Abstract
Halophilic archaeal strain YC25T was isolated from Yuncheng salt lake in Shanxi, China. Cells of strain YC25T were observed to be pleomorphic rods, stained Gram-negative, and formed red-pigmented colonies on solid media. Strain YC25T was found to be able to grow at 25–50 °C (optimum 37 °C), at 1.4–4.8 M NaCl (optimum 1.7 M), at 0–1.0 M MgCl2 (optimum 0.01 M), and at pH 5.5–9.0 (optimum pH 6.5). The cells lysed in distilled water, and the minimal NaCl concentration to prevent cell lysis was found to be 8 % (w/v). The major polar lipids of the strain were phosphatidylglycerol (PG), phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfate (PGS), sulfated galactosyl mannosyl glucosyl diether (S-TGD-1), sulfated mannosyl glucosyl diether (S-DGD-1), galactosyl mannosyl glucosyl diether (TGD-1), mannosyl glucosyl diether (DGD-1), and an unknown diglycosyl diether (DGD-2) chromatographically identical to those of Halorussus rarus CGMCC 1.10122T. The 16S rRNA gene and rpoB′ gene of strain YC25T were phylogenetically related to the corresponding genes of Halorussus rarus CGMCC 1.10122T (94.3–95.4 and 91.5 % nucleotide identity, respectively). The DNA G+C content of strain YC25T was determined to be 63.3 mol%. The phenotypic, chemotaxonomic, and phylogenetic properties suggested that strain YC25T (=CGMCC 1.12122T = JCM 18363T) represents a new species of Halorussus, for which the name Halorussus ruber sp. nov. is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inland salt lakes, natural athalassohaline environments, harbor diverse halophilic archaea, the members of the family Halobacteriaceae (Pagaling et al. 2009; Youssef et al. 2012). Some of these hypersaline environments have been long-term targets for the study of halophilic archaeal resources, and a number of novel strains representing new species have been isolated from these inland salt lakes (Corral et al. 2013; Qiu et al. 2013a, b; Zhang et al. 2013; Amoozegar et al. 2014). The family Halobacteriaceae encompasses 47 genera containing over 165 species as of February 2014 (Oren 2012; 2014; Parte 2014). During our survey on halophilic archaeal diversity of the Yuncheng salt lake in Shanxi, China, we obtained a halophilic archaeal isolate, YC25T, that was most closely related to the members of Halorussus, as judged from 16S rRNA gene sequences.
The genus Halorussus, belonging to the family Halobacteriaceae, was proposed to accommodate the species Halorussus rarus described based on two strains, TBN4T and TBN5, which were isolated from an artificial marine solar saltern in Eastern China (Cui et al. 2010). The most distinctive characteristic of Halorussus rarus was its polar lipid profile. Halorussus rarus contained phosphatidylglycerol (PG), phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfate (PGS), sulfated galactosyl mannosyl glucosyl diether (S-TGD-1), sulfated mannosyl glucosyl diether (S-DGD-1), galactosyl mannosyl glucosyl diether (TGD-1), mannosyl glucosyl diether (DGD-1), and an unknown diglycosyl diether (DGD-2; Cui et al. 2010). In this study, we characterize strain YC25T as a new species of the genus Halorussus, for which the name Halorussus ruber sp. nov. is proposed.
Materials and methods
Isolation and cultivation of halophilic archaeal strain
Strain YC25T was isolated from the brine sample taken from Yuncheng salt lake at Yuncheng, Shanxi Province, China (35°00′14″N, 111°00′19″E; elevation 323 m above sea level) and stored at 4 °C during transport to the laboratory in 2010. The pH of the brine was 7.9 and the salinity 235.6 g/L. The neutral haloarchaeal medium (NHM) was used for the isolation procedure and contained the following ingredients (g/L): yeast extract (Oxoid) 0.05, fish peptone (Sinopharm Chemical Reagent Co., Ltd.) 0.25, sodium pyruvate 1.0, KCl 5.4, K2HPO4 0.3, CaCl2 0.29, NH4Cl 0.27, MgSO4·7H2O 26.8, MgCl2·6H2O 23.0, and NaCl 184.0 (pH adjusted to 7.0–7.2 with 1 M NaOH solution). The brine was serially diluted in liquid NHM medium and spread onto NHM agar plates. The inoculated plates were incubated for 1 month at 37 °C. After this initial cultivation, colonies were successively restreaked on NHM agar plates at least three times to obtain pure colonies. The strain was routinely grown aerobically at 37 °C in NHM medium.
Phenotypic determination
Phenotypic tests were performed according to the proposed minimal standards for the description of novel taxa in the order Halobacteriales (Oren et al. 1997). Determination of morphology and growth characteristics, nutrition, miscellaneous biochemical tests, and sensitivity to antimicrobial agents was performed as described and cited previously (Cui et al. 2010). The halophilic archaeal strains Halorussus rarus CGMCC 1.10122T, Haladaptatus paucihalophilus JCM 13897T, and Haloarchaeobius litoreus CGMCC 1.10390T were selected as reference strains. These reference strains were routinely grown aerobically at 37 °C in NHM medium.
Chemotaxonomic characterization
Polar lipids were extracted using a chloroform/methanol system and analyzed using one- and two-dimensional TLC, as described previously (Cui et al. 2010). Merck silica gel 60 F254 aluminum-backed thin-layer plates were used for TLC analyses. In two-dimensional TLC, the first solvent was chloroform–methanol–water (65:25:4, by vol.) and the second solvent was chloroform–methanol–acetic acid–water (80:12:15:4, by vol.). The latter solvent mixture was also used in one-dimensional TLC. Two specific detection spray reagents, phosphate stain reagent for phospholipids and α-naphthol stain for glycolipids, were used. The general detection reagent, sulfuric acid–ethanol (1:2, by vol.), was also used to detect total polar lipids.
Phylogenetic and genomic analysis
Genomic DNA from halophilic archaeal strains was prepared as described previously (Cui et al. 2011). The 16S rRNA genes were amplified, cloned, and sequenced according to the previous protocol (Cui et al. 2009). PCR-mediated amplification and sequencing of the rpoB′ genes were performed as described previously (Minegishi et al. 2010). Multiple sequence alignments were performed using the ClustalW program integrated in the MEGA 5 software (http://www.megasoftware.net/). Phylogenetic trees were reconstructed using maximum-likelihood (ML), maximum-parsimony (MP), and neighbor-joining (NJ) algorithms in the MEGA 5 software (Tamura et al. 2011). Gene sequence similarity among halophilic archaea was calculated using the Pairwise-Distance computing function of MEGA 5. The DNA G+C content was determined from the midpoint value (T m) of the thermal denaturation method (Marmur and Doty 1962) at 260 nm with a Beckman-Coulter DU800™ spectrophotometer equipped with a high-performance temperature controller.
Results and discussion
Cells of strain YC25T were observed to be motile and pleomorphic rods when grown in NHM liquid medium (Supplementary Fig. S1). They stained Gram-negative, and the colonies were observed to be red-pigmented. Strain YC25T was found to be able to grow at 25–50 °C (optimum 37 °C), at 1.4–4.8 M NaCl (optimum 1.7 M), at 0–1.0 M MgCl2 (optimum 0.01 M), and at pH 5.5–9.0 (optimum pH 6.5). The cells lysed in distilled water, and the minimal NaCl concentration to prevent cell lysis was found to be 8 % (w/v). The strain was able to grow under anaerobic conditions using nitrate, l-arginine, and DMSO. It was found to be positive for indole formation while negative for H2S formation. Strain YC25T hydrolyzed Tween 80 but did not hydrolyze starch, gelatin, or casein. Strain YC25T was sensitive to the following antimicrobial compounds (µg per disk, unless otherwise indicated): Anisomycin (5), aphidicolin (5), novobiocin (30), bacitracin (0.04 IU per disk), rifampin (5), ciprofloxacin (5), mycostatin (100), and nitrofurantoin (300). It was resistant to the following antimicrobial compounds: trimethoprim (5), erythromycin (15), penicillin G (10 IU per disk), ampicillin (10), chloramphenicol (30), neomycin (30), norfloxacin (10), streptomycin (10), kanamycin (30), tetracycline (30), vancomycin (30), gentamicin (10), and nalidixic acid (30). The main phenotypic characteristics differentiating strain YC25T from Halorussus rarus CGMCC 1.10122T are optimum NaCl, optimum Mg2+, optimum pH, anaerobic growth with nitrate, arginine, and DMSO, utilization of specific carbon sources, gelatin and casein hydrolysis, and H2S formation (Table 1). More detailed results of phenotypic features of strain YC25T are given in the species description.
The major polar lipids of strain YC25T were phosphatidylglycerol (PG), phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfate (PGS), sulfated galactosyl mannosyl glucosyl diether (S-TGD-1), sulfated mannosyl glucosyl diether (S-DGD-1), galactosyl mannosyl glucosyl diether (TGD-1), mannosyl glucosyl diether (DGD-1), and an unknown diglycosyl diether (DGD-2) chromatographically identical to those of Halorussus rarus CGMCC 1.10122T (Supplementary Fig. S2). The major polar lipid compositions supported classification of strain YC25T in the genus Halorussus.
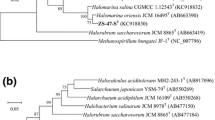
Sequence comparisons indicated that strain YC25T possessed two different 16S rRNA genes (denoted rrnA and rrnB) that differed in sequence by 4.4 %. The rrnA and rrnB genes of strain YC25T were phylogenetically related to that of Halorussus rarus CGMCC 1.10122T (94.3 and 95.4 % nucleotide identity, respectively), 16S rRNA gene similarities that are well below the recently recommended thresholds (98.2–99.0 %) to separate two prokaryotic species (Meier-Kolthoff et al. 2013). Phylogenetic tree reconstructions using the ML algorithm revealed that strain YC25T tightly clustered with Halorussus rarus (Fig. 1a). The phylogenetic position was also confirmed in other trees generated using the MP and NJ algorithms (Supplementary Fig. S3a & Fig. S4a).
Maximum-likelihood phylogenetic tree reconstructions based on 16S rRNA gene (a) and rpoB′ gene (b) sequences, showing the relationships between strain YC25T and related members within the family Halobacteriaceae. Bootstrap values (%) are based on 1,000 replicates and are shown for branches with more 70 % bootstrap support. Bar represents expected changes per site
The rpoB′ gene of strain YC25T was closely similar to the corresponding gene of Halorussus rarus (91.5 % nucleotide identity). In phylogenetic tree reconstructions using rpoB′ (Fig. 1b), strain YC25T tightly clustered with Halorussus rarus. The phylogenetic position was also confirmed in trees generated using the MP and NJ algorithms (Supplementary Fig. S3b & Fig. S4b).
The 16S rRNA gene and rpoB′ gene-based phylogenetic analysis results supported the placement of strain YC25T in the genus Halorussus.
The DNA G+C content of strain YC25T was determined to be 63.3 mol%, which is lower than that of Halorussus rarus CGMCC 1.10122T (66.1 mol%).
Based on these phenotypic, chemotaxonomic, and phylogenetic properties, a novel species of the genus Halorussus, Halorussus ruber sp. nov., is proposed to accommodate strain YC25T. Characteristics that distinguish strain YC25T from Halorussus rarus CGMCC 1.10122T are shown in Table 1.
Description of Halorussus ruber sp. nov
Halorussus ruber (ru.ber. L. adj. ruber red-colored, red)
Cells are motile, pleomorphic rods under optimal growth conditions and stain Gram-negative. Colonies on agar plates containing 1.7 M NaCl are red, elevated, and round. Chemoorganotrophic and aerobic. Growth occurs at 25–50 °C (optimum 37 °C), at 1.4–4.8 M NaCl (optimum 1.7 M), at 0–1.0 M MgCl2 (optimum 0.01 M), and at pH 5.5–9.0 (optimum pH 6.5). Cells lyse in distilled water, and the minimal NaCl concentration to prevent cell lysis is 8 % w/v. Catalase and oxidase positive. Grow anaerobically in the presence of nitrate, arginine, and DMSO. Nitrate reduction to nitrite is observed, but no gas formed from nitrate. Indole formation is positive, but H2S formation is negative. Hydrolyze Tween 80 but does not hydrolyze starch, gelatin, or casein. The following substrates are utilized as single carbon and energy sources for growth: acetate, citrate, fumarate, d-galactose, d-glucose, dl-lactate, lactose, l-malate, d-mannitol, d-mannose, d-sorbitol, and sucrose. The following substrates are utilized as single carbon, nitrogen, or energy sources for growth: l-glutamate and l-ornithine. No growth occurs on l-alanine, l-arginine, l-aspartate, d-fructose, glycerol, glycine, l-lysine, maltose, pyruvate, d-ribose, l-sorbose, starch, succinate, or d-xylose. The major polar lipids are phosphatidylglycerol (PG), phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfate (PGS), sulfated galactosyl mannosyl glucosyl diether (S-TGD-1), sulfated mannosyl glucosyl diether (S-DGD-1), galactosyl mannosyl glucosyl diether (TGD-1), mannosyl glucosyl diether (DGD-1), and an unknown diglycosyl diether (DGD-2). The DNA G+C content of strain YC25T was 63.3 mol% (T m). The type strain is YC25T (=CGMCC 1.12122T = JCM 18363T). The GenBank/EMBL/DDBJ accession numbers for the rrnA and rrnB 16S rRNA gene and rpoB′ gene sequences of strain YC25T are JQ237116, KJ913074 and KJ913075, respectively.
References
Amoozegar MA, Makhdoumi-Kakhki A, Mehrshad M, Fazeli SAS, Spröer C, Ventosa A (2014) Halorientalis persicus sp. nov., an extremely halophilic archaeon isolated from a salt lake and emended description of the genus Halorientalis. Int J Syst Evol Microbiol 64:940–944
Corral P, Gutiérrez MC, Castillo AM, Domínguez M, Lopalco P, Corcelli A, Ventosa A (2013) Natronococcus roseus sp. nov., a haloalkaliphilic archaeon from a hypersaline lake. Int J Syst Evol Microbiol 63:104–108
Cui H-L, Zhou P-J, Oren A, Liu S-J (2009) Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13:31–37
Cui H-L, Gao X, Yang X, Xu X-W (2010) Halorussus rarus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Extremophiles 14:493–499
Cui H-L, Yang X, Mou Y-Z (2011) Salinarchaeum laminariae gen. nov., sp. nov.: a new member of the family Halobacteriaceae isolated from salted brown alga Laminaria. Extremophiles 15:625–631
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P (2013) When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010) Further refinement of Halobacteriaceae phylogeny based on the full-length RNA polymerase subunit B’ (rpoB′) gene. Int J Syst Evol Microbiol 60:2398–2408
Oren A (2012) Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. Int J Syst Evol Microbiol 62:263–271
Oren A (2014) Taxonomy of halophilic archaea: current status and future challenges. Extremophiles 18:825–834
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238
Pagaling E, Wang H, Venables M, Wallace A, Grant WD, Cowan DA, Jones BE, Ma Y, Ventosa A, Heaphy S (2009) Microbial biogeography of six salt lakes in Inner Mongolia, China, and a salt lake in Argentina. Appl Environ Microbiol 75:5750–5760
Parte AC (2014) LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42:D613–D616
Qiu X-X, Mou Y-Z, Zhao M-L, Zhang W-J, Han D, Ren M, Cui H-L (2013a) Halobellus inordinatus sp. nov., from a marine solar saltern and an inland salt lake of China. Int J Syst Evol Microbiol 63:3975–3980
Qiu X-X, Zhao M-L, Han D, Zhang W-J, Cui H-L (2013b) Halorubrum rubrum sp. nov., an extremely halophilic archaeon from a Chinese salt lake. Antonie Van Leeuwenhoek 104:885–891
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Youssef NH, Ashlock-Savage KN, Elshahed MS (2012) Phylogenetic diversities and community structure of members of the extremely halophilic archaea (Order Halobacteriales) in multiple saline sediment habitats. Appl Environ Microbiol 78:1332–1344
Zhang W-J, Han D, Qiu X-X, Zhao M-L, Mou Y-Z, Cui H-L, Li Z-R (2013) Halobellus rarus sp. nov., a halophilic archaeon from an inland salt lake of China. Antonie Van Leeuwenhoek 104:377–384
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31370054), the grant from China Ocean Mineral Resources R&D Association (COMRA) Special Foundation (DY125-15-R-03), and the Qinglan Project of Jiangsu Province and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Erko Stackebrandt.
Phase-contrast micrograph of strain YC25T, thin-layer chromatograms of the polar lipids extracted from strain YC25T and some related haloarchaea, maximum-parsimony and neighbor-joining phylogenetic tree reconstructions based on 16S rRNA gene and rpoB′ gene sequences showing the relationships between strain YC25T and related members within the family Halobacteriaceae are available as supplementary materials.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, WD., Zhang, WJ., Han, D. et al. Halorussus ruber sp. nov., isolated from an inland salt lake of China. Arch Microbiol 197, 91–95 (2015). https://doi.org/10.1007/s00203-014-1058-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-014-1058-z