Abstract
Genome editing is now a routine procedure in many mammalian genetics laboratories. The ostensibly short but intense history of genome-editing approaches illustrates how a disruptive technology can universally colonize a field when this new methodology, conceived to alter mammalian genomes at specific locations, is found to efficiently and robustly deliver results. This review summarizes the early development of genome editing using nucleases, from the pioneering experiments using yeast meganucleases, to the latest prokaryotic nucleases used for precise genome manipulation. Gene-editing nucleases belong to one of three known categories: zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR) and their associated proteins (Cas) tools. All operate on the same principle; they are all capable of inducing a double-strand break (DSB) at a defined genomic sequence that is subsequently corrected by endogenous DNA repair mechanisms. DSBs can be repaired through non-homologous end joining (NHEJ), resulting in small insertions and/or deletions (INDELs) and, hence, often leading to gene disruption. Alternatively, DSBs can be repaired through homology-driven repair (HDR), in the presence of donor homologous DNA sequences, resulting in gene-editing events.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The exchange of endogenous genomic sequences for exogenous donor DNA molecules via homologous recombination (HR) is a process that has been known for decades. Pioneering studies by the late Oliver Smithies illustrated how homologous DNA molecules could recombine and correctly insert at defined mammalian chromosomal locations (Smithies et al. 1984, 1985). These findings were instrumental for envisaging and developing gene targeting methodologies in mouse embryonic stem (ES) cells, for which Smithies, together with Mario Capecchi and Martin Evans, were jointly awarded the Nobel Prize in the Physiology or Medicine in 2007 (Mak 2007). In these first experiments, the recorded efficiency of targeted integration by HR in cultured somatic cell lines was relatively low (at best 1 in 1000), even in the presence of selection (Smithies et al. 1985). Nonetheless, the method was at least ten times more efficient than attempting to insert a genomic DNA fragment into its corresponding chromosomal location by HR through direct pronuclear microinjection of mouse fertilized eggs, as demonstrated by the perseverance of Ralph Brinster and collaborators (Brinster et al. 1989).
It became quickly obvious that opening the donor DNA molecule through a double-strand break (DSB) within the region of homology considerably enhanced the frequency of HR within the gene (Kucherlapati et al. 1984; Smithies et al. 1985). A similar observation was confirmed some years later in mouse ES cells when Allan Bradley reported increased targeting efficiencies using insertion versus replacement vectors (Hasty et al. 1991). Therefore, the benefits for the integration efficiency somehow correlated with the promotion of DSBs in the donor DNA template, so exposing homologous sequences at the end of these DNA molecules, a concept that was solidly established. This observation would play a fundamental role in subsequent developments leading to the first genome-editing strategies.
Indeed, the next step towards the establishment of the first genome-editing strategies was based on using I-SceI yeast meganuclease, a rare-cutter endonuclease with a recognition site of 18 base pairs responsible for intron homing in yeast mitochondria (Jacquier and Dujon 1985). 10 years after its discovery, it was shown that I-SceI meganuclease could be used to promote HR in mammalian chromosomes with a frequency two orders of magnitude higher than spontaneous HR (Choulika et al. 1995). The authors demonstrated the efficient repair of I-SceI-induced specific DSBs with donor DNA molecules carrying regions of homology flanking the endogenous I-SceI site in mouse cells, which was probably one of the first attempts of applying genome-editing strategies based on I-SceI meganuclease to produce a DSB for repair by endogenous cellular mechanisms. Subsequently, the introduction of I-SceI restriction sites at a mouse genomic locus by HR in ES cells, and the succeeding expression of this meganuclease in the presence of a replacement vector with homologous DNA sequences surrounding the introduced I-SceI site, resulted in a significant (100-fold stimulation) increase in targeting efficiency (Cohen-Tannoudji et al. 1998). Of course, the procedure was lengthy and cumbersome, requiring two consecutive HR steps and two selection markers. It was also not very practical since the I-SceI-based integration promotion was strictly dependent on the prior introduction of I-SceI recognition sites at the locus to be modified. Moreover, recent reports have suggested that HR by meganuclease-induced DSBs may be locus dependent in mammalian cells (Fenina et al. 2012). Introducing I-SceI meganuclease sites at either end of transgenes and co-microinjecting with the meganuclease also resulted in increased efficiency of transgenesis in medaka fish embryos (Thermes et al. 2002) and Xenopus (Pan et al. 2006), but not in mammalian embryos. Recently, this limitation has been overcome using a modified meganuclease with a nuclear localization signal (NLS), which efficiently mediates germline transgenesis in mouse and porcine embryos (Wang et al. 2014).
The evident limitation of target sequences associated with I-SceI would ultimately be overcome by producing engineered meganucleases with customized recognition sites, once they became commercially available. One of these engineered meganucleases, derived from I-CreI, was used to induce targeted recombination at high frequency at the SCID gene in human cells (Grizot et al. 2009). More recently, another engineered I-CreI, cleaving a 22-bp sequence of the Rag1 gene, was applied to demonstrate effective targeting events in rat and mouse embryos (Menoret et al. 2013).
The recognition that meganucleases were the first to be used for genome-editing purposes, well before other more celebrated and recent genome-editing nucleases (zinc-finger nucleases, ZFN; transcription activator-like effector nucleases, TALEN; CRISPR-associated proteins, Cas), is often not sufficiently acknowledged; hence these introductory words in the short but intense history of genome editing in mammals.
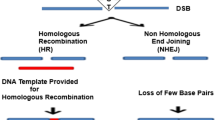
The mechanism by which the different types of nucleases enhance gene disruption or HR, in the presence of suitable donor DNA molecules, is the same; namely, they all efficiently promote DSBs at specific genomic locations. Subsequently, the endogenous cellular repair mechanisms, in the absence (non-homologous end joining, NHEJ) or presence (homology-driven repair, HDR) of adequate donor DNA molecules, with homologous sequences surrounding the DSB, will seal this breach in the genome. Depending on the repair route taken by the cell, the original DSB can result in gene disruption events, which are associated with the insertion or deletion of nucleotides (INDELs) through NHEJ, or in gene-editing events, which are associated with HDR (Fig. 1). Indeed, the quantifiable benefits of these nucleases regarding the increase in the frequency of HR events they can promote are estimated to be at least 1000-fold (Bibikova et al. 2001), as compared to previous attempts without nucleases (Smithies et al. 1985; Brinster et al. 1989).
Basic scheme illustrating the process of genome editing mediated by nucleases. This scheme is valid for all known types of genome editors. The associated nucleases trigger DNA cleavage at specific genomic locations and this generates a double-strand break (DSB) that is eventually repaired through one of the two possible routes: NHEJ or HDR, as indicated. The PAM sequence is indicated for the most common CRISPR-Cas9 system used for genome-editing purposes. DSB double-strand break, NHEJ non-homologous end joining, HDR homology-driven repair, PAM protospacer adjacent motif, INDELs insertion and deletions, HR homologous recombination
Zinc-finger nucleases (ZFN)
Even though yeast meganucleases were the first to be utilized to edit mammalian genomes (Choulika et al. 1995; Cohen-Tannoudji et al. 1998), a fundamental change in methodology occurred in 2009 that would transform how transgenic animals are produced. In a pioneering study, Guerts, Jacob and Buelow, and colleagues used zinc-finger nucleases (ZFN) to produce the world´s first knockout rats (Geurts et al. 2009). The methodological approach was based on the use of a DNA endonuclease domain from the bacterial restriction enzyme FokI, engineered with zinc-finger domains with known DNA-binding capacity, which was used to target and cleave a specific genome location. Zinc-finger nucleases (ZFN) had actually been devised some years earlier when the first chimeric proteins resulting from fusing the FokI endonuclease domain with engineered DNA-binding zinc-finger domains were shown to efficiently cleave and promote HR events at specific genomic sequences in Xenopus embryos (Bibikova et al. 2001). The technology was improved a few years after this and the engineered ZFN were shown to drive the correction of the human SCID mutation in cells by HR with an extrachromosomal DNA donor at high efficiencies (more than 18% of cells) without selection (Urnov et al. 2005). Generating the expected DSB required dimerization of the FokI endonuclease domain, indicating that the combined effect of two ZFN was required for efficient DNA cleavage at both strands (Mani et al. 2005). Soon, approaches were developed to design ZFN for the genome editing of mammalian genomes (Porteus 2008).
Zinc-finger nucleases (ZFN) were quickly converted into an efficient gene-editing tool to be applied for the generation of mammalian transgenesis. As stated earlier, the first of these reports described the generation of knockout rats (Geurts et al. 2009), a mammalian species in which the classical gene targeting approach was not possible because of the lack of equivalent rat ES cells. In fact, rat ES cells had been isolated a few months before the publication of the knockout rat study (Buehr et al. 2008). The use of ZFN for generating transgenic mammals rapidly disseminated among numerous laboratories (Remy et al. 2010) and soon ZFN were shown to drive successful genome-editing experiments in mice (Carbery et al. 2010; Meyer et al. 2010), cattle (Yu et al. 2011; Liu et al. 2013; Wei et al. 2015), and pigs (Hauschild et al. 2011; Carlson et al. 2012; Kwon et al. 2013; Qian et al. 2015).
The biggest impact of ZFN was probably in livestock species, where the absence of bona fide species-specific ES cells had traditionally limited genome alterations that required HR events (Petersen and Niemann 2015). Good examples of ZFN for livestock applications are exemplified by genome-edited cattle with increased resistance to mastitis (Liu et al. 2014a, b), or genome-edited domestic pigs carrying a warthog-derived variant of the RELA gene with the aim of transmitting the associated resilience to African swine fever (Lillico et al. 2016).
Gene therapy applications with ZFN were also explored and analyzed in tissue culture (e.g., Overlack et al. 2012), including the delivery of expression constructs for these chimeric nucleases through adeno-associated virus (AAV) particles (Ellis et al. 2013). In parallel, however, some unwanted consequences were discovered. For example, it was observed that the sequence specificity of ZFN is not strict and similar genomic sequences (known as off-targets) can also be targeted and eventually repaired with the donor DNA molecule, highlighting one of the current limitations of genome-editing nucleases (Radecke et al. 2010). Nevertheless, some pre-clinical studies using ZFN in human cells have progressed into clinical trials, as illustrated by the inactivation of the HIV co-receptor CCR5, approved by the US regulatory authorities (Maier et al. 2013; Tebas et al. 2014).
As a final note for this section, it became apparent that the ZFN methodology was not easily affordable or accessible to standard molecular biology laboratories, a fact that may have limited the success and impact of ZFN in mammalian transgenesis. Indeed, although some open platforms were established for producing ZFN (Hermann et al. 2012), most were produced commercially by the company who owned the patent for the technology (Swarthout et al. 2011).
Transcription activator-like effector nucleases (TALEN)
In 2011, a new type of genome-editing nuclease, known as transcription activator-like effector nuclease (TALEN), was reported in the field, representing a potential alternative to ZFN. Similar to that described for ZFN, TALEN were first used to efficiently produce knockout rats (Tesson et al. 2011). Whereas established and robust gene targeting in mice has been standard practice since 1987, precise genome editing in the rat was not possible for decades, likely explaining why these new technologies were first used in rats and eventually transferred to mice. Indeed, soon thereafter, the use of TALEN was extended to genome editing in mice, where numerous novel knockout models were quickly created (Panda et al. 2013; Sung et al. 2013; Wang et al. 2013a; Wefers et al. 2013). TALEN were also applied to the production of genome-edited livestock species (Carlson et al. 2012), including pigs (Xin et al. 2013), goats (Cui et al. 2015), sheep and cattle (Proudfoot et al. 2015; Wei et al. 2015), and non-human primates (Liu et al. 2014a, b). Again, similar to what was observed for ZFN, TALEN were shown to be robust and reliable tools for several biotechnological and biomedical applications in livestock, and, in particular as an efficient strategy to generate large animal models of human disease (Whitelaw et al. 2016). Agricultural applications of TALENs have included the production of genome-edited cattle genetically without horns, matching an existing natural mutant allele (Tan et al. 2013), or the generation of genome-edited cattle with an increased resistance to tuberculosis (Wu et al. 2015).
While TALEN share similarities with ZFN, they also present some attractive differences. Both ZFN and TALEN are chimeric proteins generated by fusing a different DNA-binding domain with the same endonuclease domain, from the restriction enzyme FokI. They also operate as dimers, hence one pair of ZFN or TALEN must always be considered to target a given genomic location. This is normally achieved through web-based algorithms that aid researchers to find the best target DNA sequences in both strands within the target locus (Periwal 2016). However, whereas ZFN are entirely engineered in the laboratory, with a defined DNA-binding specificity, according to an inferred code that links groups of three amino acids with three nucleotides, TALEN derive from TALE proteins, which exist in nature and are used by some plant-infecting pathogens to hijack and transform the cellular machinery to support the pathogen’s life cycle (Mussolino and Cathomen 2012). Through the systematic investigation of numerous TALE proteins, it was possible to decipher a code linking a precise dual amino acid found inside the protein with its capacity to bind to one specific nucleotide (Bochtler 2012). Unlike ZFN, which remained proprietary to a company and had to be ordered through it, engineered TALEN were easy to generate locally. Indeed, any molecular biology laboratory could easily purchase a large collection of intermediate reagents, made available as plasmids, which would be used to generate customized TALEN using a convenient step-by-step Golden Gate cloning protocol based on an innovative use of type II restriction enzymes for progressively assembling DNA fragments (Cermak et al. 2015; Sakuma and Yamamoto 2016). In theory, the Golden Gate assembly kit and protocol offers the possibility to design and easily produce any TALEN according to the target DNA sequence. In reality, however, this process can be long and cumbersome due to the numerous plasmids that are required for producing one of these engineered nucleases. In addition, whether the final TALEN will eventually target and cleave the expected DNA sequence or not unfortunately remains unpredictable, and can only be resolved by validating each newly produced TALEN in vitro and in vivo (Seruggia and Montoliu 2014).
Transcription activator-like effector nucleases (TALEN) have been also instrumental for gene therapy applications, where a number of pre-clinical strategies, including human induced pluripotent stem cells (iPSC), have been investigated successfully (e.g., Osborn et al. 2013; Ramalingam et al. 2014; Biffi 2015; Garate et al. 2015).
In the mouse genetics field, TALEN have proven their great utility as an efficient genome-editing tool in a study that aimed to correct a mutation affecting retinal function that was found associated with C57BL/6 N mice, one of the most common mouse inbred strains used currently by the International Mouse Phenotyping Consortium (IMPC; http://www.mousephenotype.org). This unexpected mutation could impair the interpretation of the eye phenotype of additional mouse mutations generated on the same genetic background (Mattapallil et al. 2012). Researchers used TALEN to correct the C57BL/6 N-specific mutation at the Crb1 locus (rd8 mutant allele) and obtained genome-edited C57BL/6 N mice without the original retinal degeneration (Low et al. 2014).
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas)
In 2013, the scientific community discovered a new genome-editing tool with a formidable capacity to drive precise genome alterations with high efficiency and, at the same time, was extraordinarily easy to implement in any molecular biology laboratory. This disruptive technology was envisaged in 2012 by at least two different groups (Jinek et al. 2012; Gasiunas et al. 2012), but it was not until 2013 when its capacity as a powerful genome-editing tool could be demonstrated. Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) are elements of an adaptive immune system discovered and investigated in prokaryotes more than two decades ago (Mojica and Montoliu 2016). The CRISPR-Cas system, or simply CRISPR tools, were first tested and used for genome editing in mammalian cells in culture (Cong et al. 2013; Mali et al. 2013), and soon after in mice (Wang et al. 2013b). It was immediately apparent that these third generation genome-editing tools would be the simplest, easiest, most robust, and efficient of all four reported nuclease methodologies, meganucleases, ZFN, TALEN, and CRISPR.
CRISPR tools are derived from an adaptive defense system found in most archaea and bacteria. In brief, the CRISPR systems require two basic components: a small molecule of RNA, which would be responsible for pairing with the corresponding homologous DNA sequence at the target site, and a DNA endonuclease, which would cleave the DNA and generate the unique DSB at the locus of choice, driven by short RNA molecule. The indicated small RNA molecule in reality corresponds to two original RNA molecules used by prokaryotes, crRNA and tracrRNA, responsible for pairing with the target sequence and for interacting with a Cas endonuclease, respectively. The two small RNA molecules were artificially merged into a synthetic guide RNA (sgRNA), or simply gRNA, by the laboratories of Jennifer Doudna and Emmanuelle Charpentier in their milestone 2012 paper, thereby further simplifying a system that was already fairly simple (Jinek et al. 2012). The most common CRISPR tools are derived from the CRISPR-Cas system found in Streptococcus pyogenes, where the relevant Cas endonuclease is Cas9. For this reason, CRISPR tools are often reported as CRISPR-Cas9. However, since we currently know the existence of numerous types of CRISPR systems in prokaryotes, each one associated with a different set of Cas proteins (Makarova et al. 2015), it is recommended to refer to these tools as CRISPR-Cas or, even simpler, CRISPR tools (Mojica and Montoliu 2016).
The mechanism by which CRISPR tools can target and cleave specific DNA sequences to generate a DSB is analogous to that of ZFN or TALEN. In essence, all three sets of tools use a binary system formed by an endonuclease and an element to direct it to a specific genomic location. As stated previously, in the case of ZFN and TALEN, the engineered chimeric proteins obtain the endonuclease domain from the bacterial restriction enzyme FokI. By contrast, the CRISPR systems use their own natural endonuclease, a Cas protein, usually Cas9. Whereas both ZFNs and TALENs depend on protein-DNA interactions to detect the planned target genomic sequences, through different amino acid/nucleotide codes, the CRISPR system relies on its own natural partner, a small RNA molecule, to match the target DNA sequence through the standard Watson-and-Crick nucleotide pairing code, which is biochemically more stable than the electrostatic interactions between DNA and protein molecules (Seruggia and Montoliu 2014).
The amazing plasticity and simplicity for generating CRISPR tools (e.g., Harms et al. 2014), along with their proven efficacy in promoting gene disruption (through NHEJ) or gene editing (through HDR, in the presence of exogenous DNA templates) has transformed the field and has positioned CRISPR as the most popular genomic-editing tool and the method of choice for gene editing. Using CRISPR reagents, any molecular biology laboratory can very easily envisage a strategy for inactivating a locus, through INDELs, or for generating large insertions or deletions, substitutions, duplications, inversions, knockins, and many other complex mutations or chromosomal rearrangements.
CRISPR tools have been used successfully for the generation of a large variety of tailored genomic alterations in many mammalian species. Some examples include strategies for introducing small INDELs (Wang et al. 2013b; Han et al. 2015), substitutions and insertions (Yang et al. 2013; Ma et al. 2014; Platt et al. 2014; Peng et al. 2015), large deletions and insertions (Xiao et al. 2013; Seruggia et al. 2015; Zhang et al. 2015; Birling et al. 2017), inversions (Xiao et al. 2013; Seruggia et al. 2015), and duplications, and other chromosomal rearrangements (Maddalo et al. 2014). The largest known deletions and gross chromosomal alterations were recently reported in rats and mice, where the excision of genomic fragments up to 24.4 Mb could be triggered by CRISPR (Birling et al. 2017). CRISPR tools have also been applied for investigating the role and relevance of defined DNA regulatory elements in non-coding genomic areas (Han et al. 2015; Seruggia et al. 2015; Guo et al. 2015; Lupiañez et al. 2015).
As expected, the use of CRISPR approaches for gene therapy applications are beginning to be explored. CRISPR tools have been used successfully to generate or correct mutations in human iPSC (e.g., Flynn et al. 2015). Moreover, CRISPR reagents encapsulated into AAV or non-viral particles have also been used to partially restore gene function in a number of animal models of human genetic disorders, such as Duchene muscular dystrophy (Nelson et al. 2016), human hereditary tyrosinemia (Yin et al. 2016) or retinitis pigmentosa (Suzuki et al. 2016). Preliminary attempts to use CRISPR for gene editing in human embryos have been reported; however, these studies produced similar results as those observed in embryos from other mammalian species, namely, mosaicism, multiple alleles created and co-existing on top of the expected allele, and some off-target modifications (Liang et al. 2015; Kang et al. 2016). Overall, these reports suggest that CRISPR-mediated genome editing may not yet be used for irreversibly modifying the human genome, should there be a good reason to do it.
Beyond basic research and rodent models, CRISPR has also had a strong impact in large animal models. CRISPR strategies have been applied for the generation of genome-edited livestock for biomedicine and biotechnology applications, including pigs (Tan et al. 2013; Hai et al. 2014; Peng et al. 2015; Lai et al. 2016), cattle (Wang 2015; Tan et al. 2016), sheep (Crispo et al. 2015; Tan et al. 2016), rabbits (Guo et al. 2016; Lv et al. 2016), and goats (Ni et al. 2014; Wang et al. 2015; Guo et al. 2016). Indeed, the arrival of CRISPR editing may have revitalized some biomedical applications such as xenotransplantation, owing to its success in inactivating all copies of porcine endogenous retrovirus (PERV) in the porcine genome (Yang et al. 2015). In addition, CRISPR tools have facilitated the generation of new pig models carrying multiple transgenic and knockout alleles (Niemann and Petersen 2016; Wang et al. 2016; Fisher et al. 2016). Some other recent examples can serve to illustrate the power of CRISPR-mediated genome editing in livestock species. CRISPR tools were used to inactivate the PRNP prion gene in the cattle genome, as a valid strategy to interrupt the dissemination of bovine spongiform encephalopathy, or mad-cow disease (Bevacqua et al. 2016). As a route to produce animals with increased resistance to tuberculosis, CRISPR strategies were used for inactivation of the NRAMP1 locus in cattle (Gao et al. 2017).
The recent use of CRISPR tools to inactivate the NANOS2 locus in the pig has led to the generation of germline ablated male pigs, which can serve as surrogates for transplantation of donor spermatogonial stem cells to expand the availability of gametes from genetically desirable sires (Park et al. 2017). Finally, very recent and exciting application of CRISPR is its use for the production of interspecies chimeras. Experiments have been designed to trigger the differentiation of human pluripotent cells in developing swine embryos that have been previously genome-edited with CRISPR tools, in an attempt to find differentiated progenies of human cells contributing to various cell lineages and organs in developing pig fetuses. The idea behind these innovative and challenging experiments is the future generation of human organs for transplantation. The first results from these interspecies chimeras have been just released (Wu et al. 2017).
The future of genome-editing nucleases
Four different genome-editing technologies are known to date: engineered meganucleases, ZFN, TALEN, and CRISPR. Other new systems that use different mechanisms or elements might yet be discovered, but if we consider only CRISPR-Cas systems, we realize that this may represent only the tip of the iceberg regarding the adaptive defense systems in prokaryotes. Most laboratories use only CRISPR tools derived from the CRISPR-Cas9 system of a single bacterium: Streptococcus pyogenes (Jinek et al. 2012). This Cas9 protein is now being engineered and new mutant versions have been generated with increased on-target efficiency and decreased off-target activity, aiming for more suitable tools for gene therapy applications (Kleinstiver et al. 2015, 2016; Slaymaker et al. 2016). Beyond Cas9, which we all mainly use, we also know there are many other CRISPR-Cas systems and types, each one with its own pros and cons, for use as genome-editing tools (Makarova et al. 2015). Many thousands of novel CRISPR-Cas systems in known cultivated species remain to be characterized. Some of these novel CRISPR systems are just now beginning to be studied, including Cpf1 and C2c2, with Cas-like proteins with different gene-editing properties and requirements (Zetsche et al. 2015; Abudayyeh et al. 2016). Even more astounding are the new CRISPR-Cas systems that have been recently discovered from uncultivated microorganisms (Burstein et al. 2017), opening this field to an almost unlimited source of potentially useful CRISPR reagents with specific properties, perhaps each one particularly suitable for a specific type of application.
Genome-editing nucleases (meganucleases, ZFN, TALEN, and CRISPR) have been extremely useful for increasing our knowledge of regular gene function through systematic functional genomic approaches, and for the generation of newer and better animal and cellular models of human disease. These improvements are irreversible and have already shaped the approaches for producing genetic alterations in mammals. Mouse functional genomic consortia now regularly use CRISPR tools to generate the remaining mouse mutants to accomplish the announced goal of creating a library of knockout animals for all murine genes. The next wave is expected to focus on gene therapy applications. ZFN and TALEN protocols have already been used in pre-clinical and clinical trials. CRISPR approaches for gene therapy are starting to be explored, with caution, due to the inherent diversity of genetic alleles created at the target locus and the possibility of modifying similar sequences at off-target genomic locations. A careful risk to benefit analysis will have to be considered before CRISPR genome-editing capacities are eventually transferred to routine clinical protocols.
References
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353(6299):aaf5573
Bevacqua RJ, Fernandez-Martín R, Savy V, Canel NG, Gismondi MI, Kues WA, Carlson DF, Fahrenkrug SC, Niemann H, Taboga OA, Ferraris S, Salamone DF (2016) Efficient edition of the bovine PRNP prion gene in somatic cells and IVF embryos using the CRISPR/Cas9 system. Theriogenology 86:1886–1896
Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S (2001) Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol 21:289–297
Biffi A (2015) Clinical translation of TALENS: treating SCID-X1 by gene editing in iPSCs. Cell Stem Cell 16:348–349
Birling MC, Schaeffer L, André P, Lindner L, Maréchal D, Ayadi A, Sorg T, Pavlovic G, Hérault Y (2017) Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE. Sci Rep 7:43331
Bochtler M (2012) Structural basis of the TAL effector-DNA interaction. Biol Chem 393:1055–1066
Brinster RL, Braun RE, Lo D, Avarbock MR, Oram F, Palmiter RD (1989) Targeted correction of a major histocompatibility class II E alpha gene by DNA microinjected into mouse eggs. Proc Natl Acad Sci USA 86:7087–7091
Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A (2008) Capture of authentic embryonic stem cells from rat blastocysts. Cell 135:1287–1298
Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF (2017) New CRISPR-Cas systems from uncultivated microbes. Nature 542:237–241
Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X (2010) Targeted genome modification in mice using zinc-finger nucleases. Genetics 186:451–459
Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC (2012) Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA 109:17382–17387
Cermak T, Starker CG, Voytas DF (2015) Efficient design and assembly of custom TALENs using the Golden Gate platform. Methods Mol Biol 1239:133–159
Choulika A, Perrin A, Dujon B, Nicolas JF (1995) Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol 15:1968–1973
Cohen-Tannoudji M, Robine S, Choulika A, Pinto D, El Marjou F, Babinet C, Louvard D, Jaisser F (1998) I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol Cell Biol 18:1444–1448
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, dos Santos-Neto PC, Nguyen TH, Crénéguy A, Brusselle L, Anegón I, Menchaca A (2015) Efficient generation of Myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PLoS ONE 10:e0136690
Cui C, Song Y, Liu J, Ge H, Li Q, Huang H, Hu L, Zhu H, Jin Y, Zhang Y (2015) Gene targeting by TALEN-induced homologous recombination in goats directs production of β-lactoglobulin-free, high-human lactoferrin milk. Sci Rep 5:10482
Ellis BL, Hirsch ML, Porter SN, Samulski RJ, Porteus MH (2013) Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Ther 20:35–42
Fenina M, Simon-Chazottes D, Vandormael-Pournin S, Soueid J, Langa F, Cohen-Tannoudji M, Bernard BA, Panthier JJ (2012) I-SceI-mediated double-strand break does not increase the frequency of homologous recombination at the Dct locus in mouse embryonic stem cells. PLoS ONE 7:e39895
Fischer K, Kraner-Scheiber S, Petersen B, Rieblinger B, Buermann A, Flisikowska T, Flisikowski K, Christan S, Edlinger M, Baars W, Kurome M, Zakhartchenko V, Kessler B, Plotzki E, Szczerbal I, Switonski M, Denner J, Wolf E, Schwinzer R, Niemann H, Kind A, Schnieke A (2016) Efficient production of multi-modified pigs for xenotransplantation by ‘combineering’, gene stacking and gene editing. Sci Rep 6:29081
Flynn R, Grundmann A, Renz P, Hänseler W, James WS, Cowley SA, Moore MD (2015) CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Exp Hematol 43:838–848
Gao Y, Wu H, Wang Y, Liu X, Chen L, Li Q, Cui C, Liu X, Zhang J, Zhang Y (2017) Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome Biol 18:13
Garate Z, Quintana-Bustamante O, Crane AM, Olivier E, Poirot L, Galetto R, Kosinski P, Hill C, Kung C, Agirre X, Orman I, Cerrato L, Alberquilla O, Rodriguez-Fornes F, Fusaki N, Garcia-Sanchez F, Maia TM, Ribeiro ML, Sevilla J, Prosper F, Jin S, Mountford J, Guenechea G, Gouble A, Bueren JA, Davis BR, Segovia JC (2015) Generation of a high number of healthy erythroid cells from gene-edited pyruvate kinase deficiency patient-specific induced pluripotent stem cells. Stem Cell Rep 5:1053–1066
Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109:E2579–E2586
Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R (2009) Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325:433
Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, Blanco FJ, Pâques F, Duchateau P (2009) Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res 37:5405–5419
Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, Lu Y, Wu Y, Jia Z, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q (2015) CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162:900–910
Guo R, Wan Y, Xu D, Cui L, Deng M, Zhang G, Jia R, Zhou W, Wang Z, Deng K, Huang M, Wang F, Zhang Y (2016) Generation and evaluation of Myostatin knock-out rabbits and goats using CRISPR/Cas9 system. Sci Rep 6:29855
Hai T, Teng F, Guo R, Li W, Zhou Q (2014) One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24:372–375
Han Y, Slivano OJ, Christie CK, Cheng AW, Miano JM (2015) CRISPR-Cas9 genome editing of a single regulatory element nearly abolishes target gene expression in mice–brief report. Arterioscler Thromb Vasc Biol 35:312–315
Harms DW, Quadros RM, Seruggia D, Ohtsuka M, Takahashi G, Montoliu L, Gurumurthy CB (2014) Mouse genome editing using the CRISPR/Cas system. Curr Protoc Hum Genet 83:1–27
Hasty P, Rivera-Pérez J, Chang C, Bradley A (1991) Target frequency and integration pattern for insertion and replacement vectors in embryonic stem cells. Mol Cell Biol 11:4509–4517
Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H (2011) Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci USA 108:12013–12017
Hermann M, Maeder ML, Rector K, Ruiz J, Becher B, Bürki K, Khayter C, Aguzzi A, Joung JK, Buch T, Pelczar P (2012) Evaluation of OPEN zinc finger nucleases for direct gene targeting of the ROSA26 locus in mouse embryos. PLoS ONE 7:e41796
Jacquier A, Dujon B (1985) An intron encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41:383–394
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Kang X, He W, Huang Y, Yu Q, Chen Y, Gao X, Sun X, Fan Y (2016) Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J Assist Reprod Genet 33:581–588
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, Aryee MJ, Joung JK (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529:490–495
Kucherlapati RS, Eves EM, Song KY, Morse BS, Smithies O (1984) Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci USA 81:3153–3157
Kwon DN, Lee K, Kang MJ, Choi YJ, Park C, Whyte JJ, Brown AN, Kim JH, Samuel M, Mao J, Park KW, Murphy CN, Prather RS, Kim JH (2013) Production of biallelic CMP-Neu5Ac hydroxylase knock-out pigs. Sci Rep 3:1981
Lai S, Wei S, Zhao B, Ouyang Z, Zhang Q, Fan N, Liu Z, Zhao Y, Yan Q, Zhou X, Li L, Xin J, Zeng Y, Lai L, Zou Q (2016) Generation of knock-in pigs carrying Oct4-tdTomato reporter through CRISPR/Cas9-mediated genome engineering. PLoS ONE 11:e0146562
Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, Huang J (2015) CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 6:363–372
Lillico SG, Proudfoot C, King TJ, Tan W, Zhang L, Mardjuki R, Paschon DE, Rebar EJ, Urnov FD, Mileham AJ, McLaren DG, Whitelaw CB (2016) Mammalian interspecies substitution of immune modulatory alleles by genome editing. Sci Rep 6:21645
Liu X, Wang Y, Guo W, Chang B, Liu J, Guo Z, Quan F, Zhang Y (2013) Zinc-finger nickase-mediated insertion of the lysostaphin gene into the beta-casein locus in cloned cows. Nat Commun 4:2565
Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, Jing B, Si C, Lin Q, Chen X, Lin H, Pu X, Wang Y, Qin B, Wang F, Wang H, Si W, Zhou J, Tan T, Li T, Ji S, Xue Z, Luo Y, Cheng L, Zhou Q, Li S, Sun YE, Ji W (2014a) TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 14:323–328
Liu X, Wang Y, Tian Y, Yu Y, Gao M, Hu G, Su F, Pan S, Luo Y, Guo Z, Quan F, Zhang Y (2014b) Generation of mastitis resistance in cows by targeting human lysozyme gene to β-casein locus using zinc-finger nucleases. Proc Biol Sci 281:20133368
Low BE, Krebs MP, Joung JK, Tsai SQ, Nishina PM, Wiles MV (2014) Correction of the Crb1rd8 allele and retinal phenotype in C57BL/6 N mice via TALEN-mediated homology-directed repair. Invest Ophthalmol Vis Sci 55:387–395
Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161:1012–1025
Lv Q, Yuan L, Deng J, Chen M, Wang Y, Zeng J, Li Z, Lai L (2016) Efficient generation of myostatin gene mutated rabbit by CRISPR/Cas9. Sci Rep 6:25029
Ma Y, Ma J, Zhang X, Chen W, Yu L, Lu Y, Bai L, Shen B, Huang X, Zhang L (2014) Generation of eGFP and Cre knockin rats by CRISPR/Cas9. FEBS J 281:3779–3790
Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A (2014) In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516:423–427
Maier DA, Brennan AL, Jiang S, Binder-Scholl GK, Lee G, Plesa G, Zheng Z, Cotte J, Carpenito C, Wood T, Spratt SK, Ando D, Gregory P, Holmes MC, Perez EE, Riley JL, Carroll RG, June CH, Levine BL (2013) Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther 24:245–258
Mak TW (2007) Gene targeting in embryonic stem cells scores a knockout in Stockholm. Cell 131:1027–1031
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Mani M, Smith J, Kandavelou K, Berg JM, Chandrasegaran S (2005) Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun 334:1191–1197
Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, Caspi RR (2012) The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6 N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci 53:2921–2927
Ménoret S, Fontanière S, Jantz D, Tesson L, Thinard R, Rémy S, Usal C, Ouisse LH, Fraichard A, Anegon I (2013) Generation of Rag1-knockout immunodeficient rats and mice using engineered meganucleases. FASEB J 27:703–711
Meyer M, de Angelis MH, Wurst W, Kühn R (2010) Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA 107:15022–15026
Mojica FJ, Montoliu L (2016) On the origin of CRISPR-Cas technology: from prokaryotes to mammals. Trends Microbiol 24:811–820
Mussolino C, Cathomen T (2012) TALE nucleases: tailored genome engineering made easy. Curr Opin Biotechnol 23:644–650
Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351:403–407
Ni W, Qiao J, Hu S, Zhao X, Regouski M, Yang M, Polejaeva IA, Chen C (2014) Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS ONE 9:e106718
Niemann H, Petersen B (2016) The production of multi-transgenic pigs: update and perspectives for xenotransplantation. Transgenic Res 25:361–374
Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, DeFeo AP, Gabriel R, Schmidt M, von Kalle C, Carlson DF, Maeder ML, Joung JK, Wagner JE, Voytas DF, Blazar BR, Tolar J (2013) TALEN-based gene correction for epidermolysis bullosa. Mol Ther 21:1151–1159
Overlack N, Goldmann T, Wolfrum U, Nagel-Wolfrum K (2012) Gene repair of an Usher syndrome causing mutation by zinc-fingernuclease mediated homologous recombination. Invest Ophthalmol Vis Sci 53:4140–4146
Pan FC, Chen Y, Loeber J, Henningfeld K, Pieler T (2006) I-SceI meganuclease-mediated transgenesis in Xenopus. Dev Dyn 235:247–252
Panda SK, Wefers B, Ortiz O, Floss T, Schmid B, Haass C, Wurst W, Kühn R (2013) Highly efficient targeted mutagenesis in mice using TALENs. Genetics 195:703–713
Park KE, Kaucher AV, Powell A, Waqas MS, Sandmaier SE, Oatley MJ, Park CH, Tibary A, Donovan DM, Blomberg LA, Lillico SG, Whitelaw CB, Mileham A, Telugu BP, Oatley JM (2017) Generation of germline ablated male pigs by CRISPR/Cas9 editing of the NANOS2 gene. Sci Rep 7:40176
Peng J, Wang Y, Jiang J, Zhou X, Song L, Wang L, Ding C, Qin J, Liu L, Wang W, Liu J, Huang X, Wei H, Zhang P (2015) Production of Human albumin in pigs through CRISPR/Cas9-mediated knockin of human cDNA into swine albumin locus in the Zygotes. Sci Rep 5:16705
Periwal V (2016) A comprehensive overview of computational resources to aid in precision genome editing with engineered nucleases. Brief Bioinform bbw052
Petersen B, Niemann H (2015) Advances in genetic modification of farm animals using zinc-finger nucleases (ZFN). Chromosome Res 23:7–15
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F (2014) CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159:440–455
Porteus M (2008) Design and testing of zinc finger nucleases for use in mammalian cells. Methods Mol Biol 435:47–61
Proudfoot C, Carlson DF, Huddart R, Long CR, Pryor JH, King TJ, Lillico SG, Mileham AJ, McLaren DG, Whitelaw CB, Fahrenkrug SC (2015) Genome edited sheep and cattle. Transgenic Res 24:147–153
Qian L, Tang M, Yang J, Wang Q, Cai C, Jiang S, Li H, Jiang K, Gao P, Ma D, Chen Y, An X, Li K, Cui W (2015) Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci Rep 5:14435
Radecke S, Radecke F, Cathomen T, Schwarz K (2010) Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol Ther 18:743–753
Ramalingam S, Annaluru N, Kandavelou K, Chandrasegaran S (2014) TALEN-mediated generation and genetic correction of disease-specific human induced pluripotent stem cells. Curr Gene Ther 14:461–472
Rémy S, Tesson L, Ménoret S, Usal C, Scharenberg AM, Anegon I (2010) Zinc-finger nucleases: a powerful tool for genetic engineering of animals. Transgenic Res 19:363–371
Sakuma T, Yamamoto T (2016) Engineering customized TALENs using the platinum gate TALEN Kit. Methods Mol Biol 1338:61–70
Seruggia D, Montoliu L (2014) The new CRISPR-Cas system: RNA-guided genome engineering to efficiently produce any desired genetic alteration in animals. Transgenic Res 23:707–716
Seruggia D, Fernández A, Cantero M, Pelczar P, Montoliu L (2015) Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis. Nucleic Acids Res 43:4855–4867
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88
Smithies O, Koralewski MA, Song KY, Kucherlapati RS (1984) Homologous recombination with DNA introduced into mammalian cells. Cold Spring Harb Symp Quant Biol 49:161–170
Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS (1985) Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317:230–234
Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW (2013) Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 31:23–24
Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo S, Chen S, Goebl A, Soligalla RD, Qu J, Jiang T, Fu X, Jafari M, Esteban CR, Berggren WT, Lajara J, Nuñez-Delicado E, Guillen P, Campistol JM, Matsuzaki F, Liu GH, Magistretti P, Zhang K, Callaway EM, Zhang K, Belmonte JC (2016) In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540:144–149
Swarthout JT, Raisinghani M, Cui X (2011) Zinc finger nucleases: a new era for transgenic animals. Ann Neurosci 18:25–28
Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, Fahrenkrug SC (2013) Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci USA 110:16526–16531
Tan W, Proudfoot C, Lillico SG, Whitelaw CB (2016) Gene targeting, genome editing: from Dolly to editors. Transgenic Res 25:273–287
Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH (2014) Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370:901–910
Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ (2011) Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 29:695–696
Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS (2002) I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev 118:91–98
Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435:646–651
Wang Z (2015) Genome engineering in cattle: recent technological advancements. Chromosome Res 23(1):17–29
Wang H, Hu YC, Markoulaki S, Welstead GG, Cheng AW, Shivalila CS, Pyntikova T, Dadon DB, Voytas DF, Bogdanove AJ, Page DC, Jaenisch R (2013a) TALEN-mediated editing of the mouse Y chromosome. Nat Biotechnol 31:530–532
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013b) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918
Wang Y, Zhou XY, Xiang PY, Wang LL, Tang H, Xie F, Li L, Wei H (2014) The meganuclease I-SceI containing nuclear localization signal (NLS-I-SceI) efficiently mediated mammalian germline transgenesis via embryo cytoplasmic microinjection. PLoS ONE 9:e108347
Wang X, Yu H, Lei A, Zhou J, Zeng W, Zhu H, Dong Z, Niu Y, Shi B, Cai B, Liu J, Huang S, Yan H, Zhao X, Zhou G, He X, Chen X, Yang Y, Jiang Y, Shi L, Tian X, Wang Y, Ma B, Huang X, Qu L, Chen Y (2015) Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci Rep 5:13878
Wang X, Cao C, Huang J, Yao J, Hai T, Zheng Q, Wang X, Zhang H, Qin G, Cheng J, Wang Y, Yuan Z, Zhou Q, Wang H, Zhao J (2016) One-step generation of triple gene-targeted pigs using CRISPR/Cas9 system. Sci Rep 6:20620
Wefers B, Panda SK, Ortiz O, Brandl C, Hensler S, Hansen J, Wurst W, Kühn R (2013) Generation of targeted mouse mutants by embryo microinjection of TALEN mRNA. Nat Protoc 8:2355–2379
Wei J, Wagner S, Lu D, Maclean P, Carlson DF, Fahrenkrug SC, Laible G (2015) Efficient introgression of allelic variants by embryo-mediated editing of the bovine genome. Sci Rep 5:11735
Whitelaw CB, Sheets TP, Lillico SG, Telugu BP (2016) Engineering large animal models of human disease. J Pathol 238:247–256
Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, Zhang Y (2015) TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci USA 112:E1530–E1539
Wu J, Platero-Luengo A, Sakurai M, Sugawara A, Gil MA, Yamauchi T, Suzuki K, Bogliotti YS, Cuello C, Morales Valencia M, Okumura D, Luo J, Vilariño M, Parrilla I, Soto DA, Martinez CA, Hishida T, Sánchez-Bautista S, Martinez-Martinez ML, Wang H, Nohalez A, Aizawa E, Martinez-Redondo P, Ocampo A, Reddy P, Roca J, Maga EA, Esteban CR, Berggren WT, Nuñez Delicado E, Lajara J, Guillen I, Guillen P, Campistol JM, Martinez EA, Ross PJ, Izpisua Belmonte JC (2017) Interspecies chimerism with mammalian pluripotent stem cells. Cell 168:473–486
Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, Zhu Z, Lin S, Zhang B (2013) Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 41:e141
Xin J, Yang H, Fan N, Zhao B, Ouyang Z, Liu Z, Zhao Y, Li X, Song J, Yang Y, Zou Q, Yan Q, Zeng Y, Lai L (2013) Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS ONE 8:e84250
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154:1370–1379
Yang L, Güell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J, Cortazio R, Wilkinson RA, Fishman JA, Church G (2015) Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350:1101–1104
Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, Gupta A, Bolukbasi MF, Walsh S, Bogorad RL, Gao G, Weng Z, Dong Y, Koteliansky V, Wolfe SA, Langer R, Xue W, Anderson DG (2016) Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 34:328–333
Yu S, Luo J, Song Z, Ding F, Dai Y, Li N (2011) Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res 21:1638–1640
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771
Zhang L, Jia R, Palange NJ, Satheka AC, Togo J, An Y, Jin H, Feng X, Zheng Y (2015) Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS ONE 10:e0120396
Acknowledgements
LM´s laboratory is funded through the Spanish Ministry of Economy and Competitiveness (MINECO) [BIO2015-70978], the Biomedical and Biological Sciences (BMBS) European Cooperation in Science and Technology (COST) action [BM1308 SALAAM], and the Spanish Biomedical Research Networking Centre on Rare Diseases (CIBERER) from the National Institute of Heath Carlos III (Instituto de Salud Carlos III, ISCIII). The authors thank Kenneth McCreath for English language editing assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Fernández, A., Josa, S. & Montoliu, L. A history of genome editing in mammals. Mamm Genome 28, 237–246 (2017). https://doi.org/10.1007/s00335-017-9699-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-017-9699-2