Abstract
Reduced ascorbic acid (AsA) can be converted into oxidized state by ascorbate oxidase (AO). In present study, AO gene was down-regulated by RNA interference in tomato via Agrobacterium-mediated transformation. The existence of the RNA interference T-DNA in transformed plants and their progenies were confirmed by polymerase chain reaction amplification and Southern blotting. AO expression levels in transgenic lines were monitored by semi-quantitative reverse transcriptase-polymerase chain reaction. The decreased AO enzyme activity and significantly improved AsA content in tomato fruit were observed to be correlated with AO gene suppression. Transgenic plants with AO suppression and AsA improvement exhibited higher photosynthesis capacity than untransformed control under drought stress. This report demonstrates the AsA oxidation pathway by RNA interference, and provides a feasible approach to improve AsA content in horticultural plants by regulating genes related to AsA metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ascorbic acid (AsA), also named vitamin C, plays an extensive role in plant and most animals. However, human and some animals like ape and cavy lack the ability to synthesize AsA as a result of a mutation in the last enzyme required for AsA biosynthesis. AsA, therefore, must be acquired regularly from dietary sources (Dowdle et al. 2007). For plants, AsA is involved in the process of cell division, cell expansion and growth. As a necessary antioxidant, AsA protects plant from oxidative damage by scavenging free radicals and reactive oxygen species (ROS) that are generated during photosynthesis, oxidative metabolism and various stresses (Stevens et al. 2007). AsA is a necessary factor of violaxanthinde-epoxidase, which can change violaxanthinde into zeaxanthin through xanthophyll cycle on thylakoid membranes, and thus is very important to protect photosynthesis and consume surplus light energy. Increasing AsA content in plants can improve tolerance to various abiotic stresses (Stevens et al. 2008).
Several AsA biosynthesis pathways have been identified in plants including mannose/l-galactose pathway, galacturonic acid pathway, gulose pathway and myo-inositol pathway (Agius et al. 2003; Lorence et al. 2004; Linster and Clarke 2008; Zhang et al. 2009). Following biosynthesis, AsA can be oxidized into monodehydroascorbate (MDHA) by ascorbate oxidase (AO) and ascorbate peroxidase (APX) in plants. MDHA is not stable and converted into AsA by monodehydroascorbate reductase, or converted into dehydroascorbate (DHA). DHA can be hydrolyzed spontaneously into diketogulonic acid, or converted into AsA by dehydroascorbate reductase using reduced glutathione as reducer (Noctor and Foyer 1998) (Fig. 1).
AO is a cell wall-localized enzyme that uses oxygen to catalyze the oxidation of ascorbate to the unstable MDHA which rapidly disproportionates to yield DHA and AsA, and thus contributes to the modulation of the AsA redox state and perception of external and internal stimuli of plant cells. Regulation of AO gene proves its essential role in AsA metabolism and stress responses. Overexpressing cucumber AO gene in tobacco leads to significantly increased AO enzyme activity and plant sensitivity to ozone (Sanmartin et al. 2003). Both sense and antisense expression of AO can improve apoplastic AsA (Pignocchi et al. 2003). Suppressed expression of the apoplastic AO gene increases salt tolerance. Under high salt stress, seeds germination ratio, photosynthesis ratio and seeds yield of antisense AO transgenic plants were all higher than that of untransformed plants and sense transgenic plants. Even under standard condition, AsA content in apoplast of sense transgenic plants was lower than that of control (Yamamoto et al. 2005). High AO activity specifically decreased the AsA content of the apoplast (Pignocchi et al. 2006). Increased AO expression in tobacco lowered the apoplast AsA redox state and enhanced sensitivity to various oxidative stresses (Fotopoulos et al. 2006). Leaves of AO overexpressing plants exhibited reduced stomatal conductance due to partial stomatal closure. AO overexpressing plants also exhibited elevated levels of hydrogen peroxide and a decline in hydrogen peroxide-scavenging enzyme activity (Fotopoulos et al. 2008).
Besides the enzymatic ROS scavenging mechanisms of superoxide dismutase (SOD), catalase (CAT), the water–water cycle (WWC), the ascorbate–glutathione cycle (AGC), and the glutathione peroxidase cycle (GPXC), plant cells have numerous non-enzymatic antioxidant molecules involved in protection against oxidative stress and damage caused by ROS (Mittler et al. 2004). Ascorbate is one of the most important non-enzymatic antioxidant molecules (Linster et al. 2008). Arabidopsis thaliana mutant vtc1 lacking efficient AsA synthesis ability (about 25% AsA content of wild-type) is sensitive to ozone. Exogenous application of AsA can improve vtc1 tolerance to ozone (Conklin et al. 1999). Thus the metabolic engineering of AsA content could alter plant responses to abiotic stress (Yamamoto et al. 2005). The AsA biosynthesis and metabolism pathways provide a possible approach for genetic engineering of high AsA accumulation and improved tolerance to abiotic stress through regulating AsA biosynthesis and metabolism related genes. In this study, tomato AO was cloned and down-regulated by RNA interference in tomato to investigate the effect of AO suppression on AsA accumulation and stress response.
Materials and Methods
Plant Material
Seeds of tomato cultivar Zhongshu No.5 (ZS5) were purchased from Vegetable and Flower Institute of Chinese Agriculture Academy Sciences (Beijing). The ZS5 was used as plant material for gene isolation and genetic transformation.
Gene Cloning and Plasmid Construction
Total RNA was isolated from fruits of tomato ZS5 using Trizol® reagent according to the manufacture’s recommended protocol (Invitrogen, USA). Three micrograms of total RNA was reverse-transcribed using the reverse transcription-polymerase chain reaction (RT-PCR) HIGH kit (Toyobo, Tokyo, Japan). The resultant first-strand cDNAs were diluted into 100 μl with RNase-free water. AO gene was cloned by RT-PCR method from fruit cDNA based on the Unigene SGN-U581990 from Solanaceae Genomics Network (SGN) database and nucleotide sequence (GenBank accession no. AY971876) from National Center for Biotechnology Information (NCBI) under following conditions: denaturation at 94°C for 4 min, followed by 30 cycles of 94°C for 1 min, 55°C for 45 s, and 72°C for 2 min, and then followed by a 6-min extension at 72°C.
To construct the RNA interference vector, a 573-bp length fragment on 5′ end of AO gene was amplified using gene specific primer pair (AOF: 5′-TTGCTATTCATTGGCATGGA-3′ and AOR: 5′-TTACCACAAATGGCTCCACA-3′) (Fig. 2). The amplified fragment was cleaned up by gel extraction kit (Promega, USA) and cloned into the pMD18-T (TaKaRa) vector and then subcloned into pHellsgaet2 using the BP Clonnase Reaction Kit (Invitrogen). Positive clones were confirmed by PCR analysis and sequencing. The resulted AO RNAi plasmid was electroported into Agrobacterium LBA4404 for tomato transformation.
Nucleotide and deduced amino acid sequences of tomato AO gene, and its phylogenetic relationship with counterparts from other plant species. The amino acids are shown above the corresponding codons. The stop codon is indicated by asterisk (*). Three regions for multicopper oxidase (cl06664) are respectively marked by grey boxes. Primer binding sites for RNA interference were underlined. Nucleotide and amino acids are numbered on the left (upper part). Fifteen AO genes from tomato (SlAO, AY971876), pepper (CaAO, AAF33751.1), tobacco (NtAO, Q40588.1), Arabidopsis (AtAO, NP 195693.1), mustard (BjAO, AF206722_1), rice (OsAO, Os06g0567200), melon (CmAO, CAA71274.1), pumpkin (CumAO, P24792.2), cucumber (CsAO, P14133.1), pea (PsAO, BAH28261.1), soybean (GmAO, BAB86897.1), Medicago truncatula (MtAO, CAA75577.1), poplar (PtAO, XP_002312838.1), maize (ZmAO, ACG30138.1) and Ricinus communis (RcAO, XP 002528975.1) were used for phylogenetic analysis. The scale bar represents the estimated evolutionary distance as 0.1 amino acid substitutions per site (lower part)
Molecular Characterization of Transgenic Plants
Regenerated transgenic plants were screened for nptII gene by PCR and Southern blot. For PCR analysis, each PCR reaction mixture contained 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 2 U Taq DNA polymerase (MBI Fermentas), 0.5 μM primer pair, and 100 ng templates. PCR was conducted with an initial denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 45 s, 60°C for 45 s and 72°C for 1 min, and a final extension at 72°C for 10 min in a PTC-100 Thermal Cycler (Bio-Rad, Hercules, CA, USA). For Southern blot analysis, about 15 μg DNA was digested with EcoRV (MBI, Fermatas), and separated on 0.8% (w/v) agarose gel in 1× TAE buffer, followed by blotting onto Amershan Hybond-N+ nylon membrane in 0.4 M NaOH. The blots were hybridized at 65°C in a phosphorus buffer (0.5 M) containing 7% SDS, 1% BSA, 1 mM EDTA. nptII gene probe was labeled with [α32P]-dCTP (Beijing Furui Company). The membranes were washed with 2× SSC, 0.5% (w/v) SDS at room temperature for 10 min and 0.2× SSC, 0.1% (w/v) SDS under 65°C for 5 min. nptII primers used for PCR analysis and probe preparation were nptII-F: 5′-AGACAATCGGCTGCTCTGAT-3′ and nptII-R: 5′-TCATTTCGAACCCCAGAGTC-3′. The transgenic plants with single copy of T-DNA insertion were propagated into T2 generation as homozygous lines for further analysis.
To determine the expression of AO gene in homozygous transgenic plants, semi-quantitative RT-PCR was performed. Total RNA was extracted from ripe fruits and transcripted into cDNA according to gene cloning method above. Semi-quantitative RT-PCR was carried out with gene specific primers AOF and AOR under the under following conditions: denaturation at 94°C for 3 min, followed by 28 cycles of 94°C for 45 min, 55°C for 45 s, and 72°C for 45 min, and then followed by a 6-min extension at 72°C. The β-actin gene was used as internal control to keep the same RNA loading. Primers for β-actin are β-actin-F: 5′-ATGGCAGACGGAGAGGATATTCA-3′ and β-actin-R: 5′-GCCTTTGCAATCCACATCTGCTG-3′.
AO Activity Determination in Tomato Fruits
AO activity in tomato fruits from homozygous transgenic plants was determined according to Esaka et al. (1990). Ripe fruits from five independent plants as well as untransformed control (ZS5) were harvested and mixed for AO activity assay. Experiment was repeated three times. Five gram of fruit tissue plus 25 ml pre-cold 50 mM phosphate buffer were ground and then centrifuged, the resulting supernatant transferred to fresh tubes and placed on ice. A volume of 3 ml reaction mixture including 2.88 ml phosphate buffer (pH 7.0), 0.5 mM AsA, and 0.12 ml exaction supernatant was used for analysis. Using reaction mixture without exaction supernatant as control, OD265 value was recorded on an ultraviolet and visible spectrophotometer (Agilent 8453). One unit is defined as that 0.01 OD value was changed within 1 min. Results were calculated using the following formula:
where ∆A denotes the absorption change range during reaction time, W is the tomato fresh weight (g), t denotes the reaction time (min), V T is the total enzyme extraction volume (ml), V S is enzyme extraction volume used for assay (ml).
Determination of AsA Content in Tomato Fruits
AsA content in tomato fruits of homozygous plants was analyzed by titration method with 2,6-dichlorophenol indophenol (Li 2000). Ripe fruits from five independent plants as well as untransformed control (ZS5) were harvested and mixed for ascorbate assay. AsA content was calculated by the following formula:
where Y 1 is the dye volume used for control titration (ml), Y 0 is the dye volume for sample titration (ml), A is the amount of AsA which demands 1 ml dye for titration (mg), B is the sample weight (g), X is the sample solution volume used for titration (ml), and Z is the total volume of sample solution (ml). The experiment was repeated three times.
Drought Treatment of Tomato Plants
Seeds from the homozygous transgenic plants with improved ascorbate content were harvested and pre-germinated and sown on pots with culture substrate (compost/perlite/sand = 1:1:1 (v/v/v)) in greenhouse. The seedlings are equally watered until four leaves stage, and then irrigation stopped. When the substrate surface became dry, the seedlings were further subjected to drought treatment for 1 week without irrigation.
After the drought treatment, the photosynthesis rate and stomatal conductance was measured with TPS-1 photosyntometer (PP Systems, UK) at noon of a fine day, under natural illumination at 32 ± 2°C. The electric conductance (EC) of tomato leaves was assayed according to Li (2000). Leaves at the same position of plants were taken and cleaned, placed at 40°C oven for 1 h. Heat treated leaves were rinsed with distilled water two times and blotted dry, and cut into 1-cm segments, and put into a beaker capable of holding electrode of conductivity meter. Twenty milliliter distilled water was added in the beaker immerging leaves at the bottom. The beaker was put in to a vacuum desiccator and pumped to remove air among cells, and then atmosphere was let in gently. The beaker was taken out and placed still for 20 min. The leaves were agitated gently before EC value was measured using DDS-307 digital conductivity meter at 20–25°C.
Statistical Analysis
All data are presented as the mean±SE and treated with ANOVA program of SAS software. Comparisons between transgenic plants and wild-type plants were performed using Duncan’s multiple range test at 5%. Values of P < 0.05 were considered statistically significant.
Results
Cloning and Analysis of Tomato AO Gene
We searched SGN and NCBI for tomato AO and identified a Unigene (SGN-U581990) and AO gene accession (GenBank accession no. AY971876). The tomato AO gene was cloned by RT-PCR method. Tomato AO gene contains a 1,737-bp open reading fame encoding 578 amino acid residues. The region from 41 to 158 bp encodes three representative multicopper oxidase motifs, which are characteristic domains of AO. The phylogenic tree based on the deduced amino acid sequence of plant AOs demonstrates that tomato AO has high identity with counterparts from other plant species. Tomato AO gene has the highest identity of 92% with pepper AO, with the second highest identity score of 72% with melon AO. Interestingly, tomato AO gene has a relatively lower identity (49%) with tobacco AO (Fig. 2).The high homology of tomato AO gene with counterparts from other plant species makes it necessary to characterize AO function in ascorbate metabolism in tomato.
RNAi Suppression of AO Gene in Tomato
To investigate the effect of AO suppression on ascorbate accumulation and stress response, 5′ end of AO gene sequence was used for RNA interference vector construction based on pHellsgate vector (Fig. 3a).There are only one copy of AO gene in tomato genome, as proved by RFLP analysis (Zou et al. 2006), which will permit specially characterizing function of AO gene through RNAi suppression.
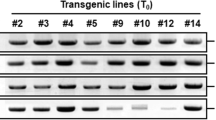
Schematic structure of T-DNA region of AO RNAi transformation vector and molecular characterizations of transgenic plants. RB right border, LB left border, P35S 35S promoter of cauliflower mosaic virus, intron PDK gene intron, Tnos nopline synthesis gene terminator, KAN kanamycin resistance coding region (nptII) containing its promoter and terminator. attR1, attR2, recombination sites from attB–attP reaction (a). Genomic DNA from young leaves was digested with EcoRV and transferred to membrane, and hybridized to nptII probe labeled with [α32P]-dCTP (b). Expression of AO gene in ripe fruits was investigated by semi-quantitative RT-PCR. actin gene was used for internal control (c). Transgenic lines used for Southern blot and semi-quantitative RT-PCR analysis are indicated above lanes. ZS5 is wild-type used as untransformed control (b, c)
A total of 10 independent transgenic plants were obtained through Agrobacterium-mediated transformation. Using the untransformed plants as negative control and plasmids as positive control, transgenic plants were PCR analyzed by nptII-specific primers (data not shown). Genomic DNA was extracted from PCR-positive plants and their progenies, and digested by EcoRV, transferred to nylon membrane by capillary method, and hybridized with nptII gene probe labeled with [α32P]dCTP. PCR and Southern blot analysis indicated the T-DNAs were integrated into tomato genome. Two transgenic lines with single copy of T-DNA integration, AO7 and AO8, were propagated to T2 generations for further characterization (Fig. 3b). In order to investigate expression profile of AO gene expression in RNAi transgenic plants, RNA was extracted from ripe fruits and transcripted into cDNA to carry out semi-quantitative RT-PCR. The RT-PCR result indicated that the expression levels of AO gene in RNAi transgenic lines, AO7 and AO8, were significantly lower than that of untransformed control (ZS5) (Fig. 3c). The two homozygous lines, AO7 and AO8, with AO suppression were selected for further analysis.
AO Enzyme Activity Decreases in Tomato Fruit
In the homozygous lines with obvious AO gene suppression, enzyme activity was analyzed to investigate the effect of AO expression changing on AO enzyme activity. The AO activity in transgenic lines of AO7 and AO8 decreased 54% and 17% compared to the untransformed control (Fig. 4a). The decrease in AO enzyme activity was found to be correlated with the variation of mRNA expression level as shown in Fig. 3. The suppression of AO expression effectively decreased the AO enzyme activity in tomato fruit.
Changes of AO enzyme activity and ascorbic acid content in tomato fruits of AO suppression plants. AO activity was decreased significantly in fruits as compared to untransformed control (ZS5) (a) accompanied by ascorbic acid content improvement in fruits (b). Different letters above columns indicate significant differences by Duncan’s multiple range test at 5% level
Improvement of AsA Content in Tomato Fruits by Suppression of AO Gene
In order to analyze the effect of AO expression suppression and AO activity decrease on AsA accumulation, the AsA content of tomato fruit was determined in ripe stage. The result indicated that the AsA content of tomato fruits of AO7 and AO8 was significantly higher than that of untransformed control. The AsA content in tomato fruits was improved up to 155% and 144% of the wild-type (Fig. 4b). The AsA accumulation was correlated with the suppression of AO gene expression and decrease in AO enzyme activity.
Alleviated Damage Under Drought Stress
ROS is frequently generated as by-products in photosynthesis, and is usually promoted to an enhanced production by various abiotic stresses. As ascorbate is an important antioxidant to scavenge ROS, plant photosynthesis indexes and electrolyte leakage were determined to investigate the effect of AO suppression and AsA accumulation on stress response. Several indexes including net photosynthetic rate (Pn), stomatal conductance (Gs) and leaf electrolyte leakage (EC) were not significantly different between wild-type and transgenic plants under normal conditions. However, after 1 week drought treatment, the Pn and Gs value of wild-type plants decreased more than onefold, and the EC value of wild-type plants increased more than twofolds, whereas the transgenic plants suffered a lesser impact under drought stress. After drought treatment, Pn and Gs of AO suppression lines, AO7 and AO8, were significantly higher than that of untransformed control, and EC of the AO suppression lines was significantly lower than that of untransformed control (Table 1). It could be considered that RNA interference of AO gene help the plants better adapt to drought stress with less affected photosynthesis capacity.
Discussion
Ascorbate can be oxidized into MDHA by AO in plants, which makes it possible to promote ascorbate accumulation in plants through suppressing expression of AO gene. In this study, the expression level of tomato AO was suppressed by RNA interference in tomato, and the AO enzyme activity was decreased and the AsA content was improved. There was a correlation between the AO expression suppression and AsA accumulation. Similar results were found in tobacco that AO activity decreased in antisense AO transgenic tobacco, leading to increasing content of AsA (Pignocchi et al. 2003). Together with our study, results indicated that suppressing enzyme in AsA metabolism pathway could improve AsA biosynthesis in plants. It should be noted that the reduced ascorbate (AsA) was determined in this study, while the total ascorbate (AsA+DHA) remained to be investigated. On the other hand, in transgenic tomato plants overexpressing ascorbate biosynthesis related genes such as GDP-d-mannose-3′,5′-epimerase gene (GME) and myo-inositol oxygenase gene (MIOX), major changing in ascorbate exists in reduced form of AsA, while the content of oxidized form of DHA remains largely unchanged (unpublished data).
Ascorbate is one of the most important of non-enzymatic antioxidant to scavenge ROS and protect plants from both abiotic and biotic stress damages (Darwish et al. 2008). The AO suppression in tomato lead to significantly increased ascorbate content in fruit, and the photosynthetic capacity and leaf electrolyte leakage under drought stress was less affected compared to wild type plants. The main reason probably lies in that the improved accumulation of ascorbate as an antioxidant protects plants from drought damage. Similarly, suppression of AO gene by antisense expression results in salt stress tolerance in tobacco, accompanied by low level of hydrogen peroxide accumulation and a high redox state of symplastic and apoplastic ascorbate which, in turn, permits stress tolerance (Yamamoto et al. 2005). Besides ascorbate as a non-enzymatic antioxidant, the enzymatic ROS scavenging mechanism in the ascorbate metabolism pathway includes APX. In APX mutant of wheat, both APX activity and photosynthesis efficiency decreased as compared to wild-type (Danna et al. 2003). Regulation of APX at the transcript level affects plant tolerance to drought stress (Fotopoulos et al. 2008). APX gene transcription was induced in plants by various abiotic stresses including drought, salt, salt–alkaline, CdCl2, and abscisic acid (Xu et al. 2009; Gao et al. 2010). In this study, transgenic plants with AO suppression has less leaf damage and improved photosynthesis ability compared to untransformed control after drought stress treatment (Table 1).
Ascorbate biosynthesis and metabolism related genes often exist in form of multiple gene family. AO gene family of three to four members has been reported in melon and Arabidopsis. RFLP analysis results demonstrated that tomato AO enzyme is encoded by a single-copy gene, similar to tobacco and pepper (Zou et al. 2006). The single copy of AO gene would permit the specific silence of AO gene to investigate gene function.
It is interesting to observe that the leaf morphology of AO RNAi lines were totally different from that of untransformed control. The compound leaves of wild type plants were converted to simple leaves in AO suppression plants. Besides its role of oxidization of AsA into MDHA, AO was frequently reported to be involved in auxin response and plant growth (Pignocchi et al. 2003, 2006). Promoter analysis revealed a cis-acting region involved in induction by auxin upstream pumpkin AO gene (Kisu et al. 1997). Application of auxin increased AO gene expression in tobacco (Pignocchi et al. 2003). Oxidation of apoplastic AsA in AO sense transgenic plants was associated with loss of the auxin response (Pignocchi et al. 2006). All these data indicate that AO expression is closely related to auxin response. On the other hand, auxin is a crucial regulator for the morphological patterning of compound leaves (Blein et al. 2008; Koenig et al. 2009). It would be very intriguing to investigate how AO is involved in the auxin patterning during leaf morphogenesis.
In conclusion, it is possible to increase AsA content through engineering AsA metabolism related genes. Modifying AsA metabolism in plant is another approach for regulating stress response as well as increasing nutrition quality.
References
Agius F, Gonzalez-Lamothe R, Caballero JL, Munoz-Blanco J, Botella MA, Valpuesta V (2003) Engineering increased vitamin C levels in plants by overexpression of a d-galacturonic acid reductase. Nat Biotechnol 21:177–181
Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P (2008) A conserved molecular framework for compound leaf development. Science 322:1835–1839
Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA 96:4198–203
Danna CH, Bartoli CG, Sacco F, Ingala LR, Santa-Maria GE, Guiamet JJ, Ugalde RA (2003) Thylakoid-bound ascorbate peroxidase mutant exhibits impaired electron transport and photosynthetic activity. Plant Physiol 132:2116–2125
Darwish SA, Li P, Ide C, Bede JC (2008) Caterpillar-specific gene expression in the legume, Medicago truncatula. Plant Mol Biol Report 26:12–31
Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N (2007) Two genes in Arabidopsis thaliana encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 52:673–689
Esaka M, Hattori T, Fujisawa K, Sakajo S, Asahi T (1990) Molecular cloning and nucleotide sequence of full-length cDNA for ascorbate oxidase from cultured pumpkin cells. Eur J Biochem 191:537–541
Fotopoulos V, Sanmartin M, Kanellis AK (2006) Effect of ascorbate oxidase over-expression on ascorbate recycling gene expression in response to agents imposing oxidative stress. J Exp Bot 57(14):3933–3943
Fotopoulos V, De Tullio MC, Barnes J, Kanellis AK (2008) Altered stomatal dynamics in ascorbate oxidase over-expressing tobacco plants suggest a role for dehydroascorbate signalling. J Exp Bot 59:729–737
Gao C, Wang Y, Liu G, Wang C, Jiang J, Yang C (2010) Cloning of ten peroxidase (POD) genes from Tamarix hispida and characterization of their responses to abiotic stress. Plant Mol Biol Report 28:77–89
Kisu Y, Harada Y, Goto M, Esaka M (1997) Cloning of the pumpkin ascorbate oxidase gene and analysis of a cis-acting region involved in induction by auxin. Plant Cell Physiol 38:631–637
Koenig D, Bayer E, Kang J, Kuhlemeier C, Sinha N (2009) Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136:2997–3006
Li H (2000) Principles and techniques of experiments in plant physiology and biochemistry. Higher Education Press, Beijing
Linster CL, Clarke SG (2008) l-Ascorbate biosynthesis in higher plants: the role of VTC2. Trends Plant Sci 13:567–573
Linster CL, Adler LN, Webb K, Christensen KC, Brenner C, Clarke SG (2008) A second GDP-l-galactose phosphorylase in Arabidopsis en route to vitamin C: Covalent intermediate and substrate requirements for the conserved reaction. J Biol Chem 283:18483–18492
Lorence A, Chevone BI, Mendes P, Nessler CL (2004) myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134:1200–1205
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49:249–279
Pignocchi C, Fletcher JM, Wilkinson JE, Barnes JD, Foyer CH (2003) The function of ascorbate oxidase in tobacco. Plant Physiol 132:1631–1641
Pignocchi C, Kiddle G, Hernández I, Foster SJ, Asensi A, Taybi T, Barnes J, Foyer CH (2006) Ascorbate oxidase-dependent changes in the redox state of the apoplast modulate gene transcript accumulation leading to modified hormone signaling and orchestration of defense processes in tobacco. Plant Physiol 141:423–435
Sanmartin M, Drogoudi PA, Lyons T, Pateraki I, Barnes J, Kanellis AK (2003) Over-expression of ascorbate oxidase in the apoplast of transgenic tobacco results in altered ascorbate and glutathione redox states and increased sensitivity to ozone. Planta 216:918–928
Stevens R, Buret M, Dufdfé P, Garchery C, Baldet P, Rothan C, Causse M (2007) Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol 143:1943–1953
Stevens R, Page D, Gouble B, Garchery C, Zamir D, Causse M (2008) Tomato fruit ascorbic acid content is linked with monodehydroascorbate reductase activity and tolerance to chilling stress. Plant Cell Environ 31:1086–1096
Xu J, Yin H, Wang W, Mi Q, Liao X, Li X (2009) Identification of Cd-responsive genes of Solanum nigrum seedlings through differential display. Plant Mol Biol Report 27:563–569
Yamamoto A, Bhuiyan NH, Waditee R, Tanaka Y, Esaka M, Ôba K, Jagendorf AT, Takabe T (2005) Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. J Exp Bot 56:1785–1796
Zhang W, Lorence A, Gruszewski HA, Chevone BI, Nessler CL (2009) AMR1, an Arabidopsis gene that coordinately and negatively regulates the mannose/l-galactose ascorbic acid biosynthetic pathway. Plant Physiol 150:942–950
Zou L, Li H, Ouyang B, Zhang J, Ye Z (2006) Cloning and mapping of genes involved in tomato ascorbic acid biosynthesis and metabolism. Plant Sci 170:120–127
Acknowledgements
This research was supported by grants of Major State Basic Research Development Program (2011CB100600), National Natural Science Foundation of China (No. 30800756), Xinjiang Corps Doctoral Fund (2008JC04).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yuyang Zhang and Hanxia Li equally contributed to this work.
Rights and permissions
About this article
Cite this article
Zhang, Y., Li, H., Shu, W. et al. Suppressed Expression of Ascorbate Oxidase Gene Promotes Ascorbic Acid Accumulation in Tomato Fruit. Plant Mol Biol Rep 29, 638–645 (2011). https://doi.org/10.1007/s11105-010-0271-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0271-4