Abstract
Key message
Hypoxia enhances lignification of carrot root.
Abstract
Hypoxia stress was thought to be one of the major abiotic stresses that inhibiting the growth and development of higher plants. The genes encoding the plant alcohol dehydrogenase (ADH-P) were induced when suffering hypoxia. To investigate the impact of hypoxia on the carrot root growth, carrot plants were cultivated in the hydroponics with or without aeration. Morphological characteristics, anatomical structure, lignin content, and the expression profiles of DcADH-P genes and lignin biosynthesis-related genes were measured. Six DcADH-P genes were identified from the carrot genome. The expression profiles of only three (DcADH-P1, DcADH-P2, and DcADH-P3) genes could be detected and the other three (DcADH-P4, DcADH-P5, and DcADH-P6) could not be detected when carrot cultivated in the solution without aeration. In addition, carrot roots had more lignin content, aerenchyma and less fresh weight when cultivated in the solution without aeration. These results suggested that hypoxia could enhance the lignification and affect anatomical structure of the carrot root. However, the expression levels of the genes related to lignin biosynthesis were down-regulated under the hypoxia. The enhancement of lignification may be the consequence of the structure changes in the carrot root. Our work was potentially helpful for studying the effect of hypoxia on carrot growth and may provide useful information for carrot hydroponics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hydroponics is a highly efficient way to cultivate vegetables without the use of soil (Kimura and Rodriguez-Amaya 2003; Yu and Matsui 1993). The production of hydroponic vegetables was about 35,000 ha in 2011 (Hickman 2011). Moreover, hydroponics was often employed in the research of plant science, especially in the research of plant root growth and development (Hickman 2011; Kimura and Rodriguez-Amaya 2003). In the environmental toxicant biomonitoring, the root elongation of higher plants has been the most commonly used index. Root elongation was also an important endpoint of plant development (Park et al. 2016; Ratsch 1983). However, roots are easily destroyed during the process of experiment when cultivated in the soils. In hydroponics, this problem will be relieved. Hydroponics is a rapid and low-cost screening tool for root characteristics at the early growth stages of many plants. One obvious advantage of hydroponics is that the subsequent and non-destructive investigation of plant root growth can be carried out (Tuberosa et al. 2002).
Hypoxia is a serious impedimental factor in plant growth and results in yield losses (Bai et al. 2011; Christianson et al. 2010; Geigenberger 2003). Deficiency of oxygen can dramatically affect the efficiency of cellular adenosine triphosphate (ATP) production. In Arabidopsis, the expression of the gene encoding alcohol dehydrogenase (ADH) was induced when suffering from hypoxia. The loss-of-function mutants of ADH genes in rice, maize, and Arabidopsis showed poor tolerance when suffering from hypoxia. ADH genes are necessary for plants to survival under hypoxia (Christianson et al. 2010; Peng et al. 2001). When plants are grown under anoxic conditions, anaerobic fermentation will increase transiently and then carbohydrate catabolism will be restricted. Cellular metabolism and plant development processes would be affected seriously, too (Fukao and Bailey-Serres 2004). To survive, plants have to make some alternation to adapt to the oxygen deficiency. The appearances of aerenchyma and adventitious roots were important alterations, which allow submerged tissues to capture oxygen (Drew et al. 2000; Geigenberger 2003; Suralta and Yamauchi 2008). Space formation within aerenchymatous organs was caused by middle lamella cell separation or cell death (Schussler and Longstreth 1996). In addition, the cell wall of wheat roots was found to thicken under hypoxia in the previous study (Albrecht and Mustroph 2003).
As the second most abundant organic component, lignin plays important roles in response to abiotic stress in plants (Boerjan et al. 2003; Moura et al. 2010). The degree of lignification, development of transfusion tissues, and epidermal accessory structure of roots are involved in the stress tolerance of plants (Cervilla et al. 2009; Nobel 2005). Lignin is the main component of plant secondary cell wall that imparts rigidity and hydrophobicity of the secondary cell wall (Boerjan et al. 2003; Bonawitz and Chapple 2010; Zhao and Dixon 2011). Previous studies demonstrated that the lignification of plants would be triggered by abiotic stresses, such as drought, low temperatures, mineral deficiency and ultraviolet-B (UV-B) (Alvarez et al. 2008; Moura et al. 2010; Rozema et al. 1997; Thomashow 1999). In the lignin biosynthesis pathway, phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), hydroxycinnamoyltransferase (HCT), p-coumaroylshikimate/quinate 3′-hydroxylase (C3′H), caffeoyl CoA O-methyltransferase (CCoAOMT), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), peroxidase (PER) and laccase (LAC) are important participants (Vanholme et al. 2010; Weng and Chapple 2010). In addition, DcPAL, DcCCoAOMT, DcCAD, and DcPER1 were reported to play positive roles in carrot root development (Wang et al. 2016). However, whether hypoxia can trigger lignification in carrot roots under hypoxia is unclear.
Carrot (Daucus carota L.) is an important root vegetable and has been one of the top ten vegetable crops due to its abundant nutritional value. Taproot, the prized part of carrot, is the edible part and main object of research (Cavagnaro et al. 2011; Xu et al. 2014b; Wang et al. 2015). The hydroponics of carrot will be a useful way to study the growth and development of carrot roots. To our knowledge, relevant studies about the carrot hydroponics remain rare.
In this study, to investigate the response of carrot roots to hypoxia, carrot plants were cultivated in the environment with or without aeration by hydroponic cultivation. The expression profiles of lignin-related genes and the DcADH-P genes were measured through quantitative real-time PCR (qRT-PCR). The lignin contents of the roots at 30 and 60 days after sowing (DAS) were measured. The microscopy method was also performed to investigate the lignin distribution and cell development. The results obtained from this study will be helpful for studying the impact of hypoxia on plant growth and provide useful information for carrot hydroponics.
Materials and methods
Experiment in hydroponics
‘Kurodagosun’ was chosen as the experimental material and cultivated in a climate-controlled chamber by hydroponics at the state key laboratory of crop genetics and germplasm enhancement in Nanjing Agricultural University (32°04′N, 118°85′E). ‘Kurodagosun’ is a widely cultivated variety and was used in the genome sequencing (Xu et al. 2014a; Wang et al. 2018). Carrot seeds were germinated in the petri dishes. After a few days, seeds with germs were transferred into 96-well plates whose ends were cut off. The edges of the plates were not used to cultivate seedlings. Plates with seeds were placed at a tank (25 × 16 × 6 cm) with two shelves. The solution (Hoagland) was renewed every 5 days. Seedlings of the carrot with two true leaves were divided into two parts and placed in two tanks. One tank was continuously aerated through rubber tubes by an air compressor and another one without air transport was used as control. The oxygen concentrations of the two conditions were about 3 and 8 mg/L, respectively. The temperature of the chamber was held at 25 °C for 16 h during daytime with 320 µmol/m/s light intensity and 18 °C for 8 h in the dark.
Plants were sampled at 30 and 60 DAS and stored at − 80 °C for molecular research. Plants at 30 and 60 DAS were also harvested to measure lignin content and perform microscopy analysis. Total of 15 carrot seedlings were used for measuring root length, root fresh weight, and root diameter in different developmental stages and different oxygen environments, respectively.
Identification, sequence structure, and phylogenetic analysis of carrot alcohol dehydrogenases
To identify carrot DcADH-P genes at the genome level, the data of carrot genome was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/genome/?term=Daucus) and CarrotDB (Iorizzo et al. 2016; Xu et al. 2014a). The candidate ADH-P genes were obtained by using HMMER 3.0 with the Hidden Markov models (HMMs) of ADH_N (PF08240) domain (Eddy 2011; Finn et al. 2013, 2015). The alcohol_DH_plants (cd08301) domain was the signature structure of plant ADH genes. Then the proteins coding by candidate genes were analyzed with InterPro (http://www.ebi.ac.uk/interpro/search/sequence-search). The analysis of intron was performed via GSDS 2.0 (Hu et al. 2014). In addition, multiple alignments of the 6 DcADH-P proteins were performed by DNAMAN version 5.2.2 (Lynnon Biosoft, Quebec, Canada). The phylogenetic tree was constructed via MEGA 6 with Neighbor-joining method under the Jones–Thornton–Taylor (JTT) model. The bootstrap replication was set as 1000 (Tamura et al. 2013).
Measurement of lignin content
Lignin content was extracted and measured according to previous description with the following modifications (Cai et al. 2006; Cervilla et al. 2009; Wang et al. 2016). Briefly, approximately 2 g carrot roots were initially ground with liquid nitrogen. The powders were then mixed with 6 mL of the 99.5% ethanol. The mixture was centrifuged at 10,000×g for 20 min. The precipitates were collected and exposed at the room temperature overnight. About 10 mg dried precipitates were then mixed with 1 mL of the 2 M HCl, and 0.1 mL of the thioglycolic acid. The mixture was then transferred into a 2-mL plastic tube. The tubes were tightly sealed and incubated at 100 °C for 8 h. After boiling, tubes were cooled on ice. Cooled mixture was then centrifuged at 12,000×g for 20 min at 4 °C to get a pellet. The obtained pellet was then washed with 1 mL of the deionized water and resuspended in 1 mL of 1 M NaOH. The mixed solution was stored in a gently agitated shaker at 25 °C for 18 h. Following the 18-h agitation, the mixture was centrifuged at 12,000×g for 20 min. The upper liquid was transferred into a new tube and mixed with 1 mL of concentrated HCl. The mixture was stored at 4 °C for 6 h to precipitate lignin thioglycolate. Finally, the mixture was centrifuged at 12,000×g for 20 min and the sediment was then dissolved in 1 mL of the 1 M NaOH. The absorbance of the 1 M NaOH at 280 nm was measured as blank control. The calibration curve was made using a commercial alkali lignin (Sigma-Aldrich).
Histochemical staining and UV fluorescence microscopy
Plants grown under conditions with or without aeration were sampled at 30 and 60 DAS for histochemical staining to investigate the lignin distribution. Phosphate buffer (pH 7.2) containing 2.5% glutaraldehyde was used to store the carrot root samples. To perform safranin-O/fast green staining, xylene was used to deparaffinize the root samples. Subsequently, ethanol was used to dehydrate the deparaffinized root samples. The dehydrated sections were then stained in 1% safranin-O for 2 h and counterstained with 0.5% fast green for 15 s. After staining, ethanol was used to remove excess stain and neutral balsam was used to mount the sections. Finally, the lignified parts were stained into red, whereas the cellulosic tissues were stained into green. When explored under the UV excitation, lignified cell walls could exhibit autofluorescence (Donaldson and Knox 2012; Jia et al. 2015). Fluorescence microscopy was used to search the lignin containing components in the root.
Total RNA isolation
A RNA extraction kit (Tiangen, Beijing, China) was used to extract the total RNA of the carrot roots according to the manufacturer’s instructions. cDNA was synthesized using a PrimerScript RT reagent kit (TaKaRa, Dalian, China). The cDNA was diluted 15-fold for qRT-PCR analysis.
Gene expression analysis
In this study, 12 genes involved in the lignin biosynthesis and 6 DcADH-P genes were chosen for qRT-PCR analysis. The 12 genes involved in the lignin biosynthesis of carrot were reported in a previous study. Primers of genes related to lignin synthesis for qRT-PCR was designed according to previous description (Wang et al. 2016). The primers of the 6 DcADH-P genes were designed through Primer 5.0 (Lalitha 2000) (Table 1). The qRT-PCR analysis was performed via MyIQ Real-Time PCR Detection System (Bio-Rad, CA, USA). The cycling conditions were maintained as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 30 s, at last a melting curve (65–95 °C, at increments of 0.5 °C) was generated. The relative gene expression profile was calculated with the 2− ΔΔCT method (Schmittgen and Livak 2008). The internal control was DcACTIN in this study (Tian et al. 2015). Each PCR reaction had three biological replicates and technical replicates.
Data analysis
Data were analyzed by using the office 2007 software. In addition, Student’s t test was used to analyze the data in this study at the 0.05 significance through SPSS 17.0 software.
Results
Growth analysis of carrot plants
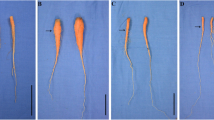
To study the response of carrot roots to hypoxia in hydroponics, carrot plants were cultivated in the conditions with or without aeration. Plants were sampled at 30 and 60 DAS, respectively. The growth status of carrot plants under the two treatments is displayed in Fig. 1. Carrots cultivated in nutrition solution had a large number of fibrous roots at the middle and tip parts of taproots. The fibrous roots seemed to originate from the cortex. These fibrous roots were long and intertwined with each other. The length, fresh weight and diameter of the carrot roots were measured (Fig. 2). Carrot plants treated with aeration grew much better than that treated without aeration. Morphological characters of carrot roots under different oxygen concentrations were obviously different. At 30 DAS, the fresh weight of the seedling roots cultivated in the higher O2 concentration (with aeration) was significantly higher than that in lower O2 concentration (without aeration). At 60 DAS, the fresh weight of the carrot root treated with or without aeration were 32.0 and 29.7 g, respectively. Moreover, the root diameter was about 29.4 and 27.0 mm, respectively.
Growth status of carrots under different oxygen environments. Carrots were cultivated under two different oxygen environments in hydroponics: no air entering (NAE) and with air entering (WAE). The samples were collected at 30 and 60 days after sowing, respectively. Black lines in the lower right corner of each plant represent 2 cm in that pixel
Morphological characteristics of carrot roots. a Root length (cm) of carrot roots; b root fresh weight (g) of carrot roots; c root diameter (mm) of carrot roots. Student’s t test was used to identify the differences under different oxygen concentrations (P < 0.05; *control versus treatment). Error bars represent standard deviation (SD). DAS days after sowing. (Colour figure online)
Identification, evolution analysis of carrot alcohol dehydrogenases
To identify the ADH-P genes from the carrot genome, HMMER 3.0 was used with the ADH_N (PF08240) domain. Initially, 69 candidate genes were detected from the carrot genome. Then, InterPro database was used to analyze the protein structure of the candidate genes. Six DcADH-P proteins were found to have NAD(P)-binding domain and a signature structure of alcohol_DH_plants (cd08301) (Supplementary Table 1). As a result, these six genes were determined and were designated as DcADH-P1, DcADH-P2, DcADH-P3, DcADH-P4, DcADH-P5, and DcADH-P6. Multiple alignments indicated that the six DcADH-P proteins have 72% sequence similarity (Supplementary Fig. 1).
In the study of gene evolution, the absence of introns was thought to be an important evidence. Here, the introns of the six DcADH-P genes were analyzed (Fig. 3b). DcADH-P1 and DcADH-P3 had 8 introns, and DcADH-P2, DcADH-P4, DcADH-P5, and DcADH-P6 had 9 introns. Furthermore, a phylogenetic tree was constructed based on the full-length amino acid sequence of six carrot DcADH-P genes with 16 homologues from other plant species (Fig. 3a). DcADH-P1, DcADH-P2, and DcADH-P3 aligned within the same sub-lineage group. DcADH-P4, DcADH-P5, and DcADH-P6 aligned into other sub-lineages.
The evolution analysis of carrot DcADH-P genes. a Phylogenetic tree of carrot DcADH-P proteins. The phylogenetic tree was constructed via MEGA 6.0 on the basis of full-length amino acid sequences of DcADH-P proteins and 16 ADH-P proteins from other species with Neighbor-joining method. Bootstrap analysis was conducted with 1000 replicates. The DcADH-P proteins were marked with solid circles. b The component of introns in carrot DcADH-P genes. The intron analysis was performed via GSDS 2.0. Blue blocks represent the CDS sequence and black blocks represent the upstream or downstream sequences. (Colour figure online)
Expression profiles of DcADH-P genes in carrot roots
According to previous reports, the expression profile of ADH-P gene was induced under the hypoxia in Arabidopsis. Here, the expression profiles of the six carrot DcADH-P genes under different oxygen environments were measured by qRT-PCR (Fig. 4). As a result, expression profiles of three carrot DcADH-P genes (DcADH-P1, DcADH-P2, and DcADH-P3) could be detected and the other three (DcADH-P4, DcADH-P5, and DcADH-P6) could not be detected. The expression profiles of DcADH-P1, DcADH-P2, and DcADH-P3 were significantly induced in carrots cultivated in the low oxygen concentration compared with that cultivated in the high oxygen concentration. In addition, the expression profiles of the three genes (DcADH-P1, DcADH-P2, and DcADH-P3) at 60 DAS were significantly higher than that at 30 DAS under low oxygen concentration environment.
Expression profile of DcADH-P genes at different growth stages and under different oxygen situations. The value on the left Y-axis indicates the relative gene expression profiles. The X-axis represents the two stages of carrot root development, corresponding to 30 and 60 days after sowing. The expression profiles of the genes were measured by qRT-PCR. The relative gene expression was calculated with the 2− ΔΔCT method. Student’s t test was used to do the statistic analysis (P < 0.05; *control versus treatment). Error bars represent standard deviation (SD) of three replicates. DAS days after sowing. (Colour figure online)
Anatomical structure of carrot roots
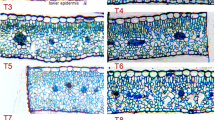
To observe the anatomical structure, carrot roots treated with or without aeration were sectioned and stained by safranin-O/fast green (Fig. 5). Roots cultivated in the environment with abundant oxygen had less aerenchyma (space) compared with plants grown with limited oxygen. The vascular pericycle was destroyed and pitted with aerenchyma under limited oxygen environment. By contrast, the vascular pericycle was complete under abundant oxygen environment. Furthermore, the secondary xylem was also destroyed and pitted with aerenchyma in carrot roots without aeration at 60 DAS. The root xylem was more tight and complete under the abundant oxygen environment. Vessels had better development under the abundant oxygen environment.
Microstructure of carrot roots under different oxygen environments and at different growth stages. Carrot roots treated with or without aeration were sectioned and stained by safranin-O/fast green. The part around the vascular cambium of a root cross section is shown. a The 30 DAS carrot root cultivated without aeration. b The 30 DAS carrot root cultivated with aeration. c The 60 DAS carrot root cultivated without aeration. d The 60 DAS carrot root cultivated with aeration. Staffs in the figure represent the magnification. Ep epidermis, Xy xylem, SP secondary phloem, Ve vessel, VC vascular cambium, Ae aerenchyma
The micrographs under the white light and fluorescence were used to determine the effect of hypoxia on carrot root lignification (Fig. 6). In the carrot roots, lignin was mainly accumulated in the cell walls of tracheary elements of the xylem. As shown in Fig. 6b, lignification of carrot roots under the low-oxygen environment was enhanced. However, the number of vessels in the carrot roots cultivated in the higher oxygen environment was more.
Transverse sections of carrot roots. a Transverse sections of carrot roots under white light. a The 30 DAS carrot root cultivated without aeration. b The 30 DAS carrot root cultivated with aeration. c The 60 DAS carrot root cultivated without aeration. d The 60 DAS carrot root cultivated with aeration. b Fluorescence micrographs of transverse sections of carrot roots. a The 30 DAS carrot root cultivated without aeration. b The 30 DAS carrot root cultivated with aeration. c The 60 DAS carrot root cultivated without aeration. d The 60 DAS carrot root cultivated with aeration. Scale bars are 50 µm in length. Ve vessel, Xy xylem, VC vascular cambium
Lignin content of carrot roots
To investigate the lignification of carrot roots, the lignin content of carrot roots at 30 and 60 DAS were measured (Fig. 7). These carrot roots under the low (without aeration) and abundant (with aeration) oxygen conditions were collected at 30 and 60 DAS, respectively. During the development, the lignin contents were significantly decreased in the roots of carrot. The content changed from 163.96 to 16.55 mg/g during the development under the low-oxygen environment. Similar results were obtained under the abundant oxygen environment (from 118.17 to 11.86 mg/g). Moreover, the lignin contents in the roots without aeration were significantly higher than that with aeration.
Lignin content of carrot roots at different growth stages and cultivated in different oxygen environments. Student’s t test was used to identify the differences under different oxygen concentrations (P < 0.05; *control versus treatment). Error bars represent standard deviation (SD) of three replicates. DAS days after sowing. (Colour figure online)
Expression profiles of lignin biosynthesis-related genes in carrot roots
To study the molecular mechanisms of lignin accumulation, genes related to lignin biosynthesis were selected for expression profiles analysis (Fig. 8). Among the 12 selected genes, most of them were differentially expressed under different oxygen environments. The expression levels also changed significantly during the development of carrot roots. During the carrot root development, the expression profiles of most genes decreased significantly in spite of the oxygen environment. However, the expression profile of DcCCR showed an opposed trend which increased during the development of carrot roots. The transcription of most genes showed higher levels in the high oxygen environment. At 30 DAS, the expression profiles of most genes were similar except DcPER1. At 60 DAS, Dc4CL, DcHCT, DcCCoAOMT, DcF5H, DcCOMT, DcCAD, and DcLAC1 increased significantly when carrot roots were surrounded with abundant oxygen. By contrast, the transcription of DcC3′H showed an opposite trend.
Expression profile of genes related to lignin biosynthesis at different growth stages and under different oxygen situations. The expression profiles of the genes were measured by qRT-PCR. The relative gene expression was calculated with the 2− ΔΔCT method. Student’s t test was used to do the statistic analysis (P < 0.05; *control versus treatment). Error bars represent standard deviation (SD) of three replicates. DAS days after sowing. (Colour figure online)
Discussion
Impact of hypoxia on expression profiles of DcADH-P genes in carrot roots
Plant ADH-P genes contribute much in the survival of plants under hypoxia. Hypoxia could induce the expression of ADH-P genes in Arabidopsis, maize, and rice (Baxter-Burrell et al. 2003; Ismond et al. 2003; Kürsteiner et al. 2003). The plant ADH gene family is a small gene family comprised two or three members (Chang and Meyerowitz 1986). Here, we found six DcADH-P genes in the carrot genome. The six DcADH-P genes did not align within the same sub-lineage group in the phylogenetic tree. The sequences of DcADH-P1, DcADH-P2, and DcADH-P3 were located near the ADH-P genes reported in other species. In contrast, the sequences of DcADH-P4, DcADH-P5, and DcADH-P6 were located far from the other three DcADH-P genes. These results suggested that there could be some duplication events among them during the evolution (Gabaldón and Koonin 2013).
To measure the expression profiles of DcADH-P genes under the different oxygen environments, we performed qRT-PCR analysis on the six DcADH-P genes. The expression profiles of only three DcADH-P genes (DcADH-P1, DcADH-P2, and DcADH-P3) could be detected and the other three genes (DcADH-P4, DcADH-P5, and DcADH-P5) could not be detected. Those results suggested that the three DcADH-P genes (DcADH-P4, DcADH-P5, and DcADH-P6) may not response to the hypoxia. The expression profiles of DcADH-P1, DcADH-P2, and DcADH-P3 in the carrot cultivated without aeration were significantly higher than that cultivated with aeration. Furthermore, the expression profiles of the three genes (DcADH-P1, DcADH-P2, and DcADH-P3) were significantly induced at 60 DAS compared with that at 30 DAS under low-oxygen environment. These results suggested that carrots cultivated in the solution without aeration suffering from hypoxia.
Impact of hypoxia on anatomical structure of carrot roots
Hypoxic environment is injurious to the growth and development of plants. Therefore, plants have to make some alterations to adapt to the low-oxygen environments. Lysigenous aerenchyma and adventitious roots were generated to adapt oxygen deficiency. Maize, soybeans, as well as some wetland species were reported to produce lysigenous aerenchyma when suffering flooding (Bacanamwo and Purcell 1999; Drew et al. 1989; Kawai et al. 1998; Morgan 1994). Lysigenous aerenchyma not only serves as a pathway for oxygen transfer, but also reduces the expenditure of oxygen by reducing the quantity of O2-consuming cells. However, the formation of lysigenous aerenchyma was the result of cell separation or cell death and dissolution (Drew et al. 2000; Schussler and Longstreth 1996). Here, safranin-O/fast green staining was used to observe the anatomical structure of carrot roots. Safranin-O/fast green staining was a useful method to investigate plant anatomical structure and tissues containing lignified cell walls which has been used in previous studies (Jia et al. 2014; Wang et al. 2016). In the present study, carrot roots cultivated in the solution without aeration had more aerenchyma (space), and the aerenchyma size was bigger. Similar results appeared in maize roots suffering hypoxia under laboratory conditions (Ober and Sharp 1996).
Impact of hypoxia on lignin content of carrot roots
Lignin biosynthesis was believed to occur during the process of normal tissue development and can be triggered under many various biotic and abiotic stresses (Bonawitz and Chapple 2010; Moura et al. 2010). However, the study on stressing situations changing lignin content is still little. Lignin is the second most abundant compound in plant organs (Boerjan et al. 2003). In vascular plants, lignin was found to be the main component of the secondary cell wall (Müse et al. 1997). In carrot roots, the lignin was mainly deposited in the cell walls of tracheary elements in the xylem (Wang et al. 2016). During maize stem development, the lignin content continuously increased (Jung and Casler 2006). Similar results were reported in the stem development of other plants (Shen et al. 2009). Here, lignin contents decreased during the root development which is consistent with the results from a previous study (Wang et al. 2016). In addition, the result of analysis on lignin content and autofluorescence under UV excitation revealed that the lignification of carrot roots cultivated in the solution without aeration was enhanced. Compared with the carrots cultivated in the soil, the lignin contents of hydroponic carrot roots were significantly increased. These results suggested that hypoxia may enhance the lignification of carrot roots.
Impact of hypoxia on expression profiles of lignin biosynthesis-related genes in carrot roots
Based on previous reports, the response of plants to hypoxia not only appeared in the biochemical and physiological reconfiguration, but also in the change of gene expression level (Christianson et al. 2010; Narsai et al. 2011). In this study, 12 genes involved in the biosynthesis of lignin were selected and analyzed. Among these genes, DcPAL, DcCCoAOMT, DcCAD, and DcPER1 were reported to be positively related to lignin accumulation during the developmental of carrot root. The expression profiles of these four genes in this study were consistent with the profiles reported in previous study, during carrot root development (Wang et al. 2016). PAL encodes the enzyme of the first step in monolignol synthesis and was also reported to be a key gene in the phenylpropanoid pathway (Dixon et al. 2002; Kao et al. 2002). In the present work, the transcription level of DcPAL was found to decrease during the root development and increase when carrot roots were treated with aeration. In Arabidopsis, the expression of PAL and 4CL were reported to be coordinately regulated (Lee et al. 1995). Herein, DcPAL and Dc4CL showed similar expression trend. 4CL is an important enzyme in lignin synthesis and is generally thought to encode the third step of phenylpropanoid pathway (Costa et al. 2005). In Populus tremuloides, the down-regulation of 4CL led to the reduction of lignin contents (Hu et al. 1999). Similar result appeared during the development of carrot roots. However, the transcription level of Dc4CL increased when in the environment with aeration (lignin content decreased). Similar results appeared in the expression trends of DcC4H, DcHCT, DcCCoAOMT, DcCCR, DcF5H, DcCOMT, DcCAD, and DcLAC1 in carrot roots in the environment with or without aeration.
Lignin is the main component of the secondary cell walls in vascular plants. The secondary cell walls biosynthesized during the development of vascular plant build a strong xylem thus providing mechanical support for the plant (Boudet 2000). However, the development of plants would be negatively affected by hypoxia.(Geigenberger 2003; Schussler and Longstreth 1996). In cotton, genes involved in cell wall synthesis were down-regulated when suffering from hypoxia (Christianson et al. 2009). The development of carrot roots would be restricted and the initial structure would also be destructed during hypoxia. These results suggested that the enhanced lignification in carrot roots may due to the decrease of contains and destroyed structure in the carrot roots when suffering from hypoxia.
Conclusion
Here, six DcADH-P genes were identified from carrot genome. Among them, three DcADH-P genes were significantly induced when carrot cultivated without aeration in hydroponics, while the other three could not be detected. This result suggested that carrot cultivated without aeration suffered from hypoxia. In addition, the carrot roots cultivated without aeration had more aerenchyma (space). Analysis of the lignification of carrot roots showed that the roots of carrot cultivated without aeration had more lignin content. The same result appeared in the autofluorescence under UV excitation analysis. Our results indicated that hypoxia could enhance the lignification of carrot root. The current study will be useful for investigating the hypoxia on carrot growth and development and may also provide information for improving carrot hydroponics.
Author contribution statement
Conceived and designed the experiments: Xiong AS and Que F. Performed the experiments: Que F, Wang GL, Fen K, Xu ZS, and Wang F. Analyzed the data: Que F and Wang GL. Contributed reagents/materials/analysis tools: Xiong AS. Wrote the paper: Que F. Revised the paper: Xiong AS and Wang GL. All authors read and approved the final manuscript.
References
Albrecht G, Mustroph A (2003) Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 217:252–260
Alvarez S, Marsh EL, Schroeder SG, Schachtman DP (2008) Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ 31:325–340
Bacanamwo M, Purcell LC (1999) Soybean root morphological and anatomical traits associated with acclimation to flooding. Crop Sci 39:143–149
Bai T, Yin R, Li C, Ma F, Yue Z, Shu H (2011) Comparative analysis of endogenous hormones in leaves and roots of two contrasting Malus species in response to hypoxia stress. J Plant Growth Regul 30:119–127
Baxter-Burrell A, Chang R, Springer P, Bailey-Serres J (2003) Gene and enhancer trap transposable elements reveal oxygen deprivation-regulated genes and their complex patterns of expression in Arabidopsis. Ann Bot 91:129–141
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44:337–363
Boudet AM (2000) Lignins and lignification: selected issues. Plant Physiol Biochem 38:81–96
Cai C, Xu C, Li X, Ferguson I, Chen K (2006) Accumulation of lignin in relation to change in activities of lignification enzymes in loquat fruit flesh after harvest. Postharvest Biol Technol 40:163–169
Cavagnaro PF, Chung SM, Manin S, Yildiz M, Ali A, Alessandro MS, Iorizzo M, Senalik DA, Simon PW (2011) Microsatellite isolation and marker development in carrot - genomic distribution, linkage mapping, genetic diversity analysis and marker transferability across Apiaceae. BMC Genom 12:386
Cervilla L, Rosales M, Rubio-Wilhelmi M, Sánchez-Rodríguez E, Blasco B, Ríos J, Romero L, Ruiz J (2009) Involvement of lignification and membrane permeability in the tomato root response to boron toxicity. Plant Sci 176:545–552
Chang C, Meyerowitz EM (1986) Molecular cloning and DNA sequence of the Arabidopsis thaliana alcohol dehydrogenase gene. Proc Natl Acad Sci 83:1408–1412
Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW (2009) Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol 51:21–37
Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW (2010) Comparisons of early transcriptome responses to low-oxygen environments in three dicotyledonous plant species. Plant Signal Behav 5:1006–1009
Costa MA, Bedgar DL, Moinuddin SG, Kim KW, Cardenas CL, Cochrane FC, Shockey JM, Helms GL, Amakura Y, Takahashi H (2005) Characterization in vitro and in vivo of the putative multigene 4-coumarate: CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66:2072–2091
Dixon RA, Achnine L, Kota P, Liu CJ, Reddy M, Wang L (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol 3:371–390
Donaldson LA, Knox JP (2012) Localization of cell wall polysaccharides in normal and compression wood of radiata pine: relationships with lignification and microfibril orientation. Plant Physiol 158:642–653
Drew MC, He CJ, Morgan PW (1989) Decreased ethylene biosynthesis, and induction of aerenchyma, by nitrogen- or phosphate-starvation in adventitious roots of Zea mays L. Plant Physiol 91:266–271
Drew MC, He CJ, Morgan PW (2000) Programmed cell death and aerenchyma formation in roots. Trends Plant Sci 5:123–127
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J (2013) Pfam: the protein families database. Nucleic Acids Res 42:D222-D230
Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR (2015) HMMER web server: 2015 update. Nucleic Acids Res 43:W30-W38
Fukao T, Bailey-Serres J (2004) Plant responses to hypoxia—is survival a balancing act? Trends Plant Sci 9:449–456
Gabaldón T, Koonin EV (2013) Functional and evolutionary implications of gene orthology. Nat Rev Genet 14:360–366
Geigenberger P (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6:247–256
Hickman G (2011) Greenhouse vegetable production statistics: a review of current data on the international production of vegetables in greenhouses. Cuesta Roble greenhouse consultants, Mariposa
Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai C-J, Chiang VL (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17:808–812
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Iorizzo M, Ellison S, Senalik D, Zeng P, Satapoomin P, Huang J, Bowman M, Iovene M, Sanseverino W, Cavagnaro P, Yildiz M, Macko-Podgórni A, Moranska E, Grzebelus E, Grzebelus D, Ashrafi H, Zheng Z, Cheng S, Spooner D, Van Deynze A, Simon P (2016) A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet 48:657–666
Ismond KP, Dolferus R, De Pauw M, Dennis ES, Good AG (2003) Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol 132:1292–1302
Jia XL, Wang GL, Xiong F, Yu XR, Xu ZS, Wang F, Xiong AS (2015) De novo assembly, transcriptome characterization, lignin accumulation, and anatomic characteristics: novel insights into lignin biosynthesis during celery leaf development. Sci Rep 5:8259
Jung H, Casler M (2006) Maize stem tissues: impact of development on cell wall degradability. Crop Sci 46:1801
Kao YY, Harding SA, Tsai CJ (2002) Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifying cells of quaking aspen. Plant Physiol 130:796–807
Kawai M, Samarajeewa PK, Barrero RA, Nishiguchi M, Uchimiya H (1998) Cellular dissection of the degradation pattern of cortical cell death during aerenchyma formation of rice roots. Planta 204:277–287
Kimura M, Rodriguez-Amaya DB (2003) Carotenoid composition of hydroponic leafy vegetables. J Agric Food Chem 51:2603–2607
Kürsteiner O, Dupuis I, Kuhlemeier C (2003) The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol 132:968–978
Lalitha S (2000) Primer premier 5. Biotech Softw Internet Rep 1:270–272
Lee D, Ellard M, Wanner LA, Davis KR, Douglas CJ (1995) The Arabidopsis thaliana 4-coumarate: CoA ligase (4CL) gene: stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol Biol 28:871–884
Morgan PW (1994) Induction of enzymes associated with lysigenous aerenchyma formation in roots of Zea mays during hypoxia or nitrogen starvation. Plant Physiol 105:861–865
Moura JCMS., Bonine CAV, De Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52:360–376
Müse G, Schindler T, Bergfeld R, Ruel K, Jacquet G, Lapierre C, Speth V, Schopfer P (1997) Structure and distribution of lignin in primary and secondary cell walls of maize coleoptiles analyzed by chemical and immunological probes. Planta 201:146–159
Narsai R, Rocha M, Geigenberger P, Whelan J, van Dongen JT (2011) Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. N Phytol 190:472–487
Nobel PS (2005) Physicochemical and environmental plant physiology. Q Rev Biol 54:507–543
Ober ES, Sharp RE (1996) A microsensor for direct measurement of O2 partial pressure within plant tissues3. J Exp Bot 47:447–454
Park J, Yoon JH, Depuydt S, Oh JW, Jo Y, Kim K, Brown MT, Han T (2016) The sensitivity of an hydroponic lettuce root elongation bioassay to metals, phenol and wastewaters. Ecotoxicol Environ Saf 126:147–153
Peng HP, Chan CS, Shih MC, Yang SF (2001) Signaling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiol 126:742–749
Ratsch H (1983) Interlaboratory root elongation testing of toxic substances on selected plant species. Environ Prot Agency Carvallis Environ Res Lab Coevallis OR EPA 600:3–85
Rozema J, van de Staaij J, Björn LO, Caldwell M (1997) UV-B as an environmental factor in plant life: stress and regulation. Trends Ecol Evol 12:22–28
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Schussler EE, Longstreth DJ (1996) Aerenchyma develops by cell lysis in roots and cell separation in leaf petioles in Sagittaria lancifolia (Alismataceae). Am J Bot 83:1266–1273
Shen H, Fu C, Xiao X, Ray T, Tang Y, Wang Z, Chen F (2009) Developmental control of lignification in stems of lowland switchgrass variety Alamo and the effects on saccharification efficiency. BioEnergy Res 2:233–245
Suralta RR, Yamauchi A (2008) Root growth, aerenchyma development, and oxygen transport in rice genotypes subjected to drought and waterlogging. Environ Exp Bot 64:75–82
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Biol 50:571–599
Tian C, Jiang Q, Wang F, Wang GL, Xu ZS, Xiong AS (2015) Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS One 10:e0117569
Tuberosa R, Sanguineti MC, Landi P, Giuliani MM, Salvi S, Conti S (2002) Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol Biol 48:697–712
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153:895–905
Wang GL, Xiong F, Que F, Xu ZS, Wang F, Xiong AS (2015) Morphological characteristics, anatomical structure, and gene expression: novel insights into gibberellin biosynthesis and perception during carrot growth and development. Hortic Res 2:e0134166
Wang GL, Huang Y, Zhang XY, Xu ZS, Wang F, Xiong AS (2016) Transcriptome-based identification of genes revealed differential expression profiles and lignin accumulation during root development in cultivated and wild carrots. Plant Cell Rep 35:1743–1755
Wang F, Wang GL, Hou XL, Li MY, Xu ZS, Xiong AS (2018) The genome sequence of ‘Kurodagosun’, a major carrot variety in Japan and China, reveals insights into biological research and carrot breeding. Mol Genet Genom 2018:1–11
Weng JK, Chapple C (2010) The origin and evolution of lignin biosynthesis. N Phytol 187:273–285
Xu ZS, Tan HW, Wang F, Hou XL, Xiong AS (2014a) CarrotDB: a genomic and transcriptomic database for carrot. Database J Biol Databases Curation 2014:1229–1245
Xu ZS, Ying H, Feng W, Xiong S, Wang GL, Xiong AS (2014b) Transcript profiling of structural genes involved in cyanidin-based anthocyanin biosynthesis between purple and non-purple carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biol 14:1–10
Yu JQ, Matsui Y (1993) Extraction and identification of phytotoxic substances accumulated in nutrient solution for the hydroponic culture of tomato. Soil Sci Plant Nutr 39:691–700
Zhao Q, Dixon RA (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci 16:227–233
Acknowledgements
The research was supported by the New Century Excellent Talents in University (NCET-11-0670); Jiangsu Natural Science Foundation (BK20130027); Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Maike Petersen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Que, F., Wang, GL., Feng, K. et al. Hypoxia enhances lignification and affects the anatomical structure in hydroponic cultivation of carrot taproot. Plant Cell Rep 37, 1021–1032 (2018). https://doi.org/10.1007/s00299-018-2288-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-018-2288-3