Abstract
The halophilic archaeal strain ZS-3T (= CGMCC 1.12866T = JCM 30239T) was isolated from a sediment sample of Zhoushan marine solar saltern, P. R. China. Phylogenetic analyses based on 16S rRNA, rpoB′ genes and the concatenation of 738 protein sequences reveal that strain ZS-3T was related to members of the genus Halorussus. The OrthoANI and in silico DDH values between strain ZS-3T and the current Halorussus members are much lower than the threshold values proposed as the species boundary (ANI 95–96% and in silico DDH 70%), suggesting that strain ZS-3T represents a novel species of Halorussus (Halorussus halophilus sp. nov.). Diverse phenotypic characteristics differentiate strain ZS-3T from current Halorussus members. Since the strain expressed diverse hydrolyzing enzyme activity, its complete genome was sequenced. The genome of strain ZS-3T was found to be 4,450,731 bp with total GC content of 61.51%, and comprises one chromosome and three plasmids. A total of 4694 protein coding genes, 43 tRNA genes and 6 rRNA genes were predicted. A CRISPR–Cas system was also detected. The genome encodes sixteen putative glycoside hydrolases, nine extracellular proteases, seventeen aminopeptidases, seven carboxypeptidases, one esterase and one nitrite reductase. The exploration of the hydrolase genes may expand our understanding of adapted mechanism of halophilic archaea surviving optimally in hypersaline environments where containing organic matter. Meanwhile, various hydrolyzing enzymes may extend this microorganism for further applications in salt-based fermentation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Halorussus (belonging to family Halobacteriaceae, order Halobacteriales, class Halobacteria) was established in 2010 and currently contains five validly published species, Hrs. amylolyticus [1], Hrs. litoreus [2], Hrs. rarus [3], Hrs. ruber [4] and Hrs. salinus [5]. These five current members of Halorussus exhibited diverse halophilic enzyme activity, three of them hydrolyzing starch, four of them hydrolyzing gelatin, two of them hydrolyzing casein and Tween 80, which reveals that Halorussus species may have the potential biotechnological applications. In this study, a novel strain ZS-3T, isolated from a sediment sample of Zhoushan marine solar saltern of China, was subjected to a polyphasic taxonomic characterization based on phenotypic, genotypic, and chemotaxonomic characteristics and identified as a novel species of the genus Halorussus, for which the name Halorussus halophilus sp. nov. is proposed.
Materials and Methods
Isolation of Halophilic Archaeal Strain and Culture Conditions
Strain ZS-3T was isolated from a sediment sample of Zhoushan marine solar saltern in Zhejiang Province, China (29° 56′ 56′′ N, 122° 20′ 20′′ E; elevation, sea level) in 2012. The isolation and cultivation of halophilic archaea were performed on a neutral haloarchaeal medium designated NHM containing (g/L): 0.05 yeast extract (Angel Yeast), 0.25 fish peptone (Sinopharm Chemical Reagent), 1.0 sodium pyruvate, 5.4 KCl, 0.3 K2HPO4, 0.29 CaCl2, 0.27 NH4Cl, 26.8 MgSO4·7H2O, 23.0 MgCl2·6H2O, 184.0 NaCl (adjusted to pH 7.0–7.2 with 1 M NaOH) [6]. Agar (2%, w/v) was added to prepare NHM plates. Strains were routinely grown aerobically at 37 °C in NHM medium.
Phenotypic Determination
Cell morphology and motility in exponentially growing liquid cultures in NHM broth at 37 °C were examined using a Nikon microscope (Ci-L) equipped with phase-contrast optics. The range of NaCl concentrations for growth was determined with modified NHM medium containing 0.9, 1.4, 1.7, 2.1, 2.6, 3.1, 3.4, 3.9, 4.3, 4.8 and 5.1 M NaCl. The range of MgCl2 concentrations for growth was studied in modified NHM medium with 0, 0.005, 0.01, 0.03, 0.05, 0.1, 0.3, 0.5, 0.7 and 1.0 M MgCl2. The temperature range for growth was examined at 10, 15, 20, 25, 30, 37, 40, 42, 45, 50, 55 and 60 °C. The pH range for growth was determined in modified NHM medium at pH 5.0–10.0 (at intervals of 0.5 pH units). Other phenotypic and physiological characteristics of strain ZS-3T were determined according to the proposed minimal standards for description of novel taxa in the order Halobacteriales [7].
Phylogenetic Analysis
The genomic DNA of strain ZS-3T was extracted and purified using the genomic DNA extraction kit (CW0552, Beijing ComWin Biotech Co., Ltd.) according to the manufacturer’s instruction. The 16S rRNA gene was amplified by PCR using the forward primer 20F (5′-ATTCCGGTTGATCCTGCCGG-3′) and reverse primer 1452R (5′-AGGAGGTGATCCAGCCGCAG-3′), then cloned and sequenced as described previously [8]. The rpoB′ gene was amplified, cloned and sequenced as described by Han et al. [9]. The 16S rRNA gene and the rpoB′ gene sequences were aligned using the ClustalW program integrated in the MEGA 6 software [10] and the phylogenetic trees were reconstructed using maximum-likelihood (ML) [11], neighbour-joining (NJ) [12] and maximum-parsimony (MP) [13] algorithms. The similarity of gene sequences was assessed by comparing the 16S rRNA gene and rpoB′ gene sequences of strain ZS-3T with those available from the EzBioCloud server [14]. The phylogenomic analysis was carried out as described by Zhao et al. [15].
Genome Sequence Analysis
The complete genome of strain ZS-3T was sequenced using a PacBio RS II platform and Illumina HiSeq 4000 platform at the Beijing Genomics Institute, China. Two libraries containing 10-kb and 350-bp inserts were constructed and the Pbdagcon (https://github.com/PacificBiosciences/pbdagcon) was used for self-correction. All reads were de novo assembled using the Celera Assembler and the assembled sequence was screened further to correct errors and identify single nucleotide polymorphisms (SNP) using the GATK, SOAPsnp and SOAPindel packages. Confirmation of circular replicons and plasmid comparisons were performed with the SOAP package mapped to the bacterial plasmid database. The final assembly generated four circular sequences without any gap. The average nucleotide identity (ANI) was calculated using the OrthoANIu algorithm by ChunLab’s online Average Nucleotide Identify calculator (https://www.ezbiocloud.net/tools/ani). The in silico DNA-DNA hybridization (DDH) values were calculated by Genome-to-Genome Distance Calculator 2.1 (GGDC) (https://ggdc.dsmz.de/ggdc.php) [16,17,18,19]. Gene prediction was performed with glimmer 3 [20] and Hidden Markov models. The best hits found using the Blast alignment tool were used for functional annotation. The databases, KEGG [21], COG [22], NR (Non-Redundant Protein Database databases), Swiss-Prot [23] and GO (Gene Ontology), were searched in order to assign or improve general function annotations. Enzymes for degrading carbohydrates and glycoconjugates were annotated using dbCAN [24]. The tRNA and rRNA genes were detected using tRNA scan-SE [25] and RNAmmer [26]. CRISPRFinder was used to screen for CRISPR arrays [27]. Putative signal sequences of hydrolytic enzymes were analysed using the predictive algorithm of the server SignalP 5.0 [28].
Chemotaxonomic Characteristics
Polar lipids of strain ZS-3T and reference halophilic archaeal strains were extracted and analyzed by one- and two-dimensional TLC according to the procedures described by Cui et al. [3]. Specific detection spray reagents for phospholipids and glycolipids were used and the general detection reagent, sulfuric acid–ethanol (1:2, by vol.), was used to detect total polar lipids [3].
Results and Discussion
Morphological and Physiological Characteristics
Cells of strain ZS-3T are motile, pleomorphic rods (0.8–1.0 × 1.0–6.0 μm, Fig. S1), aerobic, and Gram-stain-negative. Colonies are circular, smooth and red pigmented. Strain ZS-3T grows under 25–42 °C (optimum 37 °C), in the presence of 1.4–4.8 M NaCl (optimum 2.6 M), with 0–1.0 M MgCl2 (optimum 0.1 M), and at pH 5.5–9.5 (optimum 7.0). Cells lyse in distilled water and the minimal NaCl concentration to prevent cell lysis is 10% (w/v). Catalase and oxidase are positive. No growth occurs anaerobically with nitrate, l-arginine or DMSO. Strain ZS-3T does not reduce nitrate to nitrite, thus formation of gas from nitrate does not occur. Casein, gelatin and starch are hydrolysed but Tween 80 is not. Both indole formation and H2S production are negative. d-glucose, d-mannose, d-galactose, sucrose, starch, glycerol, pyruvate, dl-lactate, succinate, l-malate, fumarate and citrate can be used as carbon sources. l-glutamate and l-ornithine can support growth. No growth occurs on d-fructose, l-sorbose, d-ribose, lactose, d-xylose, maltose, d-mannitol, d-sorbitol, acetate, l-arginine, l-aspartate, l-lysine, glycine or l-alanine. Acid is produced from d-glucose and sucrose. The main phenotypic characteristics differentiating strain ZS-3T from the current members of Halorussus, Haladaptatus and Halalkalicoccus were shown in Table 1.
Phylogenetic Analysis
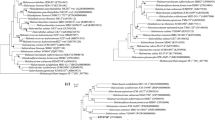
Phylogenetic analyses based on 16S rRNA and rpoB′ genes sequences using the ML, NJ and MP algorithms reveal that strain ZS-3T was related to the member of genus Halorussus, then with the members of Haladaptatus or Halalkalicoccus (Fig. 1, Fig. S3 and Fig. S4). The 16S rRNA and rpoB′ gene sequences similarities of strain ZS-3T towards the members of Halorussus, Haladaptatus and Halalkalicoccus were 91.0–92.5% and 87.4–88.3%, 91.0–91.9% and 84.2–85.3%, 89.7–89.9% and 81.1–82.9% similarities, respectively. These relatively low similarities in rRNA and rpoB′ genes motivated us to perform further polyphasic taxonomic study and complete genome analyses. The phylogenomic tree reconstruction (Fig. 2) revealed unequivocally that strain ZS-3T clustered with the current Halorussus members, confirming their positions as a new taxon within the genus Halorussus.
Maximum-Likelihood phylogenetic tree reconstructions based on 16S rRNA gene (a) and rpoB′ gene (b) sequences, showing the relationships between strain ZS-3T and related members within the order Halobacteriales. Bootstrap values (%) are based on 1000 replicates and are shown for branches with more 50% bootstrap support. Bar represents expected substitutions per nucleotide position
Maximum-Likelihood phylogenetic tree reconstruction based on the concatenation of 738 protein sequences, showing the relationships between strain ZS-3T and related members within the family Halobacteriaceae. Bootstrap values (%) are based on 1000 replicates. Haloferax mediterranei ATCC 33500T was used as an outgroup. Bar, 0.1 substitutions per amino acid position
Genome Sequence Analysis
The complete genome sequence of strain ZS-3T consists of one circular chromosome and three circular plasmids, with lengths of 3,636,252 bp (chromosome), 531,768 bp (pUJS01), 219,256 bp (pUJS02) and 63,455 bp (pUJS03), respectively. The G + C contents of the chromosome and three plasmids were 61.68%, 62.04%, 58.05%, and 59.56%, respectively (Table S1; Fig. S5). The OrthoANI and in silico DDH values between strain ZS-3T, Halorussus members and Haladaptatus species are much lower than the threshold values proposed as the species boundary (ANI 95–96% and in silico DDH 70%, Table S2). A total of 4694 protein coding genes, 43 tRNAs and 6 rRNA genes, and two 16S rRNA gene (1472 bp) were predicted. Among the 4694 ORFs, only 2759 ORFs could be classified into COG categories. The major categories were amino acid transport and metabolism (12.0%), translation, ribosomal structure and biogenesis (8.7%), transcription (8.0%), inorganic ion transport and metabolism (7.6%), energy production and conversion (6.5%), coenzyme transport and metabolism (6.3%). Approximately 38.8% of all ORFs could be assigned to a pathway using the KEGG database. Twelve CRISPR repeat regions were identified on the chromosome and one on plasmid pUJS03, and these CRISPR repeat regions contained 107 spacers, 7 types of direct repeats. There was only one predicted Cas protein operon (type I) on the chromosome (cas6, Csc3, Csc2, cas5, cas3, cas4, cas1, and cas2). Other general genomic features that distinguish strain ZS-3T from the type strains of the species of the genus Halorussus are shown in Table S3.
The types of the hydrolyzing enzymes encoded in the genome of strain ZS-3T were assessed. For primary organic carbon degradation, according to the dbCAN database we found glycosyl transferases (29), glycoside hydrolases (16), polysaccharide lyases (2), carbohydrate esterases (16), auxiliary activities (10), and carbohydrate-binding modules (4). Nine kinds of glycoside hydrolases were predicted: 1 levansucrase (EC 2.4.1.10), 3 β-1,4-glucanase / cellulase (EC 3.2.1.4), 1 endo-inulinase (EC 3.2.1.7), 3 β-glucosidase (EC 3.2.1.21), 1 β-galactosidase (EC 3.2.1.23), 2 chitinase (EC 3.2.1.14), 1 glucoamylase (EC 3.2.1.3), 3 α-amylase (EC 3.2.1.1), and 1 α-N-acetylgalactosaminidase (EC 3.2.1.49), respectively. Four of these glycoside hydrolases (1 β-1,4-glucanase / cellulase, 1 β-glucosidase and 2 chitinase) have tat signal peptide-coding sequence, they exert their function by being secreted into extracellular environment. Strain ZS-3T has be confirmed to be able to hydrolyze starch and hydroxymethyl cellulose. These activities indicate the organism is likely to be able to hydrolyze or even grow on various types of carbohydrates.
For primary protein degradation, fourteen encoded serine protease genes were annotated in the genome, eight of which have a tat signal peptide while one has a sec signal peptide and the others have no signal peptide. In all, nine out of the fourteen enzymes are predicted to be secreted outside cell to function. Strain ZS-3T was also confirmed to be able to hydrolyze casein and gelatin. Compared to other haloarchaea on the ability to hydrolyze casein, strain ZS-3T appears to encode more extracellular proteases. For example, Halobacterium salinarum NRC-1 (GCA_000006805.1) encodes two serine protease genes, Natrialba magadii ATCC 43099T (GCA_000025625.1) has nine ones and Natrinema sp. J7-1 (GCA_000493245.1) has five ones. Strain ZS-3T carries five aminopeptidases (Xaa-Pro), three putative aminopeptidases (FrvX), a methionine aminopeptidase, four leucyl aminopeptidases, and four dipeptidyl aminopeptidases. Muramoyltetrapeptide carboxypeptidase, metal-dependent carboxypeptidase, d-alanyl- d-alanine carboxypeptidase, and four Zn-dependent carboxypeptidases were also found. The presence of these various protease-related genes indicates that strain ZS-3T possesses a wide range of proteolytic activities that would be active in hypersaline environments.
An esterase was predicted in the genome of strain ZS-3T, but no Tween 80 hydrolytic activity of strain ZS-3T was detected. It may act on smaller molecule esters. We also found that strain ZS-3T had capacity to reduce nitrite, and a copper-containing nitrite reductase coding gene sequence is located in the chromosome. Such halophilic archaeal strain ZS-3T may have commercial applications in the fermentation of high-salt foods such soy sauce and fish sauce, by accelerating hydrolysis of proteins, carbohydrates, and esters, meanwhile reducing the harm of nitrite.
The sequence of strain ZS-3T determined in this study represents the first complete genome sequence reported for the genus Halorussus. The annotation and genomic analysis provide new insight into the metabolic capacity and heterotrophic lifestyle of strain ZS-3T, and opens up new possibilities for use of this microorganism or its enzymes in microbial biotechnology.
Chemotaxonomic Characteristics
Strain ZS-3T contained phosphatidylglycerol (PG), phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfate (PGS), three major glycolipids, and two minor glycolipids. The three major glycolipids (GL1, GL2 and GL4) were identical to sulfated galactosyl mannosyl glucosyl diether (S-TGD-1), sulfated mannosyl glucosyl diether (S-DGD-1), and mannosyl glucosyl diether (DGD-1), while the two minor glycolipids (GL3 and GL5) were unidentified (Fig. S2).
Description of Halorussus halophilus sp. nov.
Halorussus halophilus (ha.lo'phi.lus. Gr. n. hals, halos salt; Gr. adj. philos loving; N.L. masc. adj. halophilus salt-loving referring to the requirement for salt).
The cells are motile, pleomorphic rods (0.8–1.0 × 1.0–6.0 μm), aerobic, and Gram-stain-negative. Colonies are circular, smooth and red pigmented. Growth occurs under 25–42 °C (optimum 37 °C), in the presence of 1.4–4.8 M NaCl (optimum 2.6 M), with 0–1.0 M MgCl2 (optimum 0.1 M), and at pH 5.5–9.5 (optimum 7.0). Cells lyse in distilled water and the minimal NaCl concentration to prevent cell lysis is 10% (w/v). Catalase and oxidase are positive. No growth occurs anaerobically with nitrate, l-arginine or DMSO. The novel strain does not reduce nitrate to nitrite, thus formation of gas from nitrate does not occur. Casein, gelatin and starch are hydrolysed but Tween 80 is not. Both indole formation and H2S production are negative. d-glucose, d-mannose, d-galactose, sucrose, starch, glycerol, pyruvate, dl-lactate, succinate, l-malate, fumarate and citrate can be used as carbon sources. l-glutamate and l-ornithine can support growth. No growth occurs on d-fructose, l-sorbose, d-ribose, lactose, d-xylose, maltose, d-mannitol, d-sorbitol, acetate, l-arginine, l-aspartate, l-lysine, glycine or l-alanine. Acid is produced from d-glucose and sucrose. The polar lipids are phosphatidylglycerol (PG), phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfate (PGS), three major glycolipids, and two minor glycolipids. The three major glycolipids are GL1, GL2 and GL4, while the two unidentified minor glycolipids are GL3 and GL5. Genomic DNA G + C content is 61.5 mol%. The novel strain ZS-3T (= CGMCC 1.12866T = JCM 30239T) was isolated from Zhoushan marine solar saltern in Zhejiang Province, P. R. China. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA genes, rpoB′ gene and whole genome sequences of strain ZS-3T are KJ689292, MF443863, and CP044523 ~ CP044526, respectively.
References
Yuan P-P, Ye W-T, Pan J-X, Han D, Zhang W-J, Cui H-L (2015) Halorussus amylolyticus sp. nov., isolated from an inland salt lake. Int J Syst Evol Microbiol 65:3734–3738. https://doi.org/10.1099/ijsem.0.000487
Han D, Zhu L, Cui H-L (2019) Halorussus litoreus sp. nov., isolated from the salted brown alga Laminaria. Int J Syst Evol Microbiol 69:767–772. https://doi.org/10.1099/ijsem.0.003233
Cui H-L, Gao X, Yang X, Xu X-W (2010) Halorussus rarus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Extremophiles 14:493–499. https://doi.org/10.1007/s00792-010-0329-0
Xu W-D, Zhang W-J, Han D, Cui H-L, Yang K (2015) Halorussus ruber sp. nov., isolated from an inland salt lake of China. Arch Microbiol 197:91–95. https://doi.org/10.1007/s00203-014-1058-z
Xu J-Q, Xu W-M, Li Y, Zhou Y, Lü Z-Z, Hou J, Zhu L, Cui H-L (2016) Halorussus salinus sp. nov., isolated from a marine solar saltern. Arch Microbiol 198:957–961. https://doi.org/10.1007/s00203-016-1253-1
Hou J, Zhao Y-J, Zhu L, Cui H-L (2018) Salinirubellus salinus gen. nov., sp. nov., isolated from a marine solar saltern. Int J Syst Evol Microbiol 68:1874–1878. https://doi.org/10.1099/ijsem.0.002757
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238. https://doi.org/10.1099/00207713-47-1-233
Cui H, Zhou P, Oren A, Liu S (2009) Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13:31–37. https://doi.org/10.1007/s00792-008-0194-2
Han D, Hong L-G, Xu Q, Cui H-L (2019) Halostella limicola sp. nov., isolated from saline soil sampled at the Tarim Basin. Int J Syst Evol Microbiol 69:3299–3304. https://doi.org/10.1099/ijsem.0.003643
Tamura K, Stecher G, Peterson DS, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Felsenstein J (1981) Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416. https://doi.org/10.1093/sysbio/20.4.406
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Zhao Y-J, Tao C-Q, Zeng C-L, Zhu L, Cui H-L (2020) Salinigranumhalophilum sp. nov., isolated from marine solar salterns. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.003951
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu X-W, Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. https://doi.org/10.1093/bioinformatics/btm009
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280. https://doi.org/10.1093/nar/gkh063
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. https://doi.org/10.1093/nar/28.1.33
Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh L-SL (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32:D115–D119. https://doi.org/10.1093/nar/gkh131
Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y (2012) dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. https://doi.org/10.1093/nar/gks479
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. https://doi.org/10.1093/nar/25.5.955
Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. https://doi.org/10.1093/nar/gkm160
Grissa I, Vergnaud G, Pourcel C (2007) CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57. https://doi.org/10.1093/nar/gkm360
Armenteros AJJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. https://doi.org/10.1038/s41587-019-0036-z
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ding, Y., Han, D. & Cui, HL. Halorussus halophilus sp. nov., A Novel Halophilic Archaeon Isolated from a Marine Solar Saltern. Curr Microbiol 77, 1321–1327 (2020). https://doi.org/10.1007/s00284-020-01921-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01921-8