Abstract
Key message
The fusion gene 4CL-CCR promotes lignification and activates lignin-related MYB expression in tobacco but inhibits auxin-related gene expression and hinders the auxin absorption of cells.
Given the importance of lignin polymers in plant growth and their industrial value, it is necessary to investigate how plants synthesize monolignols and regulate the level of lignin in cell walls. In our previous study, expression of the Populus tomentosa fusion gene 4CL-CCR significantly promoted the production of 4-hydroxycinnamyl alcohols. However, the function of 4CL-CCR in organisms remains poorly understood. In this study, the fusion gene 4CL-CCR was heterologously expressed in tobacco suspension cells. We found that the transgenic suspension cells exhibited lignification earlier. Furthermore, 4CL-CCR significantly reduced the content of phenolic acids and increased the content of aldehydes in the medium, which led to an increase in lignin deposition. Moreover, transcriptome results showed that the genes related to lignin synthesis, such as PAL, 4CL, CCoAOMT and CAD, were significantly upregulated in the 4CL-CCR group. The expression of genes related to auxin, such as ARF3, ARF5 and ARF6, was significantly downregulated. The downregulation of auxin affected the expression of transcription factor MYBs. We hypothesize that the upregulated genes MYB306 and MYB315 are involved in the regulation of cell morphogenesis and lignin biosynthesis and eventually enhance lignification in tobacco suspension cells. Our findings provide insight into the function of 4CL-CCR in lignification and how secondary cell walls are formed in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignin is a major structural component of plant cell walls (Grossman and Vermerris 2019). The function of this polymer is mainly related to the terrestrial environment because it is responsible for mechanical support and protection (Ralph et al. 2019; Dong and Lin 2021). In addition to playing a central role in the development of vascular plants, lignin provides a significant contribution to the quality of a range of economical products derived from plant lignocellulosic biomass (Rais and Zibek 2019; Shi et al. 2023). Although the composition of lignin polymers is complex and varied, the main components are derived from the oxidative coupling of three p-hydroxycinnamyl alcohols (monolignols), p-coumaryl, coniferyl and sinapyl alcohols, which differ by methoxylation (Wang et al. 2016). After these monomers were incorporated into the lignin polymer, p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units were produced (Vanholme et al. 2019). Given the importance of lignin polymers in plant growth and their industrial value, it is necessary to investigate how plants synthesize monolignols and regulate the level of lignin in cell walls.
4-Coumaric acid: coenzyme A ligase (4CL) is a key enzyme in the phenylpropanoid metabolic pathway that regulates the biosynthesis of lignin and flavonoids (Li et al. 2020, 2023). Cinnamoyl-coenzyme A reductases (CCRs) have been reported as key enzymes involved in monolignol biosynthesis (Chao et al. 2019; Hu et al. 2021). In our previous study (Liu et al. 2015), 4CL and CCR were fused to generate an artificial bifunctional enzyme, 4CL-CCR. The fusing enzyme 4CL-CCR catalyzes continuous multistep reactions (Liu et al. 2017) and significantly enhances the biosynthesis of 4-hydroxycinnamyl alcohols (Vanholme et al. 2019), which promotes the synthesis of lignin. However, the function of 4CL-CCR in organisms remains poorly understood. We found that cells treated with auxin analogs, including 2,4-dichlorophenoxyacetic acid (2,4-D) or picloram, showed enhanced division and inhibited lignification, while treatment with benzylaminopurine (BA) induced lignification in bamboo (Ogita et al. 2018). In addition, the CCR gene was upregulated in response to BA treatment, and the 4CL gene was upregulated in response to both 2,4-D and BA treatments (Wiszniewski et al. 2009; Khadr et al. 2020). These findings suggest that the specific upregulation of genes encoding 4CL and CCR may be involved in lignification through coordinated transcriptional regulation and metabolic alterations.
In this study, we investigated the role of the fusion gene 4CL-CCR during the proliferation and lignification of tobacco suspension cells. Histochemical staining and microscopic observations showed that 4CL-CCR significantly accelerated the lignification of tobacco suspension cells. Moreover, gas chromatography‒mass spectrometry (GC‒MS) and high-performance liquid chromatography‒mass spectrometry (HPLC‒MS) were used to analyze the lignin composition and its precursor, phenolic acid, respectively. Furthermore, the transcriptional changes associated with lignification were examined to clarify the functions of 4CL-CCR. Our findings have provided knowledge of how secondary cell walls are formed in plants.
Materials and methods
Construction of plant expression vector
Nicotiana tabacum was cultivated in the laboratory. Pto4CL1-CCR was cloned as described previously (Liu et al. 2015). The full-length fusion Pto4CL1-CCR gene fragment was amplified using specific primers, and the sense primer and antisense primer were 5′-TCTAGAATGAATCCACAAGAAG-3′ and 5′-CCCGGGTTATTGAATCTTCACAG-3′, respectively. The specific primers were designed on both sides of the gene with Xba I and Sma I restriction enzyme sites. The PCR product was gel-purified and cloned into the pBI121 plant expression vector under the transcriptional control of the 35S promoter and then introduced into Agrobacterium tumefaciens GV3101 and confirmed by PCR and DNA sequencing (TSINGKE, Beijing, China).
Pto4CL1-CCR gene transformation into tobacco plants
The positive GV3101 clone with the recombinant vector was cultivated in YEB medium containing 50 mg/L kanamycin, 50 mg/L rifampicin, and 50 mg/L gentamicin at 28 °C overnight. The activated bacteria were transferred to antibiotic-free YEB liquid medium until the OD600 reached 0.3–0.6. Tobacco leaves were cut into small pieces (4.0 × 4.0 cm), immersed in Agrobacterium solution for 10 min, and then transferred to Murashige and Skoog (MS) solid medium supplemented with 1.5 mg/L 6-benzylamino-purine (6-BA) and 0.3 mg/L naphthylacetic acid (NAA) in the dark for 2–3 days. To induce the root, the regenerated buds at a height of 1.0 cm were transferred to rooting medium. Pieces of rooted tobacco leaves were harvested for DNA extraction. The transgenic plants were determined by PCR with pBI121-specific primers (sense primer: 5′-ACGCACAATCCCACTATCCTT-3′, antisense primer: 5′-TTCATTTTCGTCTGCCTGTGTT-3′) and southern blotting.
Establishment of a suspension culture
Well-growing and identical plants were selected to induce calli. Sterilized leaf segments were cultured on MS medium supplemented with 0.3 mg/L NAA and 1.5 mg/L 6-BA for four weeks under dark conditions. Initiated calli proliferated on MS medium supplemented with 1.0 mg/L NAA and 0.5 mg/L 6-BA for subculture for four weeks under dark conditions. Approximately 1.0 g fresh weight of friable white calli were transferred to 200 mL flasks containing 50 mL MS liquid medium supplemented with 1 mg/L 2,4-D, incubated on a shaker with constant temperature (110 rpm, 25 ℃), and stored in darkness. The suspension cultures were subcultured in fresh MS liquid medium at 10-day intervals. This procedure was repeated approximately 4–6 times. Then, the supernatant was filtered through a 210 μm mesh filter, and the filtrate was used as an initial source for the subsequent cell suspension cultures. The suspension cells were subcultured by transferring 5 mL of fresh cells into 45 mL of fresh liquid medium at 7-day intervals. The cell viability and morphology assays were performed by staining the cells using fluorescein diacetate (FDA) (Sigma, UK) by the method described previously (Namsi et al. 2019). The cell suspension growth curve was estimated and monitored using fresh weight according to the method described (Pan et al. 2020).
Identification of suspension cell lignification and lignin composition
The lignin content of suspension cells was measured according to the phloroglucinol-HCl reaction (Siegel 1953). Cells were collected at different cultivation periods (2, 4, 11 and 24 days), centrifuged to move the medium, and then observed and imaged under a fluorescence microscope (BX61TRF, Olympus, Japan). A GC‒MS system (Trace-GC Ultra) connected to a Trace-DSQ mass selective detector with EI ionization of full scan and selected ion monitoring (selected ion 1.0 mass unit) (Thermo-Finnigan, San Jose, CA) was used in this experiment. The column used was a 30 m × 0.25 mm I.D., 0.25 μm film thickness, DB-5 ms fused silica capillary column (Agilent, USA). The analysis of lignin composition with GC‒MS assays was adapted from described previously (Qi et al. 2019). In short, samples were milled to powder with liquid nitrogen, extracted and dried, and then thiaoacidolysis solution was added under nitrogen. The extraction was transferred to a capillary, and then a derivative reaction was carried out with BSTFA. Finally, the reaction solution was analyzed by GC‒MS. The data were analyzed using Xcalibur 2.1 software (Thermo-Finnigan).
Qualitative and quantitative analyses of phenolic acids in culture medium by HPLC‒MS
The content of phenolic compounds was analyzed by HPLC‒MS (Agilent, Santa Clara, CA, USA) (Slazak et al. 2020). Briefly, suspended cells were collected at different cultivation periods (2, 4, 11 and 24 days) and centrifuged to retain the medium, and then the medium was extracted and vacuum-dried, followed by resuspension in the mobile phase. The solution was separated and identified by an HPLC system and an ion trap mass spectrometer coupled with an ESI source, sequentially. HPLC separation was carried out using a reversed-phase column (ZORBAX 300SB-C18, 2.1 × 150 mm, 3.5 μm). The solvents, mobile phases, and MS parameters were the same as described previously. The data were analyzed using Xcalibur 2.1 software (Thermo-Finnigan).
RNA extraction and sequencing
A modified CTAB method (Khairul-Anuar et al. 2019) was used to extract total RNA from tobacco suspension cells at different cultivation periods (2, 4, 11 and 24 days). The degradation degree, concentration, purity and integrity of the RNA samples were measured. First, the mRNA was enriched with oligo (dT) magnetic beads and broken into short fragments by adding fragmentation buffer. Random primers were used to synthesize the first strand of cDNA. Then, the second strand cDNA was synthesized and purified, subjected to sticky end repair, and ligated. Finally, the effective concentration of the library was quantified to ensure the quality of the library. The constructed library was sequenced using the Illumina Sequencing Platform (San Diego, CA, USA).
Transcriptome analysis
After sequencing, the raw reads were filtered to obtain clean reads. The reads with adapters, a proportion of N greater than 10%, and low quality (the number of bases with Qphred < 5 accounted for more than 50% of the total reads) were removed. Subsequently, the clean reads were compared using TopHat software (http://ccb.jhu.edu/software/tophat). The comparison results were calculated, and the number of read counts of each sample compared to each gene was obtained, which were the input data of differentially expressed genes (DEGs). For biologically duplicated samples, we performed an analysis using DESeq2. The screening threshold was p < 0.05 and |log2FoldChange |> 1. Annotations from Gene Ontology (GO) (Ashburner et al. 2000) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Zheng et al. 2019) were tested for enrichment.
Results
Establishment of cell suspension cultures of tobacco
To obtain transgenic tobacco plants, the recombinant vector pBI121-4CL-CCR was transformed into A. tumefaciens strain GV3101 and then introduced into tobacco plants. To confirm whether pBI121-4CL-CCR was successfully transferred to the target strain, regenerated tobacco plants were screened by PCR (Fig. S1a). In addition, southern blotting analysis was carried out on the DNA obtained from the transgenic plants. The results indicated that there were the following positive transgenic lines were present: 4CL-CCR 2, 4CL-CCR 5–2, 4CL-CCR 10–1, 4CL-CCR 16–1 and 4CL-CCR 20–2 (Fig. S1b). The patterns of the hybridization bands differed from plant to plant, indicating independent transformation events and random integration. The positive transgenic plants were further analyzed. Friable and cream callus clumps were collected, and the optical growth condition was used to start the cell suspension culture (Fig. S1c-e). To find the best growth node, the growth curve of suspension cells was mapped by measuring the fresh weight of suspension cells. We found that the fresh weight of the 4CL-CCR plants was significantly higher than that of the WT plants (Fig. 1, ** P < 0.01). Furthermore, the suspension cell line entered the logarithmic stage on the 4th day and the plateau stage on the 11th day (Fig. 1). Therefore, suspension cells of Day 2, Day 4, Day 11 and Day 24 were used for the subsequent experiments.
4CL-CCR promoted the lignification of suspension cells in tobacco
To observe the morphological differences more closely during suspension cell growth, we used FDA staining (a fluorescent dye that markers living cells). We found that there was no significant difference in cell morphology between the 4CL-CCR group and the WT group at Days 4 and 11 (Fig. 2a). However, the cells in the 4CL-CCR group showed link fracture and a single globular aggregation when the growth of suspension cells reached a plateau at Day 24, and no significant change were observed in the WT group (Fig. 2a). Since the 4CL-CCR group showed morphological changes related to lignification (Habarugira et al. 2015; Que et al. 2018), we measured the level of lignification in the two groups of cells by phloroglucinol-HCl reactions. The results showed that the cells in the 4CL-CCR group exhibited a higher level of lignification than those in the WT group (Fig. 2b, c), and the tobacco culture medium of the 4CL-CCR group appeared pale-yellow and then gradually darkened (Fig. 2d). These results confirm that 4CL-CCR promotes the lignification of suspension cells in tobacco.
Dynamic changes in cell suspension growth rate, morphology and lignin accumulation during development. a Observation of suspension cell morphology by FDA at different time points, bar = 100 μm. b, c The degree of lignification of suspension cells was determined by phloroglucinol-HCl reaction at different time points. d The suspension cell growth showed a color difference at different culture time
Effect of 4CL-CCR on lignin synthesis in the suspension cell
To investigate the mechanism by which 4CL-CCR regulates lignin synthesis, the lignin precursor phenolic acid (p-coumaric acid, caffeic acid, ferulic acid and sinapic acid) and its derivatives (cinnamic aldehyde, caffeyl aldehyde, conifer aldehyde and sinap aldehyde) in suspension cell culture medium were measured to reflect the synthesis of lignin. These phenolic acids and aldehydes were analyzed by HPLC‒MS. We found that the phenolic acid content in the suspension cell culture medium decreased significantly in the 4CL-CCR group (Fig. 3a–d; ** P < 0.01). Although there were some differences at different time points, the phenolic acid content in the 4CL-CCR group showed a downward trend with increasing subculture time, while the phenolic acid content in the WT group showed an upward trend.
Solubility phenolic acid and aldehyde of suspension cell at different subculture time. a Quantification of the p-coumaric acid level of suspension cells in Wild Type and 4CL-CCR group. (F (1, 16) = 33.62, P < 0.0001). b Quantification of the caffeic acid level of suspension cells in Wild Type and 4CL-CCR group. (F (1, 16) = 181.5, P < 0.0001). c Quantification of the ferulic acid level of suspension cells in Wild Type and 4CL-CCR group. (F (1, 16) = 134.5, P < 0.0001). d Quantification of the sinapic acid level of suspension cells in Wild Type and 4CL-CCR group. (F (1, 16) = 65.79, P < 0.0001). e Quantification of the cinnamic aldehyde level of suspension cells in Wild Type and 4CL-CCR group. (F (1, 16) = 5562, P < 0.0001). f Quantification of the caffeyl aldehyde level of suspension cells in Wild Type and 4CL-CCR group. (F (1, 16) = 222.3, P < 0.0001). g Quantification of the conifer aldehyde level of suspension cells in Wild Type and 4CL-CCR group. (F (1, 16) = 20.44, P = 0.0003). h Quantification of the sinap aldehyde level of suspension cells in Wild Type and 4CL-CCR group. (F (1, 16) = 0.093, P = 0.7646). Two way ANOVA with Bonferronl’s multiple comparisons. Data are mean ± S.E.M. *, P < 0.05, **, P < 0.01
We also found that the contents of cinnamic aldehyde, caffeyl aldehyde, and conifer aldehyde in the suspension cell culture medium were increased significantly in the 4CL-CCR group (Fig. 3e–g; ** P < 0.01). There was no significant difference in the content of sinap aldehyde between the WT and 4CL-CCR groups (Fig. 3h), which indicates that 4CL-CCR had no catalytic effect on sinapic acid.
Moreover, the aldehyde content in the WT group was basically stable, while the aldehyde content in the 4CL-CCR group increased with increasing culture time. This indicates that there are more upstream substrates catalyzed by 4CL-CCR to form lignin precursor aldehydes.
The above results indicated that the rapid consumption of phenolic acid by 4CL-CCR prompted the cells to reabsorb phenolic acid in the medium, further catalyzed the formation of corresponding aldehydes, and finally released the aldehydes into the extracellular environment.
Identification of genes potentially involved in lignin biosynthesis
RNA-seq analysis of the suspension cells was performed in four developmental stages (Day 2, Day 4, Day 11 and Day 24) to characterize the dynamics of the transcriptome during suspension cell development and to identify genes involved in 4CL-CCR-specific lignification. Nicotiana sylvestris was used as a reference genome, and detailed information about differentially expressed genes (DEGs) can be found in the supplementary data, including Day 2 in Table S1, Day 4 in Table S2, Day 11 in Table S3, and Day 24 in Table S4. We first focused on lignin-related DEGs, and a total of 49 DEGs in 8 lignin biosynthesis gene families were obtained. As shown in Table 1, among the 3 homologous genes of phenylalanine ammonia lyase (PAL), only PAL-like expression increased at different sampling times. Increased expression was found for the 4CL family members except 4CL-like 7. The upregulated genes in the 4CL family of tobacco were mainly 4CL2 and 4CL2-like. The members of the tobacco 4CL gene family can be divided into the following broad categories: one is involved in the synthesis of lignin monomers, and the other is involved in the synthesis of flavonoids (Lee and Douglas 1996). In this experiment, 4CL2 and 4CL2-like, which were mainly upregulated genes in the 4CL family, may be involved in the synthesis of tobacco flavonoids. Presumably, tobacco cells contain enough 4CL protein to synthesize the lignin precursor due to the expression of the fusion gene 4CL-CCR (Hu et al. 2010). Therefore, the expression of the endogenous 4CL gene involved in lignin synthesis in tobacco was not changed. Because of the feedforward effect of the substrate, the expression of 4CL genes involved in flavonoid synthesis increased (Fig. 4a).
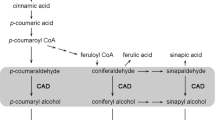
Lignin biosynthesis pathway and monolignol biosynthesis related gene expression. a The monolignol biosynthesis pathway including phenylalanine ammonia lyase (PAL), 4-coumarate-CoA ligase (4CL), p-coumarate 3-hydroxylase (C3H), cinnamoyl CoA reductase (CRR), ferulate 5-hydroxylase (F5H), cafeic acid O-methyltransferase (COMT) and cinnamyl alcohol dehydrogenase (CAD). b Expression of tobacco peroxidase and laccase in transgenic suspension cells. Gene expression was scaled using the Z-score of RPM in the heatmap. For each heatmap, the key is located at right with RPM values increasing from blue to red
In addition, several genes, such as cinnamyl alcohol dehydrogenase (CAD), caffeic acid O-methyltransferase (COMT), caffeoyl-CoA 3-O-methyltransferase (CCoAOMT) and ferulate 5-hydroxylase (F5H), are involved in the lignin biosynthesis pathway (Table 1). We also found that CAD1 and CAD6 were significantly upregulated in 4CL-CCR suspension cells. The COMT- and CCoAOMT-related genes, which are involved in transformation from H-units lignin to G-units lignin, were increased significantly (Fig. 4a). However, transcriptome results showed that the expression of F5H in the 4CL-CCR suspension cell line was downregulated compared with WT during the whole subculture period (Table 1).
Moreover, we found that the expression levels of most peroxidases were upregulated during the entire suspension cell culture period (Table 1, Fig. 4b), while only two members of laccase increased (Table 1). This result suggests that these peroxidases may play an important role in the oxidative polymerization of monolignols in transgenic suspension cells. The reason for this phenomenon may be that the oxygen content in the culture flask decreases due to the increase in the cell numbers during the cell culture period, and laccase requires oxygen to catalyze the synthesis of lignin (Duroux and Welinder 2003). Furthermore, peroxidase is involved in the polymerization of lignin to protect plants from diseases and is associated with auxin metabolism (Passardi et al. 2006; Marjamaa et al. 2009). We found that the expression of anion peroxides formed by peroxidase 16-like, peroxidase 44-like, peroxidase P7, peroxidase 11-like, peroxidase 3-like and lignin-forming anionic peroxidase-like were significantly increased in the 4CL-CCR group (Table 1). Thus, we speculated that peroxidase was involved in the oxidative polymerization of lignin monomers in the process of lignin synthesis promoted by 4CL-CCR.
4CL-CCR inhibits auxin-related gene expression
The morphology of suspension cells changed significantly during the process of suspension cell culture, and plant hormones play an important role in the regulation of cell morphology. Since the only hormone added to the culture medium was auxin analogs (2,4-D), we focused on the analysis of the differential expression of auxin-related genes, including the auxin transporter AUX1 gene and downstream genes related to the auxin response.
The transcriptome results showed that auxin transporter-like protein 2, auxin transporter-like protein 3, and auxin transporter-like protein 4, which are responsible for the regulation of auxin flow, were significantly downregulated in 4CL-CCR suspension cells (Table 2). In addition, we found that auxin response factors (ARF), such as ARF 3, ARF 5, ARF 6, ARF 17 and 7 other genes, were significantly downregulated in 4CL-CCR suspension cells (Table 2). It has been shown that the downregulation of the ARF 2.1 gene leads to lignification in P. tomentosa (Fu et al. 2019), which is consistent with the results of this study that the lignification of suspension cells increased in the 4CL-CCR group. Therefore, we speculated that the downregulation of AUX-related gene expression may inhibit the absorption of auxin in 4CL-CCR suspension cells and further accelerate lignification in the 4CL-CCR group.
4CL-CCR affects the expression of MYB-related genes
Moreover, MYB transcription factors are mainly involved in the biogenesis of secondary cell wall biosynthesis, and MYB proteins are also involved in the regulation of phenylpropanoid biosynthesis, affecting the biosynthesis of phenolic compounds. We found that the expression of the MYB306, MYB315 and MYB330 genes was significantly increased, while MYB308 and MYB-B were significantly downregulated in the 4CL-CCR group (Table 3). MYB306 in tobacco is homologous to MYB36 in Arabidopsis thaliana, and AtMYB36 regulates root endothelial cells from proliferation to differentiation, which is related to lignin synthesis (Liberman et al. 2015). This implied that MYB306 may be involved in regulating the expression of lignin synthesis in the 4CL-CCR group. In addition, MYB315 in tobacco is homologous to AtMYB85, and AtMYB85 is involved in the control of cell wall biosynthesis and activates lignin biosynthesis in fibers and/or vessels (Geng et al. 2020). This suggests that MYB315 may have a similar function in tobacco. Furthermore, overexpression of AtMYB92 induced the expression of fatty acid biosynthetic genes (To et al. 2020). Thus, MYB306 may also have similar functions mainly due to its homology with AtMYB92. Thus, we speculated that the upregulation of MYB gene expression promotes secondary cell wall formation and lignin deposition in the suspension cells of the 4CL-CCR group.
Discussion
In this study, we found that 4CL-CCR significantly promoted lignification of tobacco suspension cells, and the suspension cell culture medium of the 4CL-CCR group showed obvious color changes when the tobacco suspension cells were subcultured. Monolignols are supplied for lignification biosynthesis from the suspension cells and also from the medium in vitro (Koutaniemi et al. 2005; Takeuchi et al. 2018). We measured these phenolic acids and aldehydes in the medium by HPLC‒MS and found that the phenolic acid content in the suspension cell culture medium decreased significantly, while the aldehyde content increased significantly in the 4CL-CCR group. According to soluble phenolic acid and its derivative analysis, the most secreted aldehyde in all cell lines was coumarin aldehyde, which suggests that coumaric acid in tobacco cells is catalyzed, and a large amount of coumarin aldehyde was finally produced and effluxed. In vitro experiments confirmed that the 4CL-CCR protein could catalyze ferulic acid to coniferyl aldehyde, but only a small amount of coniferyl aldehyde was found in the medium (Xie et al. 2018). However, G-unit lignin was detected in the cell wall of the transgenic suspension cells. This indicates that the coniferous aldehyde secreted into the medium may be used to synthesize lignin polymers. In contrast, cinnamaldehyde is not catalyzed by CAD because a large amount of cinnamaldehyde is secreted into the medium. As mentioned above, conifer aldehyde is used to produce lignin.
In addition, these observations are supported by our RNA-seq analyses showing that lignin biosynthetic genes were up- or downregulated in the 4CL-CCR group. In the lignin biosynthesis pathway, the expression of CAD members was significantly upregulated due to the introduced fusion gene 4CL-CCR. The COMT and CCoAOMT genes, which are involved in transformation from H-unit lignin to G-unit lignin (Singh and Sharma 2022), were increased significantly. This suggests that the transformation of lignin monomers from H-units to G-units in transgenic tobacco relies on the conversion of phenolic acid to caffeic acid via p-coumarate 3-hydroxylase (C3H) and then to ferulic acid via COMT catalysis. It is also possible to finally generate coniferyl aldehyde and coniferyl alcohol through the conversion reaction between aldehydes. These genes showed different patterns of expression. This suggests that subfunctionalization may have caused the expression patterns of these putative paralogous genes to diversify. However, transcriptome results showed that the expression of F5H, which is another important gene related to the transition from G-units monomer to S-units monomer (Cao et al. 2022), was downregulated compared with that of WT during the whole subculture period in the 4CL-CCR suspension cell line. This indicates that the absence of S-unit monomers in the 4CL-CCR group is probably related to the decreased expression of the F5H gene.
Furthermore, auxin is very important for the normal growth of suspension cells, and it participates in regulating many metabolic adjustments of cells (Han et al. 2014). It has been confirmed that 2,4-D inhibited cell lignification of cells in tobacco and moso bamboo suspension cells (Kuboi and Yamada 1976; Ogita et al. 2012). However, we found that lignification of transgenic suspension cells occurred even in the presence of 2,4-D in the medium. Therefore, it is necessary to analyze the gene expression differences between the 4CL-CCR and WT groups in response to plant hormones, especially auxin. The results showed that the expression of auxin transport-related genes was significantly decreased in the 4CL-CCR group. It has been demonstrated that the intracellular transport of auxin mediated by AUXs responds to 75% of the auxin influx, and the other 20% is crossed by other amino acids in Arabidopsis (Maraschin Fdos et al. 2009; Lakehal et al. 2019), and the amount of auxin entering the cell through free diffusion is negligible (Rutschow et al. 2014; Bakker et al. 2022). The expression of three tobacco AUX proteins in this study and Arabidopsis AUX1 was downregulated (Xu et al. 2018). This suggests that these three AUX proteins are responsible for the uptake of auxin. Downregulation of these three genes resulted in suppression of the 2,4-D influx in transgenic tobacco suspension cells, leading to disruption of the auxin signal transduction pathway.
TIR1, an important protein in the signal transduction pathway, is auxin dose-dependent (Xiao et al. 2023). When the concentration of intracellular auxin increases, the expression of TIR1 is activated, and TIR1 mediates the ubiquitination and degradation of IAA protein (Qi et al. 2022). Then, ARFs that originally bind to IAA are released, and the released ARFs form a homologous or heterodimer activator dimer (Shimizu-Mitao and Kakimoto 2014; Lakehal et al. 2019). The dimer recognizes the target genes downstream of the auxin signal pathway target gene and binds to the cis-acting elements of the target genes; as a result, expression of the gene is activated to complete the auxin signaling pathway (Maraschin Fdos et al. 2009; Trenner et al. 2017). In this study, we found that the TIR1-like gene in transgenic tobacco suspension cells was significantly downregulated compared with that in WT cells. As a result, the low concentration of auxin in the cell inhibits the expression of the TIR1 gene, and the reduction in TIR1 protein cannot prevent IAA ubiquitination-based degradation, but the TIR1 protein will combine with ARF to form a heterodimer. ARF is a type of gene transcription factor that indirectly corresponds to auxin, and some ARFs are involved in regulating cell morphology (Niu et al. 2022). It has been shown that the loss of ARF2 significantly increased the size of seeds in Arabidopsis by increasing cell division cell expansion (Schruff et al. 2006). Therefore, when ARF exhibited no transcriptional activation function, the transgenic tobacco suspension cells finally showed a nearly circular cell shape. ARFs are also involved in regulating the biosynthesis of lignin (Hu et al. 2023). Inhibiting the expression of the ARF2.1 gene in Populus trichocarpa showed that the petioles of P. trichocarpa were excessively lignified (Fu et al. 2019). In this study, the transgenic tobacco ARF2-like protein contains the same conserved domains as P. trichocarpa and Arabidopsis PtrARF2.1 and AtARF2, which is inferred to perform the same function as PtrARF2.1 and AtARF2 (Li et al. 2014; Liu et al. 2018). In addition, the lignin-specific phloroglucinol-HCl reaction observed in the transgenic tobacco suspension cells is consistent with the excessive lignification of the petiole in the P. tomentosa mutant.
Moreover, it has been demonstrated that MYB-related genes enhance flavonoid production, which further negatively modulates the auxin response (Bernardi et al. 2019; Martínez-Rivas et al. 2023). Therefore, low concentrations of auxin cannot affect the expression of MYBs, and many upregulated MYB genes in the 4CL-CCR group are associated with decreased auxin concentrations. The subfamily domain that contains two repeats of R2R3 DNA binding is the largest domain in the MYB family (Stracke et al. 2001). Some R2R3 MYB proteins achieve mediation functions by binding AC cis-acting elements in the promoter (Luo et al. 2023). The promoter regions of multiple genes in the phenylpropanoid pathway contain AC elements (Grotewold et al. 1994; Sablowski et al. 1994). In previous studies, it was found that the R2R3 MYB protein was indeed involved in the regulation of phenylpropane biosynthesis and ultimately affected the biosynthesis of phenolic compounds, including flavonoids and lignin (Tamagnone et al. 1998; Borevitz et al. 2000).
The MYB family members of A. thaliana that had been reported to participate in the regulation of the cell wall (Shi et al. 2022), and the MYB family members that were upregulated in this study were analyzed by MEGA7. The translation product proteins of tobacco transcription factors MYB306 and MYB315 were closely related to AtMYB36 and AtMYB43, respectively. AtMYB36 is an important transcription factor that mediates root endothelial cells from the proliferation to the differentiation stage (Fernández-Marcos et al. 2017), and the sign of cell differentiation is the appearance of the casparian strip (Liberman et al. 2015). In this process of cell differentiation, MYB36 performs its feedforward effect (Liberman et al. 2015), while AtMYB43 and AtMYB85 both directly regulate the expression of genes related to lignin synthesis. In addition, AtMYB43 was found to be directly involved in activating inhibitors in Arabidopsis (Jiang et al. 2020), and this inhibitor could inhibit the expression of flavonoid genes, causing more phenylalanine metabolic intermediate products to flow to lignin (Geng et al. 2020). Therefore, MYB306 in tobacco may activate the secondary metabolism of tobacco suspension cells, MYB315 inhibits the biosynthesis of flavonoids, and more lignin accumulates in suspension cells.
However, transcriptome sequencing was performed on tobacco transgenic suspension cells in different periods, we detected lots of DEGs, but only focused on a small number of DEGs related to lignin synthesis, as well as auxin related DEGs and MYB transcription factors that may be involved in regulating lignin synthesis. There are many important issues that needs to be focused on in further studies. For example, there could be hormonal crosstalk in the regulatory cascades leading lignin biosynthesis, therefore, in addition to auxin, other plant hormones may also be involved in regulating lignin synthesis. Furthermore, it also would be interesting to see whether there are up-regulation of other transcription factors during the process of secondary growth of the cells, and lignin deposition in the cell wall area. Besides MYBs, there could be involvement of other transcription factors. In order to solve the above problems, we have made the transcriptome data available in the supplementary data for future analysis, which might aid in discovering novel and underexplored mechanisms to the formation of lignin.
Conclusion
Overexpression of the 4CL-CCR gene promotes lignification in tobacco suspension cells. The fusion gene 4CL-CCR may break the inhibitory effect of auxin on the lignification of tobacco suspension cells. The low concentration of auxin in the cell may initiate the expression of MYB306 and MYB315 in tobacco cells, respectively, turning on the secondary growth of cells and lignin deposition in the cell wall area.
Availability of data and material
Data are freely available for the readers.
Abbreviations
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- 4CL:
-
4-Coumaric acid: coenzyme A ligase
- 6-BA:
-
6-Benzylamino-purine
- ARF:
-
Auxin response factor
- BA:
-
Benzylaminopurine
- C3H:
-
4-Coumarate 3-hydroxylase
- CAD:
-
Cinnamyl alcohol dehydrogenase
- CCoAOMT:
-
Caffeoyl-CoA 3-O-methyltransferase
- CCRs:
-
Cinnamoyl-coenzyme A reductases
- COMT:
-
Acid O-methyltransferase
- DEGs:
-
Differentially expressed genes
- F5H:
-
Ferulate 5-hydroxylase
- FDA:
-
Fluorescein diacetate
- G units:
-
Guaiacyl units
- GC–MS:
-
Gas chromatography-mass spectrometry
- GO:
-
Gene ontology
- H units:
-
p-Hydroxyphenyl units
- HPLC–MS:
-
High performance liquid chromatography–mass spectrometry
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MS:
-
Murashige and Skoog
- NAA:
-
Naphthlcetic acid
- PAL:
-
Phenylalanine ammonia lyase
- S units:
-
Syringyl units
- WT:
-
Wild type
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al (2000) Gene ontology: tool for the unification of biology. Gene Ontol Consortium Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Bakker BH, Faver TE, Hupkes HJ, Merks RMH, van der Voort J (2022) Scaling relations for auxin waves. J Math Biol 85(4):41. https://doi.org/10.1007/s00285-022-01793-5
Bernardi J, Battaglia R, Bagnaresi P, Lucini L, Marocco A (2019) Transcriptomic and metabolomic analysis of ZmYUC1 mutant reveals the role of auxin during early endosperm formation in maize. Plant Sci 281:133–145. https://doi.org/10.1016/j.plantsci.2019.01.027
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12(12):2383–2394. https://doi.org/10.1105/tpc.12.12.2383
Cao Y, Yan X, Ran S, Ralph J, Smith RA, Chen X et al (2022) Knockout of the lignin pathway gene BnF5H decreases the S/G lignin compositional ratio and improves Sclerotinia sclerotiorum resistance in Brassica napus. Plant Cell Environ 45(1):248–261. https://doi.org/10.1111/pce.14208
Chao N, Jiang WT, Wang XC, Jiang XN, Gai Y (2019) Novel motif is capable of determining CCR and CCR-like proteins based on the divergence of CCRs in plants. Tree Physiol 39(12):2019–2026. https://doi.org/10.1093/treephys/tpz098
Dong NQ, Lin HX (2021) Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J Integr Plant Biol 63(1):180–209. https://doi.org/10.1111/jipb.13054
Duroux L, Welinder KG (2003) The peroxidase gene family in plants: a phylogenetic overview. J Mol Evol 57(4):397–407. https://doi.org/10.1007/s00239-003-2489-3
Fernández-Marcos M, Desvoyes B, Manzano C, Liberman LM, Benfey PN, Del Pozo JC et al (2017) Control of Arabidopsis lateral root primordium boundaries by MYB36. New Phytol 213(1):105–112. https://doi.org/10.1111/nph.14304
Fu Y, Win P, Zhang H, Li C, Shen Y, He F et al (2019) PtrARF2.1 is involved in regulation of leaf development and lignin biosynthesis in poplar trees. Int J Mol Sci. https://doi.org/10.3390/ijms20174141
Geng P, Zhang S, Liu J, Zhao C, Wu J, Cao Y et al (2020) MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol 182(3):1272–1283. https://doi.org/10.1104/pp.19.01070
Grossman A, Vermerris W (2019) Lignin-based polymers and nanomaterials. Curr Opin Biotechnol 56:112–120. https://doi.org/10.1016/j.copbio.2018.10.009
Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76(3):543–553. https://doi.org/10.1016/0092-8674(94)90117-1
Habarugira I, Hendriks T, Quillet MC, Hilbert JL, Rambaud C (2015) Effects of nuclear genomes on anther development in cytoplasmic male sterile chicories (Cichorium intybus L.): morphological analysis. Sci World J 2015:529521. https://doi.org/10.1155/2015/529521.
Han M, Park Y, Kim I, Kim EH, Yu TK, Rhee S et al (2014) Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc Natl Acad Sci U S A 111(52):18613–18618. https://doi.org/10.1073/pnas.1419525112
Hu Y, Gai Y, Yin L, Wang X, Feng C, Feng L et al (2010) Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms. Plant Cell 22(9):3093–3104. https://doi.org/10.1105/tpc.109.072652
Hu Y, Cheng H, Zhang Y, Zhang J, Niu S, Wang X et al (2021) The MdMYB16/MdMYB1-miR7125-MdCCR module regulates the homeostasis between anthocyanin and lignin biosynthesis during light induction in apple. New Phytol 231(3):1105–1122. https://doi.org/10.1111/nph.17431
Hu Z, Zong D, Zhang Q, Zhang X, Lu Y, He C (2023) PyuARF16/33 are involved in the regulation of lignin synthesis and rapid growth in populus yunnanensis. Genes (basel). https://doi.org/10.3390/genes14020278
Jiang J, Liao X, Jin X, Tan L, Lu Q, Yuan C et al (2020) MYB43 in oilseed rape (Brassica napus) positively regulates vascular lignification, plant morphology and yield potential but negatively affects resistance to sclerotinia sclerotiorum. Genes (basel). https://doi.org/10.3390/genes11050581
Khadr A, Wang YH, Zhang RR, Wang XR, Xu ZS, Xiong AS (2020) Cytokinin (6-benzylaminopurine) elevates lignification and the expression of genes involved in lignin biosynthesis of carrot. Protoplasma 257(6):1507–1517. https://doi.org/10.1007/s00709-020-01527-8
Khairul-Anuar MA, Mazumdar P, Lau SE, Tan TT, Harikrishna JA (2019) High-quality RNA isolation from pigment-rich Dendrobium flowers. 3 Biotech 9(10):371. https://doi.org/10.1007/s13205-019-1898-y
Koutaniemi S, Toikka MM, Kärkönen A, Mustonen M, Lundell T, Simola LK et al (2005) Characterization of basic p-coumaryl and coniferyl alcohol oxidizing peroxidases from a lignin-forming Picea abies suspension culture. Plant Mol Biol 58(2):141–157. https://doi.org/10.1007/s11103-005-5345-6
Kuboi T, Yamada Y (1976) Caffeic acid-O-methyltransferase in a suspension of cell aggregates of tobacco. Phytochemistry 15(3):397–400. https://doi.org/10.1016/S0031-9422(00)86831-4
Lakehal A, Chaabouni S, Cavel E, Le Hir R, Ranjan A, Raneshan Z et al (2019) A Molecular Framework for the Control of Adventitious Rooting by TIR1/AFB2-Aux/IAA-Dependent Auxin Signaling in Arabidopsis. Mol Plant 12(11):1499–1514. https://doi.org/10.1016/j.molp.2019.09.001
Lee D, Douglas CJ (1996) Two Divergent Members of a Tobacco 4-Coumarate: Coenzyme A Ligase (4CL) Gene Family (cDNA Structure, Gene Inheritance and Expression, and Properties of Recombinant Proteins). Plant Physiol 112(1):193–205. https://doi.org/10.1104/pp.112.1.193
Li C, Wang C, Meng L, Xing J, Wang T, Yang H et al (2014) Ectopic expression of a maize hybrid down-regulated gene ZmARF25 decreases organ size by affecting cellular proliferation in Arabidopsis. PLoS ONE 9(4):e94830. https://doi.org/10.1371/journal.pone.0094830
Li SS, Chang Y, Li B, Shao SL, Zhen-Zhu Z (2020) Functional analysis of 4-coumarate: CoA ligase from Dryopteris fragrans in transgenic tobacco enhances lignin and flavonoids. Genet Mol Biol 43(2):e20180355. https://doi.org/10.1590/1678-4685-gmb-2018-0355
Li F, Zhang Y, Tian C, Wang X, Zhou L, Jiang J et al (2023) Molecular module of CmMYB15-like-Cm4CL2 regulating lignin biosynthesis of chrysanthemum (Chrysanthemum morifolium) in response to aphid (Macrosiphoniella sanborni) feeding. New Phytol 237(5):1776–1793. https://doi.org/10.1111/nph.18643
Liberman LM, Sparks EE, Moreno-Risueno MA, Petricka JJ, Benfey PN (2015) MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proc Natl Acad Sci U S A 112(39):12099–12104. https://doi.org/10.1073/pnas.1515576112
Liu S, Qi Q, Chao N, Hou J, Rao G, Xie J et al (2015) Overexpression of artificially fused bifunctional enzyme 4CL1-CCR: a method for production of secreted 4-hydroxycinnamaldehydes in Escherichia coli. Microb Cell Fact 14:118. https://doi.org/10.1186/s12934-015-0309-2
Liu S, Liu J, Hou J, Chao N, Gai Y, Jiang X (2017) Three steps in one pot: biosynthesis of 4-hydroxycinnamyl alcohols using immobilized whole cells of two genetically engineered Escherichia coli strains. Microb Cell Fact 16(1):104. https://doi.org/10.1186/s12934-017-0722-9
Liu Z, Miao L, Huo R, Song X, Johnson C, Kong L et al (2018) ARF2-ARF4 and ARF5 are essential for female and male gametophyte development in Arabidopsis. Plant Cell Physiol 59(1):179–189. https://doi.org/10.1093/pcp/pcx174
Luo D, Mei D, Wei W, Liu J (2023) Identification and phylogenetic analysis of the R2R3-MYB subfamily in Brassica napus. Plants (basel). https://doi.org/10.3390/plants12040886
Maraschin Fdos S, Memelink J, Offringa R (2009) Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59(1):100–109. https://doi.org/10.1111/j.1365-313X.2009.03854.x
Marjamaa K, Kukkola EM, Fagerstedt KV (2009) The role of xylem class III peroxidases in lignification. J Exp Bot 60(2):367–376. https://doi.org/10.1093/jxb/ern278
Martínez-Rivas FJ, Blanco-Portales R, Pérez-Serratosa M, Ric-Varas P, Sánchez VG, Puche LM et al (2023) FaMYB123 interacts with FabHLH3 to regulate the late steps of anthocyanin and flavonol accumulation during ripening. Plant J. https://doi.org/10.1111/tpj.16166
Namsi A, Nury T, Khan AS, Leprince J, Vaudry D, Caccia C et al (2019) Octadecaneuropeptide (ODN) induces N2a cells differentiation through a PKA/PLC/PKC/MEK/ERK-dependent pathway: incidence on peroxisome, mitochondria, and lipid profiles. Molecules 24(18):3310. https://doi.org/10.3390/molecules24183310
Niu F, Ji C, Liang Z, Guo R, Chen Y, Zeng Y et al (2022) ADP-ribosylation factor D1 modulates Golgi morphology, cell plate formation, and plant growth in Arabidopsis. Plant Physiol 190(2):1199–1213. https://doi.org/10.1093/plphys/kiac329
Ogita S, Nomura T, Kishimoto T, Kato Y (2012) A novel xylogenic suspension culture model for exploring lignification in Phyllostachys bamboo. Plant Methods 8(1):40. https://doi.org/10.1186/1746-4811-8-40
Ogita S, Nomura T, Kato Y, Uehara-Yamaguchi Y, Inoue K, Yoshida T et al (2018) Transcriptional alterations during proliferation and lignification in Phyllostachys nigra cells. Sci Rep 8(1):11347. https://doi.org/10.1038/s41598-018-29645-7
Pan, Y., Li, L., Xiao, S., Chen, Z., Sarsaiya, S., Zhang, S., et al. (2020). Callus growth kinetics and accumulation of secondary metabolites of Bletilla striata Rchb.f. using a callus suspension culture. PLoS One 15(2):e0220084. doi: https://doi.org/10.1371/journal.pone.0220084.
Passardi F, Tognolli M, De Meyer M, Penel C, Dunand C (2006) Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 223(5):965–974. https://doi.org/10.1007/s00425-005-0153-4
Qi Q, Hu J, Qu L, Jiang X, Gai Y, Valenzuela SA et al (2019) Rapid, simplified microscale quantitative analysis of lignin H/G/S composition with GC-MS in glass ampules and glass capillaries. MethodsX 6:2592–2600. https://doi.org/10.1016/j.mex.2019.11.005
Qi L, Kwiatkowski M, Chen H, Hoermayer L, Sinclair S, Zou M et al (2022) Adenylate cyclase activity of TIR1/AFB auxin receptors in plants. Nature 611(7934):133–138. https://doi.org/10.1038/s41586-022-05369-7
Que F, Wang GL, Feng K, Xu ZS, Wang F, Xiong AS (2018) Hypoxia enhances lignification and affects the anatomical structure in hydroponic cultivation of carrot taproot. Plant Cell Rep 37(7):1021–1032. https://doi.org/10.1007/s00299-018-2288-3
Rais D, Zibek S (2019) Biotechnological and Biochemical Utilization of Lignin. Adv Biochem Eng Biotechnol 166:469–518. https://doi.org/10.1007/10_2017_6
Ralph J, Lapierre C, Boerjan W (2019) Lignin structure and its engineering. Curr Opin Biotechnol 56:240–249. https://doi.org/10.1016/j.copbio.2019.02.019
Rutschow HL, Baskin TI, Kramer EM (2014) The carrier AUXIN RESISTANT (AUX1) dominates auxin flux into Arabidopsis protoplasts. New Phytol 204(3):536–544. https://doi.org/10.1111/nph.12933
Sablowski RW, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M (1994) A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. Embo j 13(1):128–137. https://doi.org/10.1002/j.1460-2075.1994.tb06242.x
Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133(2):251–261. https://doi.org/10.1242/dev.02194
Shi Y, Man J, Huang Y, Zhang J, Zhang Z, Yin G et al (2022) Overexpression of PnMYB2 from Panax notoginseng induces cellulose and lignin biosynthesis during cell wall formation. Planta 255(5):107. https://doi.org/10.1007/s00425-022-03891-6
Shi H, Liu Y, Ding A, Wang W, Sun Y (2023) Induced defense strategies of plants against Ralstonia solanacearum. Front Microbiol 14:1059799. https://doi.org/10.3389/fmicb.2023.1059799
Shimizu-Mitao Y, Kakimoto T (2014) Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol 55(8):1450–1459. https://doi.org/10.1093/pcp/pcu077
Siegel SM (1953) On the Biosynthesis of Lignins. Physiol Plant 6(1):134–139. https://doi.org/10.1111/j.1399-3054.1953.tb08937.x
Singh S, Sharma N (2022) Biochemical and in silico molecular study of caffeic acid-O-methyltransferase enzyme associated with lignin deposition in tall fescue. Amino Acids. https://doi.org/10.1007/s00726-022-03225-6
Slazak B, Haugmo T, Badyra B, Göransson U (2020) The life cycle of cyclotides: biosynthesis and turnover in plant cells. Plant Cell Rep 39(10):1359–1367. https://doi.org/10.1007/s00299-020-02569-1
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456. https://doi.org/10.1016/S1369-5266(00)00199-0
Takeuchi M, Kegasa T, Watanabe A, Tamura M, Tsutsumi Y (2018) Expression analysis of transporter genes for screening candidate monolignol transporters using Arabidopsis thaliana cell suspensions during tracheary element differentiation. J Plant Res 131(2):297–305. https://doi.org/10.1007/s10265-017-0979-4
Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K et al (1998) The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10(2):135–154. https://doi.org/10.1105/tpc.10.2.135
To A, Joubès J, Thueux J, Kazaz S, Lepiniec L, Baud S (2020) AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana. Plant J 103(2):660–676. https://doi.org/10.1111/tpj.14759
Trenner J, Poeschl Y, Grau J, Gogol-Döring A, Quint M, Delker C (2017) Auxin-induced expression divergence between Arabidopsis species may originate within the TIR1/AFB-AUX/IAA-ARF module. J Exp Bot 68(3):539–552. https://doi.org/10.1093/jxb/erw457
Vanholme R, De Meester B, Ralph J, Boerjan W (2019) Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol 56:230–239. https://doi.org/10.1016/j.copbio.2019.02.018
Wang C, Kelley SS, Venditti RA (2016) Lignin-based thermoplastic materials. Chemsuschem 9(8):770–783. https://doi.org/10.1002/cssc.201501531
Wiszniewski AA, Zhou W, Smith SM, Bussell JD (2009) Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins. Plant Mol Biol 69(5):503–515. https://doi.org/10.1007/s11103-008-9431-4
Xiao Y, Yee C, Zhao CZ, Martinez MAQ, Zhang W, Shen K et al (2023) An expandable FLP-ON::TIR1 system for precise spatiotemporal protein degradation in Caenorhabditis elegans. Genetics. https://doi.org/10.1093/genetics/iyad013
Xie M, Zhang J, Tschaplinski TJ, Tuskan GA, Chen JG, Muchero W (2018) Regulation of Lignin Biosynthesis and Its Role in Growth-Defense Tradeoffs. Front Plant Sci 9:1427. https://doi.org/10.3389/fpls.2018.01427
Xu F, He S, Zhang J, Mao Z, Wang W, Li T et al (2018) Photoactivated CRY1 and phyB Interact Directly with AUX/IAA Proteins to Inhibit Auxin Signaling in Arabidopsis. Mol Plant 11(4):523–541. https://doi.org/10.1016/j.molp.2017.12.003
Zheng R, Zhang ZH, Zhao YX, Chen C, Jia SZ, Cao XC et al (2019) Transcriptomic Insights into the Response of the Olfactory Bulb to Selenium Treatment in a Mouse Model of Alzheimer’s Disease. Int J Mol Sci 20(12):2998. https://doi.org/10.3390/ijms20122998
Funding
This work is supported by the National Natural Science Foundation [NSF 31300498 to Y.G.] and the Beijing Higher Education Young Elite Teacher Project [YETP0755 granted to Dr. YING GAI].
Author information
Authors and Affiliations
Contributions
XJ and YG designed the study. JH and NS conducted the experiments. NS analyzed data and wrote the first draft of the paper. CL, XW and YG edited the paper. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no competing financial interests.
Additional information
Communicated by Yuree Lee.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, N., Hu, J., Li, C. et al. Fusion gene 4CL-CCR promotes lignification in tobacco suspension cells. Plant Cell Rep 42, 939–952 (2023). https://doi.org/10.1007/s00299-023-03002-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-023-03002-z