Abstract
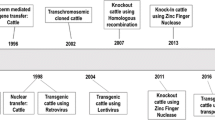
Great strides in technological advancements have been made in the past decade in cattle genome engineering. First, the success of cloning cattle by somatic cell nuclear transfer (SCNT) or chromatin transfer (CT) is a significant advancement that has made obsolete the need for using embryonic stem (ES) cells to conduct cell-mediated genome engineering, whereby site-specific genetic modifications can be conducted in bovine somatic cells via DNA homologous recombination (HR) and whereby genetically engineered cattle can subsequently be produced by animal cloning from the genetically modified cells. With this approach, a chosen bovine genomic locus can be precisely modified in somatic cells, such as to knock out (KO) or knock in (KI) a gene via HR, a gene-targeting strategy that had almost exclusively been used in mouse ES cells. Furthermore, by the creative application of embryonic cloning to rejuvenate somatic cells, cattle genome can be sequentially modified in the same line of somatic cells and complex genetic modifications have been achieved in cattle. Very recently, the development of designer nucleases—such as zinc finger nucleases (ZFNs) and transcription activator-like effector nuclease (TALENs), and clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9)—has enabled highly efficient and more facile genome engineering in cattle. Most notably, by employing such designer nucleases, genomes can be engineered at single-nucleotide precision; this process is now often referred to as genome or gene editing. The above achievements are a drastic departure from the traditional methods of creating genetically modified cattle, where foreign DNAs are randomly integrated into the animal genome, most often along with the integrations of bacterial or viral DNAs. Here, I review the most recent technological developments in cattle genome engineering by highlighting some of the major achievements in creating genetically engineered cattle for agricultural and biomedical applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Advancements in animal genomics and the development of new technologies for altering the genetic makeup of cattle have yielded unprecedented opportunities for both agricultural and biomedical applications. One of such agricultural applications is the production of genetically engineered cattle with improved disease resistance, such as mastitis-resistant dairy cattle (Donovan et al. 2005). In a pioneering study by Wall et al., mammary gland-specific expression of lysostaphin renders the cattle highly resistant to infection by Staphylococcus aureus (Wall et al. 2005). Other applications of genetically engineered cattle include increased meat production (Proudfoot et al. 2014), improved milk composition with enhanced nutrition values (Karatzas 2003; Wall et al. 1997), and safer animal products, such as the production of prion disease-free cattle by knocking out the cellular prion protein gene, PRNP (Richt et al. 2007). As already scientifically proven by the work in other livestock species (Golovan et al. 2001), cattle genome engineering may also turn out to be an indispensable approach for achieving environmentally sustainable agriculture.
Numerous biomedical applications are also being explored using genetically engineered cattle, one of which is to engineer cattle as bioreactors to produce therapeutic human recombinant proteins. Currently, most of the pharmaceutical proteins are produced by mammalian cell culture, a system that is very costly and has low production capacity (Bandaranayake and Almo 2014). With the ever-increasing demands for therapeutic human recombinant proteins to treat a variety of human diseases, more cost-effective and high-quantity production systems are needed. The use of genetically engineered dairy cattle for the expression of human genes in the lactating mammary gland promises to be one of the most cost-effective methods of producing valuable recombinant therapeutic proteins (Bosze et al. 2008). With the tremendous protein production capabilities of the cattle lactating mammary gland, it is possible for tens to hundreds of grams of recombinant therapeutic proteins to be produced daily in the milk of a genetically engineered dairy cow. Cattle also offer the advantages over most other livestock animals in having large blood volumes; therefore, and not surprisingly, the cattle blood generation system has also been explored as a bioreactor for therapeutic protein production (Matsushita et al. 2014; Sano et al. 2013).
Motivated by these agricultural and biomedical applications, extensive research efforts have been carried out in developing genetic modification technologies in cattle. Shortly after the first success in producing transgenic livestock by introducing transgenes through pronuclear (PN) injection (Hammer et al. 1985), it was realized that PN injection is extremely inefficient in introducing a transgene into the cattle genome (and into the genome of other animal species as well). Due to the lack of ES cells in cattle, the commonly used methods to effectively introduce a transgene into mouse ES cells by electroporation or lipofection were not applicable to cattle. Consequently, the major research focus in the 1990s, and to some extent the 2000s, was to develop effective techniques to introduce a transgene or other genetic modification events into the bovine genome (Chan et al. 1999). Despite the extensive research, efficiency in transgenic cattle production remained very low. Another major problem with these technologies was their inability to conduct site-specific genetic modifications in cattle, a prerequisite to engineering the genome in a designed manner. An important technical breakthrough is the cloning of cattle by somatic cell nuclear transfer (SCNT) or CT, by which genetically engineered cattle can be produced from primary somatic cells in which a transgene has been introduced (Cibelli et al. 1998). Since the genetically modified somatic cells can be fully characterized before being used as donors for animal cloning, 100 % of the calves produced from such cells could carry a desired transgene, thus greatly improving the efficiency of transgenesis. More importantly, HR-mediated site-specific genomic modifications can be carried out in cattle somatic cells, allowing for the production of cattle with defined genetic modifications, such as gene KOs and KIs (Kuroiwa et al. 2004). With this approach and the restoration of the proliferative capacity of bovine somatic cells by CT-mediated embryonic cloning, a sequential genetic modification strategy has been developed to carry out multiple site-specific genetic modifications in cattle (Kuroiwa et al. 2004). Furthermore, by incorporating animal breeding into the sequential genetic modification strategy, more complex genetic modifications can be introduced into the cattle genome (Matsushita et al. 2014).
Above all, the most significant technological advancement in genome engineering in recent years is the development of designer nucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9), which is in the process of revolutionizing the way of conducting genome engineering (Gaj et al. 2013; Kim and Kim 2014). The successful application of these innovative technologies has enabled genomic modifications of high efficiency and precision in cattle (Carlson et al. 2013). These technological developments have ushered in a new era in bovine genome engineering and are poised to fulfill many of the long envisioned agricultural and biomedical promises of genetically engineered cattle.
Technological advancement in cattle genome engineering
In this essay, I refer to “genome engineering” as the practice of introducing precise, site-specific DNA sequence changes to a genome, such as deleting or inserting a DNA sequence at a specific genomic locus or converting an endogenous DNA sequence to another at single-nucleotide precision. In the case where the deletion or insertion of a DNA sequence results in the loss of function of the target DNA sequence, I refer to it as “knock out (KO)” or “gene targeting,” and I refer to the insertion of a DNA generally as “knock in (KI)”; I also interchangeably call these events “genetic modifications.” These definitions of genome engineering are distinguished from the conventional practices of altering the genome in undefined manners, such as the random integration of a DNA into the genome, a practice widely used for much of the history for producing transgenic cattle. Genome engineering also includes the practice of introducing a naturally occurring or artificial chromosome into the cattle genome, the deletion of an entire chromosome, and the induction of site-specific translocation of chromosomal fragments within a chromosome or between chromosomes.
Genome engineering in cattle by DNA homologous recombination
Starting with the first successful transgenic cattle production by PN injection of a transgene into zygotes more than two decades ago (Krimpenfort et al. 1991), genome engineering in cattle has come a long way. In the 1990s, one of the most active research areas (which was also mirrored by the transgenic research carried out in other livestock species) was focused on improving the efficiency of transgenic bovine embryo production for yielding transgenic cattle (Robl et al. 2007); such research efforts are still ongoing, though at a much less substantial level. To overcome the extremely low efficiency in producing transgenic embryos by PN injection (on average, less than 1 % of injected embryos becomes transgenic), several ways of transgene delivery systems were explored. Among them were the techniques of sperm-mediated gene transfer (Bachiller et al. 1991) and viral vector-mediated gene transfer (Hofmann et al. 2003; Kubisch et al. 1997). While these techniques offer certain advantages over PN injection, such as the ease and low cost of delivering a transgene into the cattle genome, little improvement was made in increasing the efficiency of transgenic cattle production. More problematic, the random integration of a transgene into the animal genome results in low level (or silenced) expression of the transgene and low reproducibility in transgenic animal production. Furthermore, due to the extremely low efficiency in introducing a transgene or other genetic modification events, site-specific genetic modifications, such as gene KOs and KIs, were not possible in cattle with these techniques.
The strategy of producing animals with site-specific genetic modifications via HR-mediated gene targeting was first developed by using mouse ES cells and has been almost exclusively used in that species (Koller and Smithies 1992). This is mainly for two reasons. First, ES cells were only available in the mouse until recently, and second, only ES cells have the unique capability of self-renewal in cell culture and the potential to participate in embryogenesis to transmit to the germline when injected into, or aggregated with, a mouse embryo. These two unique properties of ES cells are essential for producing genetically modified animals: the capability of self-renewal in cell culture permits site-specific genetic modifications in ES cells via HR and selection and characterizations of the modified ES cells; the capacity to transmit genetic modifications to the germline allows the genetic modification events carried out in ES cells to be passed to an animal and its offspring. Embryonic stem cells are still not available in cattle and most other livestock species. With the ability to clone cattle by SCNT or CT, site-specific genetic modifications can now be conducted in cattle somatic cells and animals can be subsequently produced from these cells (Matsushita et al. 2014). Compared to mouse ES cells, however, somatic cells have limited life span in cell culture and low HR activity (Sedivy and Sharp 1989), two of the most desired qualities of cells for conducting HR-mediated genetic modifications. Therefore, it is very technically challenging to conduct site-directed genetic modifications in somatic cells. Despite these difficulties, James Robl and his colleagues succeeded in producing the first cattle carrying site-specific genetic modifications by conducting HR-mediated genetic modifications in bovine somatic cells followed by animal cloning by CT (Kuroiwa et al. 2004). This approach was shown to be effective in knocking out both transcriptionally active and silent genes (Robl et al. 2007). Nevertheless, due to the technical challenges with this genome engineering strategy, only a very small number of research groups has achieved cattle production with site-specific genetic modifications (Kuroiwa et al. 2004; Richt et al. 2007; Sendai et al. 2006; Wang et al. 2013; Matsushita et al. 2014; Sano et al. 2013).
Several technical considerations are believed to be important for conducting HR-mediated genetic modifications in bovine fibroblast cells. First, since HR is a very rare event in cells, an effective selection strategy is needed to identify the cells in which HR events take place. The most effective selection strategies comprise both positive and negative selection mechanisms, by which an antibiotic resistance gene (such as puromycin) and a toxic gene (such as diphtheria toxin A (DT-A)) are introduced with the gene targeting vector as the positive and negative selection marker, respectively, to select for cells in which HR has occurred (Kuroiwa et al. 2004). By using this selection strategy, only the cells that have undergone HR between the gene targeting vector and the cell genome are resistant to both the antibiotics and the toxin will survive under such selection pressures. To ensure effective positive and negative selections, strong gene promoters that can drive high-level expression of selection marker genes in bovine fibroblast cells should be used. Both the mouse phosphoglycerate kinase-1 (PGK) promoter and the SV40 promoter and thymidine kinase enhancer (ST) promoter seem sufficiently active in bovine fibroblast cells and allow for the selection of correctly targeted cells (Kuroiwa et al. 2004; Richt et al. 2007). We also recently successfully used other promoters, such as the chicken beta-actin promoter with CMV enhancer (CAG) promoter, to conduct genetic modifications in bovine fibroblast cells (Matsushita et al. 2014). The second consideration is to use isogenic DNAs as the homologous arms in gene targeting vector construction. Even though using homologous arms isolated from a nonisogenic Holstein genomic library successfully targets a Holstein cell line, it is six times more efficient to induce HR on the isogenic allele than on the other allele that displays sequence polymorphisms with the homologous arms (Kuroiwa et al. 2004). The third consideration is to develop an optimized DNA delivery method for introducing gene targeting vectors into bovine fibroblast cells. Based on our experience, while several electroporation methods, such as GenePulser II, are effective in introducing DNAs into bovine fibroblast cells, the Nucleofector system tends to result in better gene targeting efficiency in bovine fibroblast cells, as well as in the fibroblast cells from several other mammalian species that we have been working with (our unpublished data). Finally, as the ultimate goal here is to use the genetically modified cells as donors for animal cloning, an efficient animal cloning protocol is essential for effective production of genetically engineered cattle (Sullivan et al. 2004).
As discussed below, this HR-mediated gene-targeting strategy is effective not only for introducing site-specific genetic modifications at a single genomic locus but also for introducing complex genetic modifications into the cattle genome by sequential genome engineering strategies (Kuroiwa et al. 2004; Matsushita et al. 2014).
Sequential genome engineering in cattle
Complex genome engineering involving multiple genetic modification events has been routinely performed in the mouse. This can be done by sequentially modifying the mouse genome in ES cells or by breeding single-gene modified mice to achieve a genotype with complex genetic modifications. With the short gestation and sexual maturation times in mice, this can be done within a reasonable time. In contrast, the lack of ES cells in cattle and the limited life span of primary somatic cells prevent multiple rounds of genetic modifications from being conducted in cattle. Breeding is not a feasible option either, because of the long cattle reproduction cycle. To overcome such problems, a sequential genome engineering strategy that allows multiple genetic modifications to be conducted in the same line has been developed in cattle (Kuroiwa et al. 2004). This was achieved by employing embryonic cloning via CT to rejuvenate the genetically modified bovine fibroblast cells after each round of genetic modifications. In the first study of its kind, both of the alleles of the bovine immunoglobulin heavy-chain μ (bIGHM) gene and both of the alleles of the bovine prion protein (PRNP) gene were sequentially knocked out, leading to the establishment of bovine fibroblast cell lines with quadruple gene KOs (bIGHM −/− PRNP −/−) (Kuroiwa et al. 2004). By using the cell lines in which both of the bovine bIGHM alleles were knocked out as chromatin donors, bIGHM −/− cattle were cloned. In a subsequent study, this research group also produced of prion-free cattle by sequentially knocking out both of the alleles of the PRNP gene (Richt et al. 2007).
More recently, using this sequential genome engineering strategy, all of the four bovine immunoglobulin heavy (bIGH) chain alleles were sequentially knocked out (four rounds of sequential gene targeting). Furthermore, rejuvenation of the quadruple KO bovine fibroblast cells by embryonic cloning allowed the subsequent transfer of a human artificial chromosome (HAC) carrying the entire human IGH and kappa-light (IGK) chain loci in their germline configuration into these cells and led to the production of transchromosomic (Tc) cattle that express human antibodies (Kuroiwa et al. 2009; Sano et al. 2013). These Tc cattle are so far among the most extensively genetically engineered animals produced without using animal breeding (see below). The power of this sequential genome engineering strategy in engineering cattle for biomedical applications was further demonstrated by the production of large quantity of high titer tumor immunogen-specific human IgG through immunization of the Tc cattle with human tumor immunogens (Sano et al. 2013).
Animal breeding-assisted sequential genome engineering in cattle
While sequential genome engineering strategy has been very effective in creating cattle carrying up to five rounds of genetic modifications (four rounds of gene KO and one round of HAC transfer), further rounds of genetic modifications for even more complex genome engineering become prohibitive. This is due to declined cloning efficiency after repeated or serial cloning. We and others have shown that animal cloning efficiency can be reduced to a level where it becomes impossible to produce additional live offspring after serial cloning (Kubota et al. 2004; Kuroiwa et al. 2004). This phenomenon seems to be common among species, as cloning efficiency decline was also observed in other mammalian species where serial cloning was also performed (Wakayama et al. 2000). Consequently, while cells can be rejuvenated by embryonic cloning to regain proliferative capacity and can be used for further rounds of genetic modifications, only a limited number of rounds of cloning can be performed before cells lose their capacity to support normal embryogenesis and development. Therefore, unless means are developed to overcome this hurdle, this sequential genome engineering strategy may only allow for a limited number of genetic modifications to be engineered into cattle. We posited that both genetic and epigenetic alternations could be introduced into cells by repeated cloning and/or the multiple rounds of genetic modifications. To investigate whether these processes compromise genome integrity in the resultant cell lines, we employed comparative genomic hybridization (CGH) to conduct genome-wide scan for genomic alterations in three independent bovine transgenic cell lineages generated from sequential genetic modifications (Liu et al. 2011). From this study, we concluded that large genomic structural variations (! 10 kb) are less likely to arise from repeated cloning and genetic modifications. Therefore, it is more likely that epigenetic errors introduced from repeated cloning are responsible for the decline of cloning efficiency.
To reset the epigenetic state of cells after multiple rounds of genetic modifications and cloning for further rounds of genetic modifications, we recently developed an animal breeding-assisted sequential genome engineering strategy in cattle (Matsushita et al. 2014). In this new sequential genome engineering strategy, animals are produced after two or three rounds of sequential genetic modifications and are raised to sexual maturity for breeding. Adding the breeding step as one part of the sequential genome engineering strategy was based on the rationale that germline transmission would erase the epigenetic errors in a cloned genome acquired from cloning. Our epigenetic analysis on the cell lines established from fetuses produced by breeding the cloned cattle showed that their epigenetic profiles are very similar to those in the cell lines established from fetuses produced by breeding wild type (i.e., never been cloned) cattle but are distinguishable from those in the cell lines having undergone one or more rounds of cloning (our unpublished data). Therefore, cell lines established from the fetuses produced by breeding would not only carry the genetic modifications introduced from the previous rounds of sequential genome engineering but also be epigenetically reset, allowing more rounds of genetic modifications to be performed (Matsushita et al. 2014). By employing this animal breeding-assisted sequential genome engineering strategy, we recently succeeded in the production of Tc cattle carrying triple bovine immunoglobulin gene knockouts, in which both of the two bovine IGH loci, bIGHM and bIGHML1, and the bovine Ig lambda (bIGL) locus were homozygously inactivated and an HAC was incorporated, leading to the production of Tc cattle with triple bovine immunoglobulin gene homozygous KOs (in total, six genes were knocked out) (Matsushita et al. 2014).
In using this strategy, we first conducted sequential gene targeting to KO both of the bIGH loci, bIGHM and bIGHML1, in both male and female bovine cell lines. In the male cell line, we knocked out both of the alleles in the bIGHML1 locus and one of the alleles in the bIGHM locus by three rounds of sequential gene targeting and embryonic cloning and established bIGHM −/+ bIGHML −/− cell lines. In the female cell line, we sequentially knocked out both of the bIGHM alleles by two rounds of sequential gene targeting and embryonic cloning and established bIGHM −/− cell lines. To reset the epigenetic state of the above sequentially targeted cell lines through germline transmission, we produced calves from both the male bIGHM −/+ bIGHML −/− and female bIGHM −/− cell lines by CT, raised them to sexual maturity, and bred them to produce fetuses (first round of breeding). We collected day 40 fetuses and used them to establish generation zero (G0) cell lines (we call cell lines established from breeding-derived fetuses as G0 cell lines). Through genotyping analysis, we identified both male and female cell lines with the desired genotypes, bIGHM −/− bIGHML −/+ and used them for subsequent bovine bIGLJ-IGLC gene cluster deletions (see below).
Since the bIGL locus has 25 V lambda genes (17 of them being functionally expressed) (Pasman et al. 2010), it would take numerous of rounds of gene targeting and decades to individually KO each of the V lambda genes. Because the J-C genes in the bIGLJ-IGLC cluster are essential for Ig lambda gene expression, and because the bovine bIGLJ-IGLC cluster is much smaller in size (about 27 kb) than the bIGLV cluster (over 350 kb) (Pasman et al. 2010), we decided to delete the entire bIGLJ-IGLC gene cluster with Cre/loxP-mediated site-specific chromosomal recombination strategy. To integrate loxP sequences into the bovine genome to flank the bIGLJ-IGLC gene cluster, we designed and constructed two loxP KI vectors, one to be knocked in at the upstream of the bIGLJ-IGLC1gene and the other downstream of the bIGLJ5-IGLC5 gene. Our original plan was to conduct three rounds of sequential genetic modifications, with the first two rounds to KI the loxP sequences upstream and downstream of the bIGLJ- IGLC1gene cluster, respectively, and the third round to transfect the resultant cell line with a Cre-expressing plasmid to induce the Cre/loxP-mediated chromosomal recombination event to delete the bIGLJ-IGLC1 gene cluster. However, to reduce the rounds of embryonic cloning, we tested whether co-transfected the second loxP KI vector with a Cre-expressing vector can achieve the KI of the second loxP and the Cre/loxP-mediated chromosomal recombination between the two loxP sequences with a single round of genetic modification. Reducing one round of embryonic cloning would significantly lessen the time required for bIGLJ-IGLC1 gene cluster deletion; it would also significantly improve animal cloning efficiency. By using this co-transfection strategy, we successfully deleted the bIGLJ-IGLC gene cluster (Matsushita et al. 2014). After the bIGLJ-IGLC gene cluster deletion step, we conducted embryonic cloning by using the engineered cells as chromatin donors and established multiple fetal cell lines. Genotyping analysis showed that all of these cell lines carried the bIGLJ-IGLC gene cluster deletion as well as the homozygous KOs in the bIGHM and bIGHML1 loci (bIGHM −/− bIGHML1 −/+ bIGL −/+). To our knowledge, this is the first success in achieving site-specific large chromosome region deletions with the Cre/loxP system in cattle.
To reset the epigenetic state of the above sequentially engineered cells after two rounds of genetic modification and two rounds of embryonic cloning (G2 cells), we used them as chromatin donors for CT and produced female and male calves with the bIGHM −/− bIGHML1 −/+ bIGL −/+ genotype. We then raised the male and female calves to sexual maturity and crossbred them (second round of breeding) for fetus production. From the fetuses collected at 40-day gestation, both female and male cell lines with the desired triple KO (TKO) genotype bIGHM −/− IGHML1 −/− IGL −/−) were established (G0 cells). To achieve the goal of producing TKO Tc cattle, we subsequently transferred a newly engineered HAC into the above established TKO bovine fibroblast cells by micro-cell mediated chromosome transfer (MMCT) and established HAC/TKO fibroblast cell lines. We then used these cell lines as chromatin donors for CT and successfully produced multiple HAC/TKO cattle. Our characterization studies showed that bIGLJ-IGLC1 gene cluster deletion resulted in the total loss of bovine lambda light chain antibody production and encouraged high levels of fully human antibody production in these Tc cattle (Matsushita et al. 2014). These Tc animals are by far the most extensively genetically engineered cattle produced to date (in total, six alleles were knocked out with the addition of a HAC).
Epigenetic issues related to breeding-assisted sequential genome engineering strategy
The above case demonstrated that this breeding-assisted sequential genome engineering strategy is robust and straightforward in producing cattle with complex genetic modifications and, in theory, poses no limitations on the rounds of genetic modifications to be achieved. However, due to the long reproduction cycle of cattle, it takes a minimum of 2 years to reset the epigenetic state of a cloned cell line through breeding: it takes about a year from using a cloned cell line to produce cloned calves, about a year to raise the calves to sexual maturity, and about 2 months to produce fetuses for establishing new cell lines. This will become very time and cost prohibitive if more extensive genetic modifications need to be carried out. We posited that one of the possible ways to overcome this limitation is to slow or totally prevent the decline of cloning efficiency caused by repeated cloning. As mentioned above, by employing extensive genomic analysis, we first ruled out that a loss of genetic integrity during the multiple rounds of genetic modifications and embryonic cloning processes is the cause for the decline of cloning efficiency (Liu et al. 2011). Therefore, we focused our efforts on identifying the epigenetic errors that might be introduced by cloning, particularly the ones that accumulate during each round of cloning. Our goal is to use such epigenetic errors as epigenetic readouts for testing different cloning procedures for improving cloning and to use them as epigenetic markers for cell line qualifications. From such studies, several important observations were made. First, the decline of cloning efficiency among the cell lines produced from the same parental cell line at any round of cloning varies significantly. For example, when cell lines established from the fetuses cloned from the same parental cell line (either a G0 or a previously cloned cell line) were used as chromatin donors for animal cloning, some of the cell lines may become “unclonable,” i.e., no single live calf can be produced even with large number of embryo transfers (up to more than 100), while others produce multiple live calves, albeit at a relatively lower cloning efficiency than those from the parental cell line. Second, the decline of a cell line after a round of cloning seems to be permanent or irreversible. We found that no cell line established from the fetuses cloned from an unclonable cell line becomes clonable. These observations prompted us to conduct genome-wide epigenetic analyses, such as methylome analysis, to compare the epigenetic profiles between the clonable and unclonable cell lines that were derived from the same parental cell line. While research is still in progress, we have identified several epigenetic changes that are commonly found in the unclonable cell lines (our unpublished data).
Genome engineering in cattle by designer nucleases
One of the most innovative technological advancements in genome engineering in recent years is the development of designer nucleases, such as ZFNs, TALENs, and CRIPSR/Cas9 systems, which have greatly improved the efficiency and versatility in genome engineering. Common features of the designer-nuclease-mediated genome engineering technologies include the following: (1) designer nucleases can be programmed to introduce site-specific double- or single-strand breaks (DSBs or SSBs) with high efficiency, and (2) the subsequent DNA repair pathways, either the homology-directed repair (HDR) or nonhomologous end joining (NHEJ) can be employed to generate desired genetic modifications (Carroll 2014). These designer nucleases, initially developed in laboratory animals, have quickly been adopted for applications in cattle and other livestock species. Because many excellent reviews have been written on these designer nucleases (Gaj et al. 2013; Gupta and Musunuru 2014; Hsu et al. 2014; Kim and Kim 2014) and because the principle in applying them for genome engineering is the same across species, to avoid redundancy, readers are encouraged to read these reviews, and I will focus this section on summarizing the major successes in using these new technologies in the past few years and provide my thoughts on the impact of such technology on the future practice of bovine genome engineering.
-
1.
Genome engineering in cattle by ZFNs
Among the designer nucleases, ZFNs are the first of this kind of genome engineering tools developed in recent years and, consequently, are the first employed in engineering the cattle genome. Recently, a variant of ZFN, called ZFNickases in which the catalytic activity of FokI in one of the ZFN monomer in the ZFN dimer is abolished for introducing SSBs in a target DNA, was developed (Ramirez et al. 2012). As the SSBs provide bias toward HR-mediated gene modifications over NHEJ, ZFNickases are believed to have lower off-target events (Ramirez et al. 2012).
So far, several successful cases have been established in using ZFNs or ZFNickases to engineer the cattle genome for several agricultural and biomedical applications. One of such research efforts is to improve meat production in cattle. It has been long known that natural mutations in the MSTN gene found in the Belgian Blue and Piedmontese cattle breeds cause increased muscle mass compared to other breeds of cattle (McPherron and Lee 1997; Kambadur et al. 1997). To improve muscle mass production in Chinese yellow cattle, ZFNs were designed to target the bovine MSTN gene, which encodes myostatin protein, a member of the transforming growth factor-b superfamily that negatively regulates muscle growth. The authors demonstrated that cattle cloned from the MSTN-targeted Chinese yellow cattle fibroblast cells exhibit increased muscle growth (Luo et al. 2014). Another instance in using ZFNs to engineer the cattle genome is the production of dairy cattle with improved resistance to mastitis (Liu et al. 2014). To achieve high level of human lysozyme expression in the mammary gland, the authors chose to KI the human lysozyme gene into the bovine ″-casein gene (CSN2) locus to allow its expression to be under the control of the endogenous regulatory sequences of ″-casein gene. As ″-casein is one of the major milk proteins in cattle, this approach would render high-level expressions of human lysozyme in the cattle mammary gland. Compared to previous attempts in other livestock species where the human lysozyme gene was randomly integrated in the genome, the concentration of human lysozyme expressed in the milk of these transgenic cattle was reported in the range of 23 to 31 μg/ml (Liu et al. 2014). Very importantly, it was demonstrated that these genetically engineered cows are highly resistant to infection by intra-mammary infusion of mastitis-causing bacteria (Liu et al. 2014). The same research group also previously succeeded in using ZFNickases to knock in a modified lysostaphin gene from Staphylococcus simulans into the bovine ″-casein locus as another approach to create mastitis resistant cattle (Liu et al. 2013). Another example in using ZFNs to engineer the cattle genome is to KO the bovine ″-lactoglobulin gene to produce ″-lactoglobulin-free milk. As ″-lactoglobulin is one of the major allergens in cow milk (Selo et al. 1998), ″-lactoglobulin-free milk will greatly benefit human health. By employing a ZFN-mediated gene-targeting strategy, high gene targeting efficiency was achieved in bovine fibroblast cell lines, resulting in the KO of the bovine ″-lactoglobulin gene (Yu et al. 2011). Eight cattle were cloned from the bi-allelically targeted fibroblast colonies, but due to the high rate of calf loss from animal cloning, only one calf survived that harbors in-frame nucleotide deletions on both of the ″-lactoglobulin gene alleles (Yu et al. 2011).
Based on the studies spanning many organisms, it seems that ZFNs tend to be several orders of magnitude more efficient than conventional HR (i.e., without using any designer nucleases) in conducting site-specific genetic modifications (Carroll 2011; Wu et al. 2007). However, there are several serious drawbacks in using ZFNs in genome engineering. First, as there is no general code to follow for designing a zinc figure protein to recognize a DNA sequence, selection strategies such as phage display need to be used for selecting zinc figure proteins to recognize a specific DNA sequence of interest. Consequently, the design and validation of ZFNs is very labor intensive. Using pre-existing zinc figure modules to assemble ZFNs significantly alleviated this problem, but the design and validation of ZFNs are still challenging (Kim et al. 2009). These problems are further compounded by the time-consuming process of assembling ZFNs (Durai et al. 2005). Second, due to the requirements for particular DNA sequence patterns by ZFs, available target sites for ZFN design are limited (Sander et al. 2010). Third, it is often observed that there are significant variations in gene-targeting efficiency among the ZFNs designed for the same genomic locus (Kandavelou and Chandrasegaran 2009).
-
2.
Genome engineering in cattle by TALENs
With the decoding of the DNA recognition codes employed by transcription activator-like effector (TALE) proteins, TALENs can be easily programmed to target a DNA sequence of interest (Cermak et al. 2011; Li and Yang 2013). In great contrast to ZFNs, which have considerable constraints on what DNA sequences can be recognized and targeted, TALENs can be engineered to recognize any DNA sequence that starts with a thymine at the 5″ end (Wright et al. 2014). This flexibility in DNA sequence recognition places TALENs as the most versatile designer nucleases for genome engineering. TALENs have also been demonstrated to have much reduced off-target activity and nuclease-associated cytotoxicities than ZFNs (Mussolino et al. 2011). Due to these and other advantages over ZFNs, TALENs have been quickly applied to many organisms, including cattle.
In one of the first demonstrations of using TALENs to target the livestock genomes, Scott Fahrenkrug and his colleagues reported that TALENs are very efficient in inducing site-specific DNA sequence insertions and deletions (indels) in the bovine genome both in fibroblast cells and in early embryos (Carlson et al. 2012). In addition, although only demonstrated in the pig, co-transfection of two TALEN-pairs targeting the same chromosome into porcine fibroblast cells resulted in large chromosomal deletions and inversions (Carlson et al. 2012). It is conceivable that such chromosomal alterations can be achieved in cattle as well. Most encouraging is the results showing that microinjection of TALEN messenger RNAs (mRNAs) into the cytoplasm of bovine zygotes produced MSTN gene KO cattle (also demonstrated in sheep) (Proudfoot et al. 2014). This success opened up the possibility of conducting more complex site-specific genetic modifications in cattle without using animal cloning.
One of the most significant breakthroughs using designer nucleases to engineer the cattle genome for agricultural application is the achievement of nonmeiotic allele introgression via TALEN-mediated gene editing (Tan et al. 2013). These researchers demonstrated that, through TALEN-mediated homology-directed repair (HDR), naturally occurring alleles can be efficiently introduced from one breed to another. With this approach, they first succeeded in introducing the naturally occurring SNP and 11-bp deletion in the MSTN gene from Piedmontese cattle and Belgian Blue cattle (McPherron and Lee 1997; Kambadur et al. 1997), respectively, into the genome of Wagyu cattle. They also succeeded in introgressing the POOLED allele from Celtic breed (Pc allele) into horned dairy breed. Although no attempt was made in this study to use these allele-introgressed cells for cattle production, it is conceivably a very reachable goal. This study demonstrated that, by using TALEN-mediated gene editing, a SNP or an allele can be interchangeably transgressed among breeds or even between species with high efficiency (estimated to be 105-fold more efficient than conventional HR-mediated approach). This study is of great significance in that, in contrast to meiotic allele introgressions through breeding, it avoids whole-genome admixtures that often result in the loss of other desired genetic information (therefore desired traits) and the gain of undesired genetic information (therefore undesired traits). Furthermore, nonmeiotic allele introgression can be achieved marker-free—no foreign DNA, such as antibiotic resistant genes or bacterial/viral DNA sequences that are commonly introduced by conventional gene targeting processes, is introduced.
We also recently succeeded in employing TALEN-mediated HDR to KI functional genes into the bovine genome in a sequence-specific manner. To genetically engineer cattle for high-level recombinant therapeutic proteins in the mammary gland, we developed a universal KI strategy to integrate any gene of interest into the endogenous bovine ″-casein locus (Lee et al. 2013). This universal gene KI strategy composes two components: one is a TALEN pair designed to introduce DSBs in the bovine ″-casein locus and the other is the KI vector (also called donor vector) carrying a gene of interest flanked by the two universal homologous arms isolated from the ∼500 bp bovine genomic DNA immediately 5′ and 3′, respectively, of the translation start codon of the bovine ″-casein gene. We call such gene-targeting strategy “universal,” as the same TALEN pair can be used to introduce DSBs in the ″-casein locus and that the two universal homologous arms in the donor vector allow any gene of interest to be cloned into this vector and subsequently knocked in the ″-casein locus. With this gene KI strategy, the coding sequence of a gene of interest is inserted immediately before the start codon of the bovine ″-casein gene, thus allowing for the expression of the knocked in gene to be totally under the control of the endogenous cis-elements of the bovine ″-casein gene. We incorporated polyadenylation signals at the end of the coding sequence of the gene to be knocked in, for its independent mRNA maturation and protein translation. As ″-casein is the major protein expressed in cattle mammary gland, high-level expression of the knocked in gene in the milk can be expected.
We are currently employing this universal KI strategy to engineer cattle to produce high quantities of recombinant therapeutic proteins in the mammary gland. Among the genes of interest, human erythropoietin (hEPO) gene in bovine fibroblast cells was the first gene we knocked in (Lee et al. 2013). We demonstrated that co-transfection of the TALEN constructs and the donor vector into bovine fibroblasts is highly effective in knocking the hEPO gene into the bovine ″-casein locus without using any selection method: among the ten single-cell derived colonies established from the transfected cells by limiting dilution, two of them (20 %) harbor the desired KI event. We conducted animal cloning by SCNT by using these two cell lines as nuclear donors and have established multiple pregnancies. Another therapeutic protein that we are interested in expressing in the bovine mammary gland is the human serum albumin (hSA). Due to increasingly needs in therapeutic and laboratory applications, the worldwide annual demand for hSA is estimated to be at least 500 t (He et al. 2011). However, as the current production of hSA is from pooled human plasma of human donors and is dependent on voluntary donations, the supply of human plasma is not stable and often experiences severe shortages. Therefore, our work may provide a viable solution to this product shortage problem.
Very recently, a similar TALEN-mediated HDR approach was employed to replace the bovine serum albumin (BSA) gene with two hSA minigenes, one for liver-specific expression and the other for mammary gland-specific expression (Moghaddassi et al. 2014). These researchers demonstrated that co-transfection of TALENs and the hSA minigene donor constructs resulted in targeted integration of the hSA minigene into the bovine genome. However, a neomycin-resistant cassette had to be used in the donor vector to aid the establishment of correctly targeted bovine fibroblast cell lines. As acknowledged by the authors, deletion of the neomycin-resistant cassette before using such cells for cattle production is necessary. Very recently, as demonstrated by the KO of MSTN gene, TALENs are also effective in gene targeting in bovine aortic endothelial cells (Xu et al. 2013). Nevertheless, no attempt was made to produce genetically engineered cattle by these two studies.
The above studies, although not numerous, have nevertheless demonstrated the power of using TALENs to engineer cattle genome. With the ease of design and assembly of TALEN constructs, the seemingly limitless target sequences in a genome, and the high efficiency and specificity in inducing site-specific genetic modifications, it is reasonable to expect that this technology will be quickly employed in many laboratories interested in engineering the cattle genome.
-
3.
Genome engineering in cattle by CRISPR/Cas9 system
CRISPR/Cas9 system is the newest addition to the designer nuclease family as a genome engineering tool. So far, there is only one report on using CRISPR/Cas9 system to engineer the bovine genome (Heo et al. 2014). This study demonstrated that the CRISPR/Cas9 system is highly efficient in editing the bovine genome in both bovine embryos and induced pluripotent stem cells. The paucity of publications on the use of the CRISPR/Cas9 system in cattle genome engineering may largely be due to cattle’s long reproduction cycle and the fairly recent introduction of CRISPR/Cas9 as a genome engineering tool (less than 2 years). We have been successful in applying the CRISPR/Cas9 system to conduct genome engineering in cattle and several other mammalian species, such as goats, sheep, and Syrian hamsters (Fan et al. 2014). Without a doubt, other research groups are also making progress with this system in engineering the cattle genome. Therefore, it is expected that a string of scientific publications on using this powerful technology to engineer the cattle genome will ensue in the coming years.
Conclusions
The last decade has witnessed great technological advancements in cattle genome engineering. The establishment of sequential genome engineering technology in cattle a decade ago has led to the production of cattle with the most complex genetic modifications ever accomplished. However, due to the high level of technical difficulty involved and the low animal production efficiency by animal cloning, this technology had not been widely adopted by many researchers. The recent development of designer nucleases and their successful application to cattle mark a new era in cattle genome engineering. First and foremost is the capacity to engineer cattle genome at single-nucleotide precision without the necessity of introducing extraneous DNA sequences. This is a revolutionary technological step that has fundamentally changed how genome engineering can be performed; it will also help to redefine the meaning of genetically engineered animals and how the public thinks about using genetically engineered cattle (and other livestock) for agricultural and biomedical applications. For the first time, concerns about the introduction of extraneous DNA sequences, such as bacterial and viral DNA sequences, by the old transgenic techniques can be scientifically addressed. Consequently, this technological advancement will also have important implications for regulatory approval of products produced from genetically engineered cattle. Second, as demonstrated by the ability to produce gene KO cattle by microinjecting the designer nucleases into bovine zygotes, complex cattle genome engineering can potentially be achieved without using animal cloning. The capacity to engineer the bovine genome directly in zygotes with designer nucleases would eliminate the problems associated with animal cloning, including low efficiency and animal welfare issues. Finally, the huge improvement of efficiency in engineering cattle genome brought about by designer nucleases and the simplicity of their utilization will undoubtedly make designer nucleases the technology of choice for cattle genome engineering.
In summary, with the recent technological advancements in cattle genome engineering, one can expect that the long envisioned agricultural and biomedical applications by genetically engineered cattle will soon be realized in the marketplace.
Abbreviations
- SCNT:
-
Somatic cell nuclear transfer
- CT:
-
Chromatin transfer
- ES:
-
Embryonic stem cells
- HR:
-
Homologous recombination
- KO:
-
Knock out
- KI:
-
Knock in
- PN:
-
Pronuclear
- CGH:
-
Comparative genomic hybridization
- ZFNs:
-
Zinc finger nucleases
- TALENs:
-
Transcription activator-like effector nuclease
- CRISPR/Cas9:
-
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9
- HDR:
-
Homology-directed repair
- NHEJ:
-
Nonhomologous end joining
- PGK:
-
Phosphoglycerate kinase-1
- ST SV40:
-
Promoter and thymidine kinase enhancer
- CAG:
-
Chicken beta-actin promoter with CMV enhancer
- HAC:
-
Human artificial chromosome
- Indel:
-
Insertion or deletion of DNA base pair(s)
- hEPO:
-
Human erythropoietin
- hSA:
-
Human serum albumin
- BSA:
-
Bovine serum albumin
References
Bachiller D, Schellander K, Peli J, Ruther U (1991) Liposome-mediated DNA uptake by sperm cells. Mol Reprod Dev 30:194–200
Bandaranayake AD, Almo SC (2014) Recent advances in mammalian protein production. FEBS Lett 588:253–60
Bosze Z, Baranyi M, Whitelaw CB (2008) Producing recombinant human milk proteins in the milk of livestock species. Adv Exp Med Biol 606:357–93
Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC (2012) Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A 109:17382–7
Carlson DF, Tan W, Hackett PB, Fahrenkrug SC (2013) Editing livestock genomes with site-specific nucleases. Reprod Fertil Dev 26:74–82
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–82
Carroll D (2014) Genome engineering with targetable nucleases. Annu Rev Biochem 83:409–39
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82
Chan AW, Kukolj G, Skalka AM, Bremel RD (1999) Timing of DNA integration, transgenic mosaicism, and pronuclear microinjection. Mol Reprod Dev 52:406–13
Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, Ponce De Leon FA, Robl JM (1998) Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science 280:1256–8
Donovan DM, Kerr DE, Wall RJ (2005) Engineering disease resistant cattle. Transgenic Res 14:563–7
Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33:5978–90
Fan Z, Li W, Lee SR, Meng Q, Shi B, Bunch TD, White KL, Kong IK, Wang Z (2014) Efficient gene targeting in golden Syrian hamsters by the CRISPR/Cas9 system. PLoS One 9:e109755
Gaj T, Gersbach CA, Barbas CF 3rd (2013) Zfn, Talen, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405
Golovan SP, Meidinger RG, Ajakaiye A, Cottrill M, Wiederkehr MZ, Barney DJ, Plante C, Pollard JW, Fan MZ, Hayes MA, Laursen J, Hjorth JP, Hacker RR, Phillips JP, Forsberg CW (2001) Pigs expressing salivary phytase produce low-phosphorus manure. Nat Biotechnol 19:741–5
Gupta RM, Musunuru K (2014) Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest 124:4154–61
Hammer RE, Pursel VG, Rexroad CE Jr, Wall RJ, Bolt DJ, Ebert KM, Palmiter RD, Brinster RL (1985) Production of transgenic rabbits, sheep and pigs by microinjection. Nature 315:680–3
He Y, Ning T, Xie T, Qiu Q, Zhang L, Sun Y, Jiang D, Fu K, Yin F, Zhang W, Shen L, Wang H, Li J, Lin Q, Sun Y, Li H, Zhu Y, Yang D (2011) Large-scale production of functional human serum albumin from transgenic rice seeds. Proc Natl Acad Sci U S A 108:19078–83
Heo, Y., Quan, X., Xu, Y., Baek, S., Choi, H., Kim, N. & KIM, J. (2014) CRISPR/Cas9 nuclease-mediated gene knock-in in bovine pluripotent stem cells and embryos. Stem Cells Dev. doi:10.1089/scd.2014.0278
Hofmann A, Kessler B, Ewerling S, Weppert M, Vogg B, Ludwig H, Stojkovic M, Boelhauve M, Brem G, Wolf E, Pfeifer A (2003) Efficient transgenesis in farm animals by lentiviral vectors. EMBO Rep 4:1054–60
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–78
Kambadur R, Sharma M, Smith TP, Bass JJ (1997) Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res 7:910–6
Kandavelou K, Chandrasegaran S (2009) Custom-designed molecular scissors for site-specific manipulation of the plant and mammalian genomes. Methods Mol Biol 544:617–36
Karatzas CN (2003) Designer milk from transgenic clones. Nat Biotechnol 21:138–9
Kim H, Kim JS (2014) A guide to genome engineering with programmable nucleases. Nat Rev Genet 15:321–34
Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS (2009) Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19:1279–88
Koller BH, Smithies O (1992) Altering genes in animals by gene targeting. Annu Rev Immunol 10:705–30
Krimpenfort P, Rademakers A, Eyestone W, Van Der Schans A, Van Den Broek S, Kooiman P, Kootwijk E, Platenburg G, Pieper F, Strijker R et al (1991) Generation of transgenic dairy cattle using ‘in vitro’ embryo production. Biotechnology (N Y) 9:844–7
Kubisch HM, Larson MA, Eichen PA, Wilson JM, Roberts RM (1997) Adenovirus-mediated gene transfer by perivitelline microinjection of mouse, rat, and cow embryos. Biol Reprod 56:119–24
Kubota C, Tian XC, Yang X (2004) Serial bull cloning by somatic cell nuclear transfer. Nat Biotechnol 22:693–4
Kuroiwa Y, Kasinathan P, Matsushita H, Sathiyaselan J, Sullivan EJ, Kakitani M, Tomizuka K, Ishida I, ROBL JM (2004) Sequential targeting of the genes encoding immunoglobulin-mu and prion protein in cattle. Nat Genet 36:775–80
Kuroiwa Y, Kasinathan P, Sathiyaseelan T, Jiao JA, Matsushita H, Sathiyaseelan J, Wu H, Mellquist J, Hammitt M, Koster J, Kamoda S, Tachibana K, Ishida I, Robl JM (2009) Antigen-specific human polyclonal antibodies from hyperimmunized cattle. Nat Biotechnol 27:173–81
Lee S, Park H, Kong I, Wang Z (2013) 30 a transcription activator-like effector nuclease (Talen)-mediated universal gene knock-in strategy for mammary glands-specific expression of recombinant proteins in dairy cattle. Reprod Fertil Dev 26:129–129
Li T, Yang B (2013) TAL effector nuclease (TALEN) engineering. Methods Mol Biol 978:63–72
Liu GE, Hou Y, Robl JM, Kuroiwa Y, Wang Z (2011) Assessment of genome integrity with array CGH in cattle transgenic cell lines produced by homologous recombination and somatic cell cloning. Genome Integr 2:6
Liu X, Wang Y, Guo W, Chang B, Liu J, Guo Z, Quan F, Zhang Y (2013) Zinc-finger nickase-mediated insertion of the lysostaphin gene into the beta-casein locus in cloned cows. Nat Commun 4:2565
Liu X, Wang Y, Tian Y, Yu Y, Gao M, Hu G, Su F, Pan S, Luo Y, Guo Z, Quan F, Zhang Y (2014) Generation of mastitis resistance in cows by targeting human lysozyme gene to beta-casein locus using zinc-finger nucleases. Proc Biol Sci 281:20133368
Luo J, Song Z, Yu S, Cui D, Wang B, Ding F, Li S, Dai Y, Li N (2014) Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases. PLoS One 9:e95225
Matsushita H, Sano A, Wu H, Jiao JA, Kasinathan P, Sullivan EJ, Wang Z, Kuroiwa Y (2014) Triple immunoglobulin gene knockout transchromosomic cattle: bovine lambda cluster deletion and its effect on fully human polyclonal antibody production. PLoS One 9:e90383
Mcpherron AC, Lee SJ (1997) Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci U S A 94:12457–61
Moghaddassi S, Eyestone W, Bishop CE (2014) TALEN-mediated modification of the bovine genome for large-scale production of human serum albumin. PLoS One 9:e89631
Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T (2011) A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39:9283–93
Pasman Y, Saini SS, Smith E, Kaushik AK (2010) Organization and genomic complexity of bovine lambda-light chain gene locus. Vet Immunol Immunopathol 135:306–13
Proudfoot, C., Carlson, D. F., Huddart, R., Long, C. R., Pryor, J. H., King, T. J., Lillico, S. G., Mileham, A. J., Mclaren, D. G., Whitelaw, C. B. & Fahrenkrug, S. C. 2014. Genome edited sheep and cattle. Transgenic Res. doi:10.1007/s11248-014-9832-x
Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, Mccaffrey AP, Cathomen T, Scharenberg AM, Joung JK (2012) Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res 40:5560–8
Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, Vargas F, Sathiyaseelan J, Wu H, Matsushita H, Koster J, Kato S, Ishida I, Soto C, Robl JM, Kuroiwa Y (2007) Production of cattle lacking prion protein. Nat Biotechnol 25:132–8
Robl JM, Wang Z, Kasinathan P, Kuroiwa Y (2007) Transgenic animal production and animal biotechnology. Theriogenology 67:127–33
Sander JD, Reyon D, Maeder ML, Foley JE, Thibodeau- Beganny S, LI X, Regan MR, Dahlborg EJ, Goodwin MJ, Fu F, Voytas DF, Joung JK, Dobbs D (2010) Predicting success of oligomerized pool engineering (OPEN) for zinc finger target site sequences. BMC Bioinform 11:543
Sano A, Matsushita H, Wu H, Jiao JA, Kasinathan P, Sullivan EJ, Wang Z, Kuroiwa Y (2013) Physiological level production of antigen-specific human immunoglobulin in cloned transchromosomic cattle. PLoS One 8:e78119
Sedivy JM, Sharp PA (1989) Positive genetic selection for gene disruption in mammalian cells by homologous recombination. Proc Natl Acad Sci U S A 86:227–31
Selo I, Negroni L, Creminon C, Yvon M, Peltre G, Wal JM (1998) Allergy to bovine beta-lactoglobulin: specificity of human IgE using cyanogen bromide-derived peptides. Int Arch Allergy Immunol 117:20–8
Sendai Y, Sawada T, Urakawa M, Shinkai Y, Kubota K, Hoshi H, Aoyagi Y (2006) alpha1,3-Galactosyltransferase-gene knockout in cattle using a single targeting vector with loxP sequences and cre-expressing adenovirus. Transplantation 81:760–6
Sullivan EJ, Kasinathan S, Kasinathan P, Robl JM, Collas P (2004) Cloned calves from chromatin remodeled in vitro. Biol Reprod 70:146–53
Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, FAHRENKRUG SC (2013) Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci U S A 110:16526–31
Wakayama T, Shinkai Y, Tamashiro KL, Niida H, Blanchard DC, Blanchard RJ, Ogura A, Tanemura K, Tachibana M, Perry AC, Colgan DF, Mombaerts P, Yanagimachi R (2000) Cloning of mice to six generations. Nature 407:318–9
Wall RJ, Kerr DE, Bondioli KR (1997) Transgenic dairy cattle: genetic engineering on a large scale. J Dairy Sci 80:2213–24
Wall RJ, Powell AM, Paape MJ, Kerr DE, Bannerman DD, Pursel VG, Wells KD, Talbot N, Hawk HW (2005) Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat Biotechnol 23:445–51
Wang S, Zhang K, Ding F, Zhao R, Li S, Li R, Xu L, Song C, Dai Y, Li N (2013) A novel promoterless gene targeting vector to efficiently disrupt PRNP gene in cattle. J Biotechnol 163:377–85
Wright DA, LI T, Yang B, Spalding MH (2014) TALEN-mediated genome editing: prospects and perspectives. Biochem J 462:15–24
Wu J, Kandavelou K, Chandrasegaran S (2007) Custom-designed zinc finger nucleases: what is next? Cell Mol Life Sci 64:2933–44
Xu L, Zhao P, Mariano A, Han R (2013) Targeted myostatin gene editing in multiple mammalian species directed by a single pair of TALE nucleases. Mol Ther Nucleic Acids 2:e112
Yu S, Luo J, Song Z, Ding F, Dai Y, Li N (2011) Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res 21:1638–40
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editors: Natalay Kouprina and Vladimir Larionov
Rights and permissions
About this article
Cite this article
Wang, Z. Genome engineering in cattle: recent technological advancements. Chromosome Res 23, 17–29 (2015). https://doi.org/10.1007/s10577-014-9452-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-014-9452-6