSL esterases/lipases, AAE7, LTP1), defense responses (2 JRL 19, STO, PRP6), detoxification (LGL), and nutrient metabolism (TAT, Td, MTP1, CCH-like) (Fig. 4; Table 3). Candidate genes triggering BER development were involved in transcription regulation (ZAT11 and ZAT12), hormone metabolism (TIFY10-like), oxidation–reduction reaction (Rboh), protein metabolism (SOBIR1), and cell wall metabolism (PME 3) (Fig. 4; Table 4). The expression levels of each gene potentially inhibiting or triggering BER obtained with the GeneChip® Tomato Genome Array were highly positively correlated with the expression levels obtained for each gene using RT-PCR (Table 5).
Discussion
According to the rate of increase in BER incidence observed from 26 to 40 DAP, susceptibility to BER was lower in Apogee- and ABA-, medium in water-, and highest in GA4 + 7-treated fruit. Therefore, these treatments provided an efficient way to manipulate fruit susceptibility to BER in order to identify candidate genes and gene networks regulating or responding to BER development.
Candidate Genes Potentially Inhibiting BER
Transcription Regulation
Among genes up-regulated in Apogee and down-regulated in GA4 + 7, the polymerase similar to radical-induced cell death-one 2 (SRO2) is required for plant defenses against salt and oxidative stresses (Borsani and others 2005; Jaspers and others 2009). Transcription factors up-regulated in ABA and down-regulated in GA4 + 7 include the dehydration-responsive element-binding protein (DREB) that triggers abiotic stress tolerance in many plant species (Gupta and others 2014); the repressor of RNA polymerase III transcription MAF1 that is highly expressed to overcome oxidative stress and growth-limiting conditions (Boguta 2013); ethylene-responsive factors (ERF109 and ERF010) that have been shown to control the expression of important stress-responsive genes required for plant response to a range of abiotic and biotic stresses (Singh and others 2002); and the C repeat/dehydration-responsive element-binding factor 1 (CBF1) that has been shown to enhance tomato plant resistance to water deficit stress (Hsieh and others 2002). Another transcription factor gene up-regulated in both Apogee and ABA-treated fruit and down-regulated in GA4 + 7-treated fruit included the NAC-NOR that is involved in developmental programs, plant defense, and abiotic stress responses to dehydration and salinity (Olsen and others 2005). The NAC domain was identified based on consensus sequences from Petunia NAM and Arabidopsis ATAF1/2 and CUC2 proteins (the domain was named from their first letters of the genes) (Aida and others 1997). Studies have shown that NAC expression is induced by ROS and its overexpression triggers stress-related genes involved in plant drought and salt tolerance (Fujita and others 2004; Zheng and others 2009). NAC proteins also trigger the expression of glyoxalase I family genes that catalyze the first step in the glyoxalase system (Fujita and others 2004), a critical two-step detoxification system for methylglyoxal that is produced naturally as a byproduct of normal metabolism, but is highly toxic due to its reactions with proteins, nucleic acids, and other cellular components (Veena and Sopory 1999). These results indicate that Apogee and ABA triggered the expression of a range of transcription factors up-regulating gene networks involved in stress responses that resulted in fruit tissue becoming more resistant to BER compared to GA4 + 7-treated fruit. Other studies suggested that fruit susceptibility to BER is mainly dependent on its ability to deal with reactive radicals naturally produced in response to stresses such as salinity, drought, high light intensity, heat, and ammonia nutrition (Ho and White 2005; Saure 2014). Bioactive GAs may increase fruit susceptibility to stress and BER development, while Apogee and ABA could have the opposite effect (Saure 2014).
Hormones
Genes up-regulated in ABA and down-regulated in GA4 + 7 code for methyl jasmonate esterase (MeJA- esterase), vacuolar auxin transport facilitator (WAT1), ABA-responsive element-binding protein (AREB), and ABA receptor PYL9. MeJA-esterase hydrolyzes methyl jasmonate to jasmonic acid (Wu and others 2008) that is involved in plant responses to biotic and abiotic stresses (Mithöfer and others 2004), suggesting a crosstalk communication between ABA, GA4 + 7, and jasmonic acid that regulates stress responses and fruit susceptibility to BER. The WAT1 gene is known to be involved in cellular auxin homeostasis (Ranocha and others 2013). Auxin distribution at the cellular level is known to trigger specific cellular responses, some of which may regulate fruit resistance to BER (Saure 2001, 2005). The AREB gene family plays a critical role in plant responses to abiotic stresses and enhancing drought and salt tolerance (Li and others 2014). The ABA receptor PYL9 is required for ABA-mediated responses through inhibition of the activity of group-A protein phosphatase type 2Cs (PP2Cs), and its expression confers enhanced sensitivity to ABA (Nakagawa and others 2014). The observed high or low PYL9 expression in response to ABA or GA4 + 7 is probably a crosstalk mechanism to enhance or reduce fruit tissue sensitivity to ABA and susceptibility to BER, respectively. A candidate gene up-regulated in both Apogee and ABA and down-regulated in GA4 + 7 is the ABA and environmental stress-inducible dehydrin gene TAS14, which increases in expression upon plant exposure to osmotic stress and ABA (Godoy and others 1990) and may be involved in cellular responses to abiotic stress conditions (Godoy and others 1994; Parra and others 1996). The results show that most of the hormone-related genes potentially inhibiting BER are involved in biotic and abiotic stress responses. It is possible that these hormone-related genes are part of the transcription factor’s stress-related responses to inhibit BER development, as described above, or upstream as part of the response to the signals that activated different mechanisms inhibiting BER. Our results also suggest that ABA treatment increased fruit tissue sensitivity to ABA, possibly enhancing ABA responses related and not related with the inhibition of BER development.
Oxidation–Reduction
Genes up-regulated in ABA and down-regulated in GA4 + 7 included the dehydrogenase TIC32 and the alcohol dehydrogenase class-3-like (Class-III ADH). The TIC32 is part of a translocon involved in protein import into the chloroplast (Hörmann and others 2004). Studies attempting to knockout TIC32 activity resulted in early seed abortion, suggesting that TIC32 plays an important role in cell metabolism (Hörmann and others 2004). Therefore, the observed negative correlation between TIC32 expression and the rate of increase in BER incidence suggests that down-regulation in TIC32 expression could possibly be involved in the increase in fruit susceptibility to BER. The Class-III ADH proteins play an important role in formaldehyde detoxification in plants (Achkor and others 2003). Formaldehyde is considered a toxic compound because of its ability to react with proteins, nucleic acids, and lipids (Achkor and others 2003). It is possible that down-regulation of Class-III ADH in tomato could favor BER development in the fruit.
Protein Metabolism
Genes up-regulated in Apogee and down-regulated in GA4 + 7 included F-box/LRR-repeat protein 2-like (FBXL2-like), wound-induced proteinase inhibitor I prepropeptide (PIIF), and signal recognition particle 7s (SRP 7s). FBXL2-like is part of a ubiquitin protein ligase complex that functions in phosphorylation-dependent ubiquitination. Negative correlation between FBXL2-like levels and BER incidence suggests that ubiquitination regulated by the FBXL2-like protein may play a role in determining fruit susceptibility to BER, possibly by affecting targeted protein degradation via the proteasome, changing protein cellular location, affecting their activity, and/or promoting or inhibiting protein interactions (Mukhopadhyay and Riezman 2007). Down-regulation of PIIF suggests a higher proteinase activity in fruit with higher susceptibility to BER, which could lead to unwanted levels of protein breakdown, cell death, and BER symptoms in fruit tissue. The SRP 7s is a ribonucleoprotein that directs the traffic of proteins within the cell and allows them to be secreted through translocation channels in the plasma membrane (Noriega and others 2014). The negative correlation between SRP 7s levels and BER incidence suggests that down-regulation of protein trafficking and secretion into the apoplast could have affected membrane and/or other apoplastic metabolism, leading to higher fruit susceptibility to BER.
Genes up-regulated in ABA and down-regulated in GA4 + 7 included ubiquitin-like domain-containing CTD phosphatase (UBLCP1), two calcineurin B-like molecules (CBL), interacting protein kinase 1 and 2 (CIPK1 and 2), RAB GTPase homolog A5A (RABA5a), and dnaJ homolog subfamily C member 2-like (DNAJC2). UBLCP1 dephosphorylates 26S nuclear proteasomes, thereby decreasing their proteolytic activity that may prevent assembly of the core and regulatory particles into mature 26S proteasomes (Zheng and others 2004). Knockdown of UBLCP1 in cells promotes 26S proteasome assembly and selectively enhances nuclear proteasome activity (Zheng and others 2004), potentially leading to protein breakdown that could increase fruit susceptibility to BER. CIPKs have been shown to be involved in acquired tolerance and acclimation under environmental stresses such as salinity, drought, and chilling (Yuasa and others 2012), which also suggests a potential role in reducing fruit susceptibility to BER under stress conditions. RABA5a belongs to the Rab protein family involved in intracellular vesicle trafficking and protein transport. Studies have shown that high expression of the Rab gene AtRabG3e increased plant tolerance to salt and osmotic stresses and reduced accumulation of reactive oxygen species in plants (Mazel and others 2004). These results imply that vesicle trafficking plays an important role in plant adaptation to stress, beyond the housekeeping function in intracellular vesicle trafficking (Mazel and others 2004). The DNAJC2 homolog subfamily C member 2-like acts both as a chaperone and chromatin regulator (Kaschner and others 2015), suggesting that proper protein folding and chromatin structure can also help the fruit to avoid BER development.
Candidate genes up-regulated in both Apogee and ABA and down-regulated in GA4 + 7 included the stromal cell-derived factor 2-like (SDF2), F-box PP2-A13-like (PP2-A13), and methionine sulfoxide reductase B5-like (MSR-B5). SDF2 is involved in endoplasmic reticulum (ER) protein quality control and unfolded protein response (UPR) triggered under stress conditions (Schott and others 2010). Under normal growth conditions, SDF2 is highly expressed in fast growing, differentiating cells and meristematic tissues that require enhanced protein biosynthesis and secretion (Schott and others 2010). Therefore, down-regulation of the SDF2 gene in GA4 + 7-treated fruit could limit protein biosynthesis and secretion at the time of fast growth in distal tissues, triggering cell damage and BER symptoms. PP2-A13 is a component of ASK-cullin-F-box E3 ubiquitin ligase complexes that may mediate the ubiquitination of target proteins (Dinant and others 2003). In plants, many physiological processes such as hormone and defense responses employ F-box protein to direct negative regulators to the ubiquitin-mediated degradation pathway (del Pozo and Estelle 2000). In this case, GA4 + 7 down-regulation of PP2-A13 possibly resulted in metabolic processes that enhanced fruit susceptibility to BER. MSR-B5 catalyzes the reduction of methionine sulfoxide to methionine in proteins, playing a protective role against oxidative stress by restoring activity to proteins that have been inactivated by methionine oxidation (Rouhier and others 2006). As previously mentioned, down-regulation of oxidative stress-alleviating mechanisms in GA4 + 7-treated fruit potentially enhanced BER symptom development.
Lipid
Lipid-related genes up-regulated in ABA and down-regulated in GA4 + 7 code for two GDSL esterases/lipases and one acetate/butyrate-CoA ligase (AAE7). GDSL esterase/lipases have been reported to be up-regulated by biotic and abiotic stresses, nutrient deficiency, as well as chemical and hormonal treatments (Chepyshko and others 2012). The enzymes have hydrolytic activity and are mainly involved in the regulation of plant development, morphogenesis, synthesis of secondary metabolites, and defense responses (Chepyshko and others 2012). Therefore, the negative correlation between GDSL esterases/lipases expression and fruit susceptibility to BER suggests that these genes are possibly triggering defense responses and/or synthesis of secondary metabolites that can inhibit BER development in the fruit.
A candidate lipid-related gene up-regulated in both Apogee and ABA treatments and down-regulated in GA4 + 7 was the Lipid Transfer Protein 1 (LTP1). Although the mechanisms are still not well understood, high LTP expression has been reported to increase plant tolerance to drought, salt, and elevated H2O2 levels (Kader 1996; Guo and others 2013; Safi and others 2015). Considering that BER incidence is also enhanced by such plant stress conditions, high LTP expression can potentially lead to inhibition of BER incidence in the fruit.
Defense Response
Genes up-regulated in ABA and down-regulated in GA4 + 7 included two jacalin-related lectin 19s (JRL 19) and a salt tolerance protein (STO). The JRLs are carbohydrate binding proteins involved in signaling pathways required to overcome plant environmental stress conditions (Song and others 2014), and JRL genes have been associated with abiotic stress tolerance, such as salt stress, or even responses to multiple stresses (Claes and others 1990; Garcia and others 1998; Zhang and others 2000; Filho and others 2003; Subramanyam and others 2006). Studies have also suggested that JRLs are a part of the salicylic acid, jasmonic acid, and ABA-dependent defense signaling pathways (Xiang and others 2011; Song and others 2014). STO is a B-box type Zn finger protein reported to confer salt tolerance in plants grown in high-salt medium (Lagercrantz and Axelsson 2000; Nagaoka and Takano 2003; Griffiths and others 2003). The STO protein interacts with CEO1/RCD1, a protein required to protect against oxidative damage (Belles-Boix and others 2000; Fujibe and others 2004). Therefore, the observed JRL 19 and STO up-regulation in ABA-treated fruit and down-regulation in GA4 + 7-treated fruit suggests cross communication between these growth regulators leading to defense response pathways inhibiting and triggering BER in the fruit, respectively.
A gene coding for a pathogenesis-related protein 6 (PRP6) was up-regulated in Apogee- and ABA-treated fruit and down-regulated in GA4 + 7-treated fruit. The PRP proteins have been reported to be up-regulated in response to pathogen infection leading to plant stress defense responses (El-Kereamy and others 2011), suggesting that PRP6 up-regulation in fruit with lower susceptibility to BER could have activated defense mechanisms inhibiting BER development.
Detoxification
A gene up-regulated in both Apogee- and ABA-treated fruit and down-regulated in GA4 + 7-treated fruit was the lactoylglutathione lyase-like (LGL), also known as Glyoxalase I (Gly I), which is a detoxifying enzyme required for the glutathione-based detoxification of methylglyoxal, formed primarily as a byproduct of carbohydrate and lipid metabolism (Singla-Pareek and others 2003). Methylglyoxal is a potent mutagenic and cytotoxic compound known to arrest growth, react with DNA and protein, and increase sister chromatid exchange (Singla-Pareek and others 2003). Up-regulation of Gly I has been shown to take place in tomato in response to salt, osmotic stress, and phytohormonal stimuli (Espartero and others 1995), as well as to confer salinity and drought stress tolerance in plants (Singla-Pareek and others 2003; Bhomkar and others 2008; Alam and others 2014).
Nutrient
Genes up-regulated in Apogee and down-regulated in GA4 + 7 included a tyrosine aminotransferase (TAT) and a threonine deaminase (Td). TAT proteins have been reported to be involved in tocopherol synthesis (Riewe and others 2012), which are important antioxidants and scavengers of lipid radicals and ROS in plants (Holländer-Czytko and others 2005). The Td catalyzes the conversion of threonine to 2-ketobutyrate that is used to synthesize isoleucine, which together with other branched-chain amino acids are thought to play an important role as osmolytes in plant abiotic stress tolerance (Joshi and others 2010).
A gene up-regulated in ABA and down-regulated in GA4 + 7 was the metal tolerance protein 1 (MTP1), a vacuolar cation efflux transporter shown to play an essential role in metal homeostasis and tolerance in plants (Blaudez and others 2003; Lin and Aarts 2012).
A gene up-regulated in both Apogee and ABA and down-regulated in GA4 + 7 was the copper (Cu) transport protein Copper Chaperone-like (CCH-like), which is required to maintain proper cellular copper homeostasis and avoid toxicity (Shin and others 2012). Intracellular Cu must be accurately regulated to avoid toxicity caused by the free Cu ion that can generate ROS such as superoxide, hydrogen peroxide, and hydroxyl radicals that damage proteins, lipids, and DNA (Brewer 2010). Therefore, higher expression of the Cu transport protein Cu chaperone-like in response to Apogee and ABA may have favored cellular Cu homeostasis, inhibiting cell damage and BER development, as compared to GA4 + 7-treated fruit.
Candidate Genes Potentially Triggering BER
Transcriptional Regulation
Candidate transcription factors down-regulated in Apogee and up-regulated in GA4 + 7, as well as down-regulated in both Apogee and ABA and up-regulated in GA4 + 7 were the putative zinc fingers ZAT11-like and ZAT12-like, respectively. The zinc finger proteins ZAT11 and ZAT12 are part of a gene family known to be involved in an intricate oxidative stress-induced programed cell death (PCD) network in plants (Qureshi and others 2013). Therefore, high ZAT11-like and ZAT12-like expressions can potentially be involved in PCD leading to BER symptoms in water- and GA4 + 7-treated fruit with higher susceptibility to BER.
Hormone
A gene down-regulated in Apogee- and ABA-treated fruit and up-regulated in GA4 + 7-treated fruit was the putative TIFY10A-like gene coding for a protein that represses JA responses (Grunewald and others 2009). JA is required for appropriate plant stress responses and an increase in TIFY expression can lead to impaired cellular response and potentially cell death (Mithöfer and others 2004). Expression of the Arabidopsis JA signaling repressor TIFY10A has been shown to be stimulated by auxin (Grunewald and others 2009), suggesting that high levels of auxin could have inhibited the JA stress response pathway in fruit with higher BER susceptibility.
Oxidation–Reduction
A gene coding for the Respiratory Burst Oxidase Homolog (Rboh) was down-regulated in Apogee-treated fruit and up-regulated in GA4 + 7-treated fruit. Although studies have suggested that ROS produced by Rboh play a role in abiotic stress responses (Sagi and others 2004; Torres and Dangl 2005), high Rboh expression can potentially increase fruit susceptibility to BER by stimulating ROS accumulation and cell death in fruit tissue. Indeed, ROS accumulation leads to tissue oxidative stress and cell death in plants (Dat and others 2003; Van Breusegem and Dat 2006). For example, transgenic plants with low levels of antioxidants and mutants unable to stop the initiation or propagation of ROS-driven cell death have high susceptibility to cell death events (Mittler and Rizhsky 2000; Lorrain and others 2003). It has been suggested that when ROS levels exceed their cellular scavenging capacity, ROS build up to phytotoxic levels that attack and damage proteins, lipids, and DNA, resulting in cell death and necrotic tissue phenotypes (Van Breusegem and Dat 2015).
Our results also indicate that the l -ascorbate oxidase (AO) homolog gene was down-regulated in Apogee-treated fruit and up-regulated in GA4 + 7-treated fruit. The AO enzyme converts reduced ascorbic acid into oxidized ascorbic acid (Zhang and others 2011). Because reduced ascorbic acid is an important antioxidant to counteract oxidative damage by scavenging free radicals and reactive oxygen species (ROS), increased ascorbic acid oxidation by high AO expression can increase fruit tissue susceptibility to oxidative stress, cell death, and possibly BER. Accordingly, studies have shown that transgenic plants with suppressed AO expression have higher levels of reduced ascorbic acid, which increases plant resistance to oxidative stress under high drought and salt conditions (Yamamoto and others 2005; Stevens and others 2007; Zhang and others 2011).
Protein Metabolism
An ABA down- and GA4 + 7 up-regulated gene possibly triggering BER was the leucine-rich repeat receptor-like protein kinase SOBIR1, whose overexpression activates cell death, whereas mutation in the gene suppresses cell death in plants (Gao and others 2009; Liu and others 2013). In that case, high SOBIR1 expression in GA4 + 7-treated fruit can potentially increase tissue susceptibility to cell death leading to BER symptoms.
Cell Wall
A putative pectin methylesterase 3 (PME 3) was down-regulated in both Apogee and ABA and up-regulated in GA4 + 7. High PME expression has been reported to increase Ca2+ binding to the cell wall, reducing Ca2+ availability to other cellular functions and increasing fruit susceptibility to BER (Freitas and others 2012b). Therefore, the high PME 3 expression may help explain higher fruit susceptibility to BER in response to GA4 + 7 treatment; however, PME 3 and PME1.9 showed opposite changes in expression suggesting differential regulation of PME isoforms during BER development.
Mechanisms Inhibiting and Triggering BER
The identified candidate genes potentially inhibiting BER have widely different cellular functions, including transcription regulation, hormone metabolism, oxidation–reduction reactions, protein metabolism, lipid metabolism, defense responses, cell detoxification, and nutrient metabolism. Although the candidate genes inhibiting BER had a wide range of cellular functions, most of the identified genes have functional annotations related with an increase in fruit tissue resistance to oxidative stress and toxic compounds. Therefore, comparing fruit with higher and lower susceptibility to BER, it was possible to conclude that mechanisms inhibiting BER development were possibly involved in increasing fruit tissue resistance to oxidative stress and toxic compounds. Indeed, studies have shown that fruit susceptibility to BER is enhanced by abiotic stress conditions, such as salinity, drought, high light intensity, heat, and ammonia nutrition (Ho and White 2005), which increase ROS and other toxic compounds that eventually can lead to cell death and BER symptoms (Saure 2014).
The identified candidate genes potentially triggering BER are involved in transcription regulation triggering programmed cell death, repression of JA responses possibly leading to cell death, increases in production of ROS, conversion of reduced ascorbic acid into oxidized ascorbic acid, and increases in cell wall bound Ca2+. Accordingly, since most of the genes inhibiting BER are involved in oxidative stress response, it was expected that most of the genes triggering BER would be possibly leading to higher levels of fruit oxidative stress. The production of reactive oxygen species takes place naturally during normal cell metabolism and can be enhanced by adverse environmental conditions such as drought, salt and osmotic stresses, and high temperatures (Saure 2014). Therefore, cells have developed different mechanisms to overcome oxidative stresses. In that case, the main difference observed between a fruit with higher and lower susceptibility to BER seems to be related to mechanisms triggered to overcome or increase oxidative stress in the fruit. These results agree with other studies suggesting that bioactive GAs not only can reduce Ca2+ accumulation in the fruit, but also increase tissue susceptibility to oxidative stress and the risk of BER, while GA inhibitors may have the opposite effect (Saure 2014). These results also agree with other studies showing higher ROS levels in fruit tissue with BER symptoms (Aktas and others 2005; Turhan and others 2006; Mestre and others 2012).
Calcium deficiency disorders in plants are believed to be triggered by mechanisms leading to cell membrane damage (Saure 2014). ROS are known to disintegrate cellular membranes (Mestre and others 2012), whereas Ca2+ has been shown to protect cellular membranes by binding to phospholipids and proteins on the membranes and by increasing the activity of enzymes leading to ROS detoxification (Mestre and others 2012). In our study, treating the plants from 0 to 40 DAP with water, GA4 + 7, Apogee, or ABA resulted in 21.0 (±2.4), 19.9 (±2.4), 30.2 (±2.9), or 32.3 (±2.3) µg Ca2+ g−1 fruit fresh weight, respectively. These results suggest that Apogee and ABA inhibited BER development in tomato fruit by increasing fruit Ca2+ content and possibly by increasing fruit tissue resistance to ROS and other toxic compounds. Water- and GA4 + 7-treated fruit had similar Ca2+ content, but different susceptibility to BER, suggesting that GA4 + 7-treated fruit had higher susceptibility to BER possibly due to higher levels of fruit oxidative stress. These results suggest that neither Ca2+ nor ROS alone fully explain BER development, but there is possibly an interaction between Ca2+ and ROS concentrations in fruit tissue involved in BER susceptibility. This interaction may also be influenced by mechanisms regulating cellular Ca2+ and ROS partitioning, which can change the cellular Ca2+/ROS interaction in the tissue (Turhan and others 2006; Freitas and others 2011b, 2012a, b). In that case, future studies should determine the role of Ca2+/ROS interactions at different cellular locations on BER development.
It is very possible that not all the genes inhibiting and triggering BER in tomato fruit are present on the GeneChip® Tomato Genome Array, which contains 10,209 tomato probe sets that interrogate more than 9200 Solanum lycopersicum genes. Therefore, more detailed studies can be accomplished using whole-genome sequencing technologies to explore a wider range of genes potentially regulating fruit susceptibility to BER. Studies with transgenic plants containing knockouts or up-regulated candidate genes can help identify the key genes regulating BER development in fruit tissue. Identified genes regulating BER can then be used as markers for the selection of new tomato cultivars less susceptible to BER in breeding programs. Since the mechanisms leading to Ca2+ deficiency disorders are believed to be highly conserved across different plant species, identified key genes regulating BER can be studied in other plant species and also used to select other crop plants more resistant to these physiological disorders.
References
Achkor H, Díaz M, Fernández MR, Biosca JA, Parés X, Martínez MC (2003) Enhanced formaldehyde detoxification by overexpression of glutathione-dependent formaldehyde dehydrogenase from Arabidopsis. Plant Physiol 132:2248–2255
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Aktas H, Karni L, Chang DC, Turhan E, Bar-Tal A, Aloni B (2005) The suppression of salinity-associated oxygen radicals production in pepper (Capsicum annuum) fruit by manganese, zinc and calcium in relation to its sensitivity to blossom-end rot. Physiol Plant 123:67–74
Alam MM, Nahar K, Hasanuzzaman M, Fujita M (2014) Exogenous jasmonic acid modulates the physiology, antioxidant defense and glyoxalase systems in imparting drought stress tolerance in different Brassica species. Plant Biotechnol Rep 8:279–293
Belles-Boix E, Babiychuk E, Van Montagu M, Inze D, Kushnir S (2000) CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett 482:19–24
Bhomkar P, Upadhyay CP, Saxena M, Muthusamy A, Prakash NS, Pooggin M, Hohn T, Sarin NB (2008) Salt stress alleviation in transgenic Vigna mungo L. Hepper (blackgram) by overexpression of the glyoxalase I gene using a novel Cestrum yellow leaf curling virus (CmYLCV) promoter. Mol Breeding 22:169–181
Blaudez D, Kohler A, Martin F, Sanders D, Chalot M (2003) Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 15:2911–2928
Boguta M (2013) Maf1, a general negative regulator of RNA polymerase III in yeast. BBA-Gene Regul Mech 1829:376–384
Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123:1279–1291
Brewer GJ (2010) Copper toxicity in the general population. Clin Neurophysiol 121:459–460
Brown RGS, Kawaide K, Yang YY, Rademacher W, Kamiya Y (1997) Daminozide and prohexadione have similar modes of action as inhibitors of the late stages of gibberellin metabolism. Physiol Plant 101:309–313
Chepyshko H, Lai CP, Huang LM, Liu JH, Shaw JF (2012) Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: new insights from bioinformatics analysis. BMC Genomics 13:309–328
Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, Caplan A (1990) Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 2:19–27
Dat JF, Pellinen R, Beeckman T, Van De Cotte B, Langebartels C, Kangasjarvi J, Inze´ D, Van Breusegem F (2003) Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J 33:621–632
del Pozo JC, Estelle M (2000) F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol Biol 44:123–128
Dinant S, Clark AM, Zhu Y, Vilaine F, Palauqui JC, Kusiak C, Thompson GA (2003) Diversity of the superfamily of phloem lectins (Phloem Proein 2) in Angiosperms. Plant Physiol 131:114–128
El-Kereamy A, El-Sharkawy I, Ramamoorthy R, Taheri A, Errampalli D, Kumar P, Jayasankar S (2011) Prunus domestica pathogenesis-related protein-5 activates the defense response pathway and enhances the resistance to fungal infection. PLoS ONE 6:1–11
Espartero J, Sanchez-Aguayo I, Pardo JM (1995) Molecular characterization of glyoxalase-I from a higher plant; upregulation by stress. Plant Mol Biol 29:1223–1233
Filho GAS, Ferreira BS, Dias JM, Queiroz KS, Branco AT, Bressan-Smith RE, Oliveira JG, Garcia AB (2003) Accumulation of SALT protein in rice plants as a response to environmental stress. Plant Sci 164:623–628
Freitas ST, Mitcham EJ (2012c) Factors involved in fruit calcium deficiency disorders. Hortic Rev 40:107–146
Freitas ST, Shackel KA, Mitcham EJ (2011a) Abscisic acid triggers whole-plant and fruit-specific mechanisms to increase fruit calcium uptake and prevent blossom end rot development in tomato fruit. J Exp Bot 62:2645–2656
Freitas ST, Padda M, Wu Q, Park S, Mitcham EJ (2011b) Dynamic alterations in cellular and molecular components during blossom-end rot development in tomatoes expressing sCAX1, a constitutively active Ca2+/H+ antiporter from Arabidopsis. Plant Physiol 156:844–855
Freitas ST, Jiang CZ, Mitcham EJ (2012a) Mechanisms involved in calcium deficiency development in tomato fruit in response to gibberellins. J Plant Growth Regul 31:221–234
Freitas ST, Handa AK, Wu Q, Park S, Mitcham EJ (2012b) Role of pectin methylesterase in cellular calcium distribution and blossom-end rot development in tomato fruit. Plant J 71:824–835
Freitas ST, McElrone AJ, Shackel KA, Mitcham EJ (2014) Calcium partitioning and allocation and blossom-end rot development in tomato plants in response to whole-plant and fruit-specific abscisic acid treatments. J Exp Bot 65:235–247
Fujibe T, Saji H, Arakawa K, Yabe N, Takeuchi Y, Yamamoto KT (2004) A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol 134:275–285
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, Zhang Y (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6:34–44
Garcia AB, Engler JA, Claes B, Villarroel R, Van Montagu M, Gerats T, Caplan A (1998) The expression of the salt-responsive gene salT from rice is regulated by hormonal and developmental cues. Planta 207:172–180
Godoy JA, Pardo JM, Pintor-Toro JA (1990) A tomato cDNA inducible by salt stress and abscisic acid: nucleotide sequence and expression pattern. Plant Biol 15:695–705
Godoy JA, Lunar R, Torres-Schumann S, Moreno J, Rodrigo RM, Pintor-Toro JA (1994) Expression, tissue distribution and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Mol Biol 26:1921–1934
González-Aguilera KL, Saad CF, Montes RAC, Alves-Ferreira M, Folter S (2016) Selection of reference genes for quantitative real-time RT-PCR studies in tomato fruit of the genotype MT-Rg1. Front Plant Sci 7:1–8
Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131:1855–1867
Grunewald W, Vanholme B, Pauwels L, Plovie E, Inzé D, Gheysen G, Goossens A (2009) Expression of the Arabidopsis jasmonate signaling repressor JAZ1/TIFY10A is stimulated by auxin. Eur Mol Biol Organ J 10:923–928
Guo L, Yang H, Zhang X, Yang S (2013) Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J Exp Bot 64:1755–1767
Gupta K, Jha B, Agarwal K (2014) A dehydration-responsive element binding (BREB) transcription factor from the succulent Halophyte Salicornia brachiate enhances abiotic stress tolerance in transgenic tobacco. Mar Biotechnol 16:657–673
Ho LC, White PJ (2005) A cellular hypothesis for the induction of blossom-end rot in tomato fruit. Ann Bot 95:571–581
Holländer-Czytko H, Grabowski J, Sandorf I, Weckermann K, Weiler EW (2005) Tocopherol content and activities of tyrosine aminotransferase and cystine lyase in Arabidopsis under stress conditions. J Plant Physiol 162:767–770
Hörmann F, Küchler M, Sveshnikov D, Oppermann U, Li Y, Soll J (2004) Tic32, an essential component in chloroplast biogenesis. J Biol Chem 279:34756–34762
Hsieh TH, Lee JT, Charng YY, Chan MT (2002) Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol 130:618–626
Jaspers P, Blomster T, Brosché M, Salojärvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F, Overmyer K, Kangasjärvi J (2009) Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 60:268–279
Joshi V, Joung JG, Fei Z, Jander G (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39:933–947
Kader JC (1996) Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47:627–654
Kaschner LA, Sharma R, Shrestha OK, Meyer A, Craig EA (2015) A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with ribosome. BBA-Mol Cell Res 1853:1035–1045
Lagercrantz U, Axelsson T (2000) Rapid evolution of the family of CONSTANS LIKE genes in plants. Mol Biol Evol 17:1499–1507
Li C, Yue J, Wu X, Xu C, Yu J (2014) An ABA-responsive DER-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J Exp Bot 65:5415–5427
Lin YF, Aarts MG (2012) The molecular mechanism of zinc and cadmium stress response in plants. Cell Mol Life Sci 69:3187–3206
Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y (2013) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol 161:2146–2158
Lorrain S, Vailleau F, Balagué C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8:263–271
Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134:118–128
Mestre TC, Garcia-Sanchez F, Rubio F, Martinez V, Rivero RM (2012) Glutathione homeostasis as an important and novel factor controlling blossom-end rot development in calcium-deficient tomato fruits. J Plant Physiol 169:1719–1727
Mithöfer A, Schulze B, Boland W (2004) Biotic and heavy metal stress response in plants: evidence for common signals. FEBS Lett 566:1–5
Mittler R, Rizhsky L (2000) Transgene-induced lesion mimic. Plant Mol Biol 44:335–344
Mukhopadhyay D, Riezman H (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315:201–205
Nagaoka S, Takano T (2003) Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J Exp Bot 54:2231–2237
Nakagawa M, Kagiyama M, Shibata N, Hirano Y, Hakoshima T (2014) Mechanism of high-affinity abscisic acid binding to PYL9/RCAR1. Genes Cells 19:386–404
Noriega TR, Tsai A, Elvekrog MM, Petrov A, Neher SB, Chen J, Bradshaw N, Puglisi JD, Walter P (2014) Signal recognition particle-ribosome binding is sensitive to nascent chain length. J Biol Chem 289:19294–19305
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Parra MM, Pozo O, Luna R, Godoy JA, Pintor-Toro JA (1996) Structure of the dehydrin tas14 gene of tomato and its developmental and environmental regulation in transgenic tobacco. Plant Mol Biol 32:453–460
Qureshi MK, Sujeeth N, Gechev TS, Hille J (2013) The zinc finger protein ZAT11 modulates paraquat-induced programmed cell death in Arabidopsis thaliana. Acta Physiol Plant 35:1863–1871
Ranocha P, Dima O, Nagy R, Felten J, Corratgé-Faillie C, Novák O, Morreel K, Lacombe B, Martinez Y, Pfrunder S, Jin X, Renou JP, Thibaud JB, Ljung K, Fischer U, Martinoia E, Boerjan W, Goffner D (2013) Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat Commun 4:1–9
Riewe D, Koohi M, Lisec J, Pfeiffer M, Lippmann R, Schmeichel J, Willmitzer L, Altmann T (2012) A tyrosine aminotransferase involved in tocopherol synthesis in Arabidopsis. Plant J 71:850–859
Rouhier N, Santos CV, Tarrago L, Rey P (2006) Plant methionine sulfoxide reductase A and B multigenic families. Photosynth Res 89:247–262
Safi H, Saibi W, Alaoui MM, Hmyene A, Masmoudi K, Hanin M, Brini F (2015) A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant Physiol Biochem 89:64–75
Sagi M, Davydov O, Orazova S, Yesbergenova Z, Ophir R, Stratmann JW, Fluhr R (2004) Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 16:616–628
SAS Institute (2002) Statistical Analysis System, v.8.2. SAS Institute, Cary
Saure MC (2001) Blossom-end rot of tomato (Lycopersicon esculentum Mill.)—a calcium or a stress-related disorder? Sci Hort 90:193–208
Saure MC (2005) Calcium translocation to fleshy fruit: its mechanism and endogenous control. Sci Hort 85:1–25
Saure MC (2014) Why calcium deficiency is not the cause of blossom-end rot in tomato and pepper fruit—a reappraisal. Sci Hort 174:151–154
Schott A, Ravaud S, Keller S, Radzimanowski J, Viotti C, Hillmer S, Sinning I, Strahl S (2010) Arabidopsis stromal-derived Factor2 (SDF2) is a crucial target of the unfolded protein response in the endoplasmic reticulum. J Biol Chem 285:18113–18121
Shin LJ, Lo JC, Yeh KC (2012) Copper chaperone antioxidant protein 1 is essential for copper homeostasis. Plant Physiol 159:1099–1110
Singh KB, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defence and stress responses. Curr Opin Plant Biol 5:430–436
Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. PNAS 100:14672–14677
Song M, Xu W, Xiang Y, Jia H, Zhang L, Ma Z (2014) Association of jacalin-related lectins with wheat responses to stress revealed by transcriptional profiling. Plant Mol Biol 84:95–110
Stevens J, Senaratna T, Sivasithamparam K (2007) Salicylic acid induces salinity tolerance in tomato (Lycopersicon esculentum cv. Roma): associated changes in gas exchange, water relations and membrane stabilization. Plant Growth Regul 49:77–83
Subramanyam S, Sardesai N, Puthoff DP, Meyer JM, Nemacheck JA, Gonzalo M, Williams CE (2006) Expression of two wheat defense-response genes, Hfr-1 and Wci-1, under biotic and abiotic stresses. Plant Sci 170:90–103
Suzuki K, Shono M, Egawa Y (2003) Localization of calcium in the pericarp cells of tomato fruits during the development of blossom-end rot. Protoplasma 222:149–156
Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8:397–403
Turhan E, Karni L, Aktas H, Deventurero G, Chang DC, Bar-Tal A, Aloni B (2006) Apoplastic antioxidants in pepper (Capsicum annuum L.) fruit and their relationship to blossom-end rot. J Hortic Sci Biotechnol 81:661–667
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121
Van Breusegem F, Dat JF (2006) Reactive oxygen species in plant cell death. Plant Physiol 141:384–390
Veena VSR, Sopory SK (1999) Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confers tolerance in transgenic tobacco under stress. Plant J 17:385–395
Wu J, Wang L, Baldwin IT (2008) Methyl jasmonate-elicited herbivore resistance: does MeJA function as a signal without being hydrolyzed to JA? Planta 227:1161–1168
Xiang Y, Song M, Wei Z, Tong J, Zhang L, Xiao L, Ma L, Ma Z, Wang Y (2011) A jacalin-related lectin-like gene in wheat is a component of the plant defense system. J Exp Bot 62:1–13
Yamamoto A, Bhuiyan MN, Waditee R, Tanaka Y, Esaka M, Oba K, Jagendorf AT, Takabe T (2005) Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. J Exp Bot 56:1785–1796
Yuasa T, Ishibashi Y, Iwaya-Inoue M (2012) A flower specific calcineurin B-like molecule (CBL)-interacting protein kinase (CIPK) homolog in tomato cultivar micro-tom (Solanum lycopersicum L.). Am J Plant Sci 3:753–763
Zhang W, Peumans WJ, Barre A, Astoul CH, Rovira P, Rougé P, Proost P, Truffa-Bachi P, Jalali AA, Van Damme EJM (2000) Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 210:970–978
Zhang Y, Li H, Shu W, Zhang C, Zhang W, Ye Z (2011) Suppressed expression of ascorbate oxidase gene promotes ascorbic acid accumulation in tomato fruit. Plant Mol Biol Rep 29:638–645
Zheng H, Ji C, Gu S, Shi B, Wang J, Xie Y, Mao Y (2004) Cloning and characterization of a novel RNA polymerase II C-terminal domain phosphatase. Biochem Biophys Res Commun 331:1401–1407
Zheng X, Chen B, Lu G, Han B (2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379:985–989
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
de Freitas, S.T., Martinelli, F., Feng, B. et al. Transcriptome Approach to Understand the Potential Mechanisms Inhibiting or Triggering Blossom-End Rot Development in Tomato Fruit in Response to Plant Growth Regulators. J Plant Growth Regul 37, 183–198 (2018). https://doi.org/10.1007/s00344-017-9718-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-017-9718-2