Abstract
Recombinant protein pharmaceutical agents have been widely used for cancer treatment. Although tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) has broad-spectrum antitumor activity, its clinical applications are limited because most tumor cells eventually develop resistance to TRAIL-induced apoptosis through various pathways. Prostate apoptosis response-4 (Par-4) selectively induces apoptosis in cancer cells after binding to the cell surface receptor, GRP78. In this study, TRAIL was fused with the core domain of Par-4 (SAC) to produce a novel recombinant fusion protein. To obtain solubly expressed fusion protein, a small ubiquitin-related modifier (SUMO) was added to the N-terminus of the target protein. Cytotoxicity assays showed that the purified fusion protein exhibited more significant antitumor activity on cancer cells than that by native TRAIL. The connection order and linker sequence of the fusion proteins were optimized. In vitro cytotoxicity assay showed that the SAC-TRAIL fusion protein, which contained a flexible linker (G4S)3, optimally inhibited the proliferation of cancer cells. Immunofluorescence assays demonstrated that SAC-TRAIL could efficiently and specifically bind to cancer cells. Additionally, circular dichroism assays showed that the secondary structure of the recombinant protein with a flexible linker (G4S)3 has both a lower α-helix and higher random coiling, which facilitates the specific binding of SAC-TRAIL to the receptor. Collectively, these results suggest that the novel recombinant fusion protein SAC-(G4S)3-TRAIL is a potential therapeutic agent for cancer.
Key points
• Improved tumor growth suppression and apoptosis induction potency of SAC-TRAIL.
• Enhanced targeting selectivity of SAC-TRAIL in cancer cells.
• Lower α-helix and higher random coiling in SAC-TRAIL with flexible linker (G4S)3.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although cancer treatment strategies continue to advance, issues such as drug resistance and incomplete treatment still exist (Mun et al. 2018). The current strategies include chemotherapy, radiotherapy, immunotherapy, and targeted therapy; however, they fail to completely eliminate cancer cells (Sudhakar 2009). In contrast to conventional therapies, recombinant proteins can improve biological function or develop new functions of proteins; their use in the treatment of various diseases, including cancer, has become a popular topic of research (Baeshen et al. 2015; Wilding et al. 2019; Sanchez-Garcia et al. 2016).
TRAIL is a promising drug candidate because of its extensive apoptosis-inducing effects on cancer cells while simultaneously leaving normal cells unharmed (Johnstone et al. 2008). TRAIL is a member of the TNF superfamily and a type II transmembrane protein composed of 281 amino acids; its soluble portion, which retains complete TRAIL protein activity, is located 114–281 amino acids from the C-terminus (Idriss and Naismith 2000). TRAIL can bind to death receptor 4/5 (DR4/5) expressed on cancer cell surfaces and activate a series of signaling pathways to promote apoptosis of cancer cells (Walczak et al. 1997). Although TRAIL shows an attractive prospect for treating cancer, it induces drug resistance and does not substantially damage tumor cells with high NF-κB activity (Seo et al. 2012). Researchers are, therefore, exploring a wide range of strategies, including those involving recombinant fusion proteins, to realize the theoretical effects of TRAIL in tumor treatment. Single-chain variable fragments of TRAIL (scFv-TRAIL) were constructed to target tumor-associated antigens or immune cell antigens, such as epidermal growth factor (EGFR), human epidermal growth factor 2 (HER2), CD25 and PD-L1 (Dubuisson and Micheau 2017; Snajdauf et al. 2021). A humanized leucine zipper (LZ)-TRAIL chimera was designed to improve the antitumor activity and half-life of recombinant protein (Rozanov et al. 2015).
Although Par-4, a tumor suppressor, was originally discovered in apoptotic prostate cancer cells, it is widely expressed in various other cells as well (Sells et al. 1994). Par-4 is a multi-domain protein and has a leucine-zipper domain (LZ) at the C-terminus, two nuclear localization sequences (NLS1 and NLS2), a SAC (selective for apoptosis of cancer cells) domain, and a nuclear export sequence (Hebbar et al. 2012). Studies on the deletion of the Par-4 sequence have identified the 59-amino acid-long SAC domain as its core component (El-Guendy et al. 2003). Par-4 is able to enter the nucleus, where the SAC domain can activate caspase, inhibit the activity of NF-κB, and selectively induce apoptosis of cancer cells without having cytotoxic effects on normal cells and tissues (Chaudhry et al. 2012; Meynier et al. 2015). Therefore, Par-4 is a promising candidate for cancer therapy.
Recombinant fusion proteins have been broadly applied for disease treatment, as they can improve biological function and enhance the protein targeting (Wang et al. 2017). Biological activities of fusion proteins are influenced by the order in which subunits are connected, as this can remarkably affect the overall protein structure (Sachdev and Chirgwin 1998; Yu et al. 2015). Linker sequence length and flexibility also affect the biological activity and expression status of fusion proteins (Arai et al. 2001; Huang et al. 2013).
In this study, the anticancer proteins TRAIL and SAC were fused using a flexible linker in Escherichia coli. The connection order and linker sequence of the fusion units were optimized to improve the antitumor activity of the fusion protein. The physicochemical properties and secondary structure changes of recombinant protein were analyzed using Antheprot software and circular dichroism, respectively. The demonstrated selectivity and cytotoxicity of the fusion proteins against human cancer cells support their use in the treatment of tumors.
Methods
Cell lines and cell culture
The human ovarian carcinoma cells SK-OV-3, human breast cancer cells MCF-7, and human embryonic kidney cells HEK-293 were purchased from cell bank of Chinese Academy of Science (Shanghai, China). MCF-7 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS; Gibco) in incubator with 5% CO2 at 37 °C. Other cell lines were cultured in RPMI 1640 medium.
Construction of expression plasmids
The genes encoding TRAIL (GenBank No. OG143333) and SAC (GenBank No. AB108448) were stored in our laboratory. In order to obtain the soluble recombinant protein in E. coli system, the fusion protein was attached to the C-terminal of SUMO fusion tag. Overlapping PCR was used to amplify the fusion genes. Target genes were inserted into pET28a(+) vector between the Nco I and Xho I endonuclease sites, respectively. All primers used in this study are listed in Table S1 and Table S2.
Optimization of the linker between SAC and TRAIL
To determine the effect of different connection orders on the activity of the recombinant fusion protein, SAC-TRAIL and TRAIL-SAC were constructed with a flexible linker (G4S)2. To further improve the biological activity of the recombinant protein, the linker of the fusion protein was optimized based on the flexibility and length. The different linkers were set as follows: flexible linker A: (GGGGS)2, B: (GGGGS)3, and C: (GGGGS)4; rigid linker D: PKPSTPPGGS and E: (PKPSTPPGGS)2; semi-rigid linker F: GGGSPKPSTPPGGGS and G: GGGS(PKPSTPP)2GGGS.
Expression and purification of fusion proteins
The recombinant strains were incubated in 200 mL Luria–Bertani (LB) medium containing kanamycin (50 mg/mL) at 37 °C until OD600 was reached 0.6–08; then, 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce the expression of recombinant protein at 18 °C for 16 h. The cell culture was harvested through centrifugation and washed with PBS buffer (pH 7.4), then resuspended with PBS and disrupted by ultrasonication. Soluble expression level of target protein was analyzed by 12% SDS-PAGE under reduction condition.
To purify the recombinant fusion protein, the supernatant was loaded onto the Ni–NTA Sepharose column. The target protein with His6-tag was eluted with PBS containing different concentrations of imidazole. In order to remove the imidazole and NaCl, the eluent was dialyzed twice at 4 °C in PB buffer (pH 7.4). Then, fusion protein was digested with SUMO protease at a ratio of 1000:1 (w/w) for 1 h at 37 °C to get rid of SUMO protein, and the target protein was purified via CM Sepharose FF. The purity of recombinant protein was analyzed by ImageJ software and protein concentration was measured by Bradford assay.
Structural characterization
Antheprot software was applied to analyze the physicochemical properties of recombinant protein with different linker. The linker sequence mainly affects the C-terminal of SAC and the N-terminal of TRAIL domain; thus, the hydrophobicity, antigenicity, and solvent accessibility of this local domain were analyzed.
Circular dichroism (CD) was used to detect the secondary structure of recombinant protein. The purified recombinant proteins were dialyzed in 20 mM PB buffer (pH 7.4) and then diluted to the same concentration. CD measurement wavelength was between 190 and 260 nm. The results were analyzed by CDNN and Chirascan software.
Cell cytotoxicity assay
CCK-8 assay was performed to detect the cytotoxicity of fusion protein on cancer cells SK-OV-3 and MCF-7 at 96-well plate. 104 cells were added to each well and incubated at 37 °C for 24 h. Cells were treated with different concentrations of recombinant protein. After incubation for 72 h, 10 μL of CCK-8 was added to each well and incubated at 37 °C for 2 h. Finally, the absorbance was measured at 450 nm by microplate reader.
Flow cytometry analysis
Flow cytometry analysis was performed to test the apoptosis-inducing potency of recombinant protein on tumor cells. SK-OV-3, MCF-7, and HEK-293 cells were seeded into six-well plates. After 24 h, fresh medium containing purified target proteins at different concentrations were added to each well. After incubating for 2 days, the cells were washed twice with ice-cooled PBS buffer and collected by centrifugation. Cells were resuspended with 100 μL binding buffer. Then, 5 μL of FITC-labeled Annexin-V and 5 μL of propidium iodide staining solution were added sequentially and incubated for 10 min in the dark. Finally, 300 μL of PBS buffer was added. Cell apoptosis was detected by flow cytometry and analyzed by Flow Jo software.
Immunofluorescence assay
Immunofluorescence assay was used to detect the localization of fusion protein in different cells. The purified protein was dialyzed against bicarbonate buffer (pH 9.0) for three times at 4 °C, and then FITC was added to cross-link with fusion protein (protein: FITC = 1 mg: 15 μg). Finally, NH4Cl was used to terminate the reaction with a final concentration of 50 mM. Cells were seeded into confocal dishes at a density of 1 × 105/dish. The FITC-labeled fusion protein was added to a final concentration of 10 μg/mL. After 48 h, the nucleus was stained with Hoechst 33342 (10 μg/mL) at 37 °C for 10 min and washed with PBS for three times. The samples were observed by laser scanning confocal microscopy.
Statistical analysis
All quantitative data were measured by at least three independent experiments. The experimental data were statistically processed by GraphPad Prism 9.0 software, and the data results were presented as mean ± standard error (mean ± SE). The data was considered to be statistically significant when P < 0.05.
Results
Construction and expression of recombinant proteins
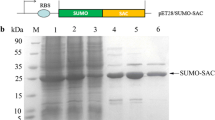
TRAIL was expressed as inclusion body in the E. coli expression system, and a small ubiquitin-related modifier (SUMO) tag was used to enhance the soluble expression of the target protein. To determine the effect of different connection orders on the activity of the recombinant protein, fusion proteins TRAIL-SAC and SAC-TRAIL with a flexible linker (G4S)2 were designed. A schematic diagram of the recombinant protein construction is shown in Fig. 1a. IPTG (0.1 mM) was used to induce the expression of recombinant protein at 18 °C. SDS-PAGE was performed to verify the expression level and solubility of the recombinant protein. The results showed that the recombinant proteins SUMO-TRAIL, SUMO-TRAIL-SAC, and SUMO-SAC-TRAIL were mostly solubilized in E. coli BL21 (DE3) (Fig. 1b, c, and d). Analysis using ImageJ software quantified the soluble expressed SUMO-TRAIL, SUMO-TRAIL-SAC, and SUMO-SAC-TRAIL as accounting for 30.1%, 37.0%, and 34.8% of the total soluble protein, respectively.
The expression of fusion proteins. a Schematic of expression vectors. pET28a(+) was used to construct the expression vectors. b The expression of SUMO-TRAIL. M, protein molecular weight marker. Lanes 1–3, cell lysate, soluble fraction, and insoluble fraction from Escherichia coli harboring pET28a; lanes 4–6, cell lysate, soluble fraction, and insoluble fraction from Escherichia coli harboring pET28a/SUMO-TRAIL. c The expression of SUMO-TRAIL-SAC. Lanes 1–3, cell lysate, soluble fraction, and insoluble fraction from Escherichia coli harboring pET28a/SUMO-TRAIL-SAC. d The expression of SUMO-SAC-TRAIL. Lanes 1–3, cell lysate, soluble fraction, and insoluble fraction from Escherichia coli harboring pET28a/SUMO-SAC-TRAIL
Preparation of recombinant proteins
Nickel column affinity chromatography was used for purification of the SUMO-TRAIL-SAC and SUMO-SAC-TRAIL due to the His6-tag that located at the N-terminal of SUMO protein. The fusion proteins were efficiently eluted by 500 mM imidazole (Fig. 2a and c). After dialyzed against PBS to remove imidazole, the sample was incubated with SUMO protease to remove the SUMO tag from the target protein, and the mixture was purified using a nickel column. However, the SUMO tag and target protein were both well bound to the nickel column, and the proteins were simultaneously eluted by imidazole. This may be due to the spatial proximity of several histidines on the TRAIL protein surface; thus, target proteins without the His6-tag are also able to bind to the nickel column (Hymowitz et al. 2000). A series of tests were performed to optimize the purification process, and the weak cation exchange chromatography was determined to be the most optimal method for removal of the SUMO tag from the cleavage mixture. The mixture was dialyzed with PB buffer (pH 7.4) to remove NaCl before purifying the recombinant TRAIL-SAC and SAC-TRAIL with a CM Sepharose Fast Flow column (Fig. 2b and d). TRAIL was also purified using a similar method (Fig. 2e and f). The purity of TRAIL, TRAIL-SAC, and SAC-TRAIL was 97.8%, 98.0%, and 94.3%, respectively.
The purification of recombinant proteins. a Purification of SUMO-TRAIL-SAC. Lanes 1–3, supernatant, precipitation, and flow-through fraction from E. coli harboring pET28a/SUMO-TRAIL-SAC; lanes 4–7, elutes from Ni–NTA by 50, 100, 200, and 500 mM imidazole. b Purification of TRAIL-SAC. Lane 1, SUMO-TRAIL-SAC cleaved by SUMO protease; lane 2, flow-through fraction from CM Sepharose column; lanes 3–4, elutes by 200 and 500 mM NaCl. c Purification of SUMO-SAC-TRAIL. Lanes 1–4, cell lysate, supernatant, precipitation, and flow-through fraction from E. coli harboring pET28a/SUMO-SAC-TRAIL; lanes 5–8, elutes from Ni–NTA by 50, 100, 200, and 500 mM imidazole. d Purification of SAC-TRAIL. Lane 1, SUMO-SAC-TRAIL cleaved by SUMO protease; lane 2, flow-through fraction from CM Sepharose column; lanes 3–4, elutes by 200 and 500 mM NaCl. e Purification of SUMO-TRAIL. Lanes 1–4, cell lysate, supernatant, precipitation, and flow-through fraction from E. coli harboring pET28a/SUMO-TRAIL; lanes 5–8, elutes from Ni–NTA by 50, 100, 200, and 500 mM imidazole. f Purification of TRAIL. Lane 1, SUMO-TRAIL cleaved by SUMO protease; lane 2, flow-through fraction from CM Sepharose column; lanes 3–4, elutes by 200 and 500 mM NaCl
In vitro cytotoxic activity of recombinant proteins
Effects of recombinant proteins on the viability of various cells were detected using CCK-8 assay. As shown in Fig. 3, dose-dependent cytotoxicity of TRAIL-SAC was observed against the human ovarian cancer cells SK-OV-3 and the breast cancer cells MCF-7. The IC50 values of TRAIL-SAC and TRAIL against SK-OV-3 cells were 0.67 nM and 3.44 μM, respectively (Fig. 3a and c). The IC50 values of TRAIL-SAC and TRAIL to MCF-7 cells were 61.98 nM and 2.86 μM, respectively (Fig. 3b and c). Overall, antitumor activity of TRAIL was remarkably enhanced after fusion with SAC, and SK-OV-3 cells were more susceptible to TRAIL-SAC than MCF-7 cells. Differences in cancer cell line susceptibility to TRAIL and TRAIL-SAC were attributable to variations in cancer cell receptor compositions (Sanlioglu et al. 2005). In addition, recombinant TRAIL and TRAIL-SAC showed almost no damage to normal cells HEK-293 (Fig. 3d). These results indicated the specificity of TRAIL-SAC toward cancer cells.
Cytotoxicity of recombinant protein to different cells. Proliferation inhibition effects of TRAIL-SAC on SK-OV-3 cells (a) and MCF-7 cells (b) were measured by CCK-8 analysis after incubation with different concentrations of recombinant protein for 72 h. c Proliferation inhibition effect of TRAIL on SK-OV-3 and MCF-7 cells. d Cytotoxicity of TRAIL and TRAIL-SAC on HEK-293 cells. Proliferation inhibition effect of SAC-TRAIL on SK-OV-3 cells (e) and MCF-7 cells (f). Data are presented as mean ± SD (n = 3)
The results of the proliferation inhibition assay showed that the IC50 values of SAC-TRAIL for SK-OV-3 and MCF-7 cells were 0.46 nM and 44.33 nM, respectively, which was markedly improved compared with TRAIL-SAC (Fig. 3e and f). This was attributable to the smaller spatial site hindrance between the two domains of SAC-TRAIL, which allows for better targeting ability. In addition, SAC-TRAIL caused almost no toxicity to HEK-293 cells (Fig. S1). SAC-TRAIL, which has the stronger inhibitory effect on tumor cells, was selected for utilization in subsequent experiments.
Optimization of the linker between SAC and TRAIL
Previous studies have shown that the character of the linker between fusion proteins has a significant influence on the soluble expression level and bioactivity of fusion protein (Lebendiker and Danieli 2014). The amino acid composition, length, and flexibility of the linker are important factors influencing the physical and chemical properties of connected domains (Yu et al. 2015). To obtain a highly active fusion protein, linker sequences with different properties were used to integrate SAC with TRAIL. Proteins fused with various linkers were purified using Ni–NTA column and weak cation exchange resin (Fig. 4a and b). The SDS-PAGE results indicated that the differences in apparent molecular weights of the fusion proteins, attributable in part to different linkers, affect the secondary structure of the recombinant fusion protein. The properties of SAC-TRAIL with different linkers are generalized in Table 1. In addition, the protein mass was calculated and summarized for each preparative step. The final yield of SAC-(G4S)3-TRAIL was 4.01% (Table S3).
Purification of SAC-TRAIL with different linkers and their cytotoxicity to cancer cells. a Purification of SUMO-tagged fusion protein by Ni–NTA. b Purification of fusion protein by CM Sepharose column. c Cytotoxicity of SAC-TRAIL with different linkers (10 ng/mL) on SK-OV-3 cells. d Cytotoxicity of SAC-TRAIL with different linkers (1 μg/mL) on MCF-7 cells. The different linkers were set as follows: flexible linker A: (GGGGS)2, B: (GGGGS)3, and C: (GGGGS)4; rigid linker D: PKPSTPPGGS and E: (PKPSTPPGGS)2; semi-rigid linker F: GGGSPKPSTPPGGGS and G: GGGS(PKPSTPP)2GGGS. *P < 0.05; **P < 0.01, compared with the fusion protein with linker (G4S)2
Co-incubating 10 ng/mL recombinant proteins with SK-OV-3 cells inhibited the proliferation of tumor cells to varying degrees, and optimal antitumor activity was demonstrated by proteins fused with the flexible linker (G4S)3 (Fig. 4c). Similar results were found by co-incubating 1 μg/mL of recombinant fusion protein with MCF-7 cells (Fig. 4d). SAC-TRAIL with different linkers caused almost no toxicity to HEK-293 cells even at the concentration of 1 μg/mL (Fig. S2).
Linker analysis based on physicochemical properties
Antheport software was used to analyze the differences in physical and chemical properties of fusion proteins with different linker sequences. As the linker sequence primarily affects the C-terminal domain of SAC and the N-terminal domain of TRAIL, the study focused on examining the physicochemical properties of this partial domain. Flexible linkers were found to have higher hydrophobicity and lower solvent accessibility (Fig. S3a and S3b). The optimal antitumor activity of the flexible linker (G4S)3 may be attributable to its higher hydrophobicity, which brings the active centers of SAC and TRAIL relatively closer to facilitate binding of the recombinant fusion protein to receptors on cancer cell surfaces. In contrast, the recombinant protein with rigid linkage (PKPSTPPGGS) showed the least favorable antitumor activity against both tumor cell types, probably due to its high solvent accessibility, thereby increasing the spatial distance between SAC and TRAIL. There were no significant differences in the antigenicity of each recombinant protein (Fig. S3c).
Circular dichroism was used to observe the effect of different linkers on the secondary structure of the recombinant fusion protein. Recombinant proteins with different linkers have typical negative absorption peaks at 222 nm and 208 nm (Fig. 5), indicating that the secondary structure of recombinant protein SAC-TRAIL is mainly α-helical. CDNN analysis showed that the fusion protein with a flexible linker (G4S)3 contained relatively low proportions of α-helix (55.3%) and high random coiling (20.2%) (Table S4). The comparatively higher antitumor activity of fusion protein with a flexible linker (G4S)3 is likely caused by their possession of fewer α-helices, which reduced the rigidity and enhanced the receptor binding specificity of the recombinant protein.
SAC-(G 4 S) 3 -TRAIL induced apoptosis of cancer cells
Flow cytometry was used to detect the apoptosis-inducing effect of recombinant fusion protein. The apoptosis rate of SK-OV-3 cells treated with 20 ng/mL SAC-(G4S)3-TRAIL was 42.71%, whereas 20 ng/mL TRAIL had almost no effect on SK-OV-3 cells (Fig. 6a and Fig. S4a). Further, 0.25 μg/mL TRAIL caused 25.98% apoptosis in the MCF-7 cells. At the same concentration, SAC-(G4S)3-TRAIL induced apoptosis in 97.8% of MCF-7 cells (Fig. 6b and Fig. S4b). Neither TRAIL nor SAC-(G4S)3-TRAIL induced apoptosis in normal cells HEK-293 at a concentration of 0.25 μg/mL (Fig. S5). Thus, SAC-(G4S)3-TRAIL strongly induces apoptosis in cancer cells while causing almost no damage to normal cells.
Flow cytometry and immunofluorescence analyses of cells treated with recombinant protein. SK-OV-3 cells (a) and MCF-7 cells (b) were incubated with TRAIL and SAC-TRAIL for 48 h and then stained by Annexin-V/PI. c Targeting selectivity of SAC-TRAIL on different cells. FITC-labeled TRAIL or SAC-TRAIL was incubated with SK-OV-3, MCF-7, and HEK-293 cells for 48 h. Cell nuclei were stained with Hoechst 33342 for 20 min. The confocal images were captured at × 1000 magnification
Specific selectivity of SAC-(G 4 S) 3 -TRAIL toward cancer cells
Immunofluorescence analysis was performed to observe the targeted binding ability of SAC-(G4S)3-TRAIL to tumor cells (Fig. 6c). The confocal microscopy results showed that SAC-(G4S)3-TRAIL could specifically bind with SK-OV-3 cells after incubation for 48 h, and relatively fewer fluorescence was observed in MCF-7 cells. This is consistent with the results of the CCK-8 assay showing that the ovarian cancer cells SK-OV-3 are more susceptible than the breast cancer cells MCF-7 to SAC-(G4S)3-TRAIL. Notably, FITC-labeled TRAIL was rarely detected on the SK-OV-3 and MCF-7 cells. In addition, neither protein was observed in HEK-293 cells. In summary, our results suggest that SAC-(G4S)3-TRAIL binds specifically to cancer cells SK-OV-3 and MCF-7.
Discussion
Recent studies have demonstrated that TRAIL is a broad-spectrum anticancer agent with substantial clinical benefits (Cousin et al. 2016). Although TRAIL can selectively induce apoptosis of tumor cells, it has a short half-life and certain tumor cells exhibit drug resistance (Han et al. 2016). Previous studies have confirmed that the sensitivity of cancer cells to TRAIL is increased by the overexpression of pro-apoptotic factors or downregulation of the expression of anti-apoptotic factors (Chu et al. 2006; Hassanzadeh et al. 2018).
Sequence deletion analysis of Par-4 revealed that its core structural domain, SAC, retained its entire anticancer activity (El-Guendy et al. 2003). SAC has wide inhibitory effects on the survival of various cancer cells while exerting no obvious effect on normal cells (Jagtap et al. 2014; Lee et al. 2010; Zhang et al. 2018). Overexpression of the gene encoding Par-4 can enhance the sensitivity of tumor lymphocytes to TRAIL treatment (Boehrer et al. 2006). In this study, we fusion expressed TRAIL with SAC to improve the anticancer effects of TRAIL against cancer cells SK-OV-3 and MCF-7.
Although approximately one-third of the recombinant protein was expressed by the E. coli expression system, most of the heterologous proteins produced in E. coli were insoluble (Ki and Pack 2020; Sanchez-Garcia et al. 2016). In the initial study, TRAIL and SAC were fused in different orders by flexible linker (G4S)2, but most of the recombinant proteins were presented as inclusion bodies. Soluble expression of recombinant proteins could not be achieved by reducing the induction temperature and inducer concentration. Recent studies have shown that SUMO tag can facilitate correct folding of proteins while improving protein expression levels. After the fusion protein is digested by SUMO protease, a native protein without any amino acid residue redundancies may be obtained (Jiao et al. 2018; Lin et al. 2017; Zhang et al. 2018) . Thus, the SUMO tag was fused to the N-terminus of TRAIL-SAC to enhance the soluble expression level of fusion protein.
Ovarian cancer, the most lethal malignant gynecologic tumor, has a reported sensitivity to TRAIL (Abdollahi 2004; Zhao et al. 1995). However, breast cancer cells are less susceptible to TRAIL-induced cell death (Sanlioglu et al. 2005). CCK-8 analysis demonstrated that TRAIL showed a stronger inhibitory effect on human ovarian cancer cells SK-OV-3 and human breast cancer cells MCF-7 when fused with the SAC domain, whereas it had no obvious cytotoxicity to normal cells HEK-293. The IC50 values of TRAIL and TRAIL-SAC to SK-OV-3 cells were 3.44 μM and 0.67 nM, respectively.
It has also been reported that Par-4 or SAC can specifically bind to GRP78 on the tumor cell surface to selectively induce apoptosis of cancer cells by promoting endoplasmic reticulum stress-mediated signaling pathways (Shrestha-Bhattarai and Rangnekar 2010). The fusion of TRAIL with SAC remarkably enhanced the targeting ability of the recombinant protein. In addition, the cytotoxic effects of the recombinant fusion protein against SK-OV-3 cells were significantly higher than MCF-7 cells. This is probably due to the high expression of TRAIL-R4 on the surface of MCF-7 cells, which results in reduced sensitivity of MCF-7 cells to TRAIL fusion protein (Sanlioglu et al. 2005). The IC50 values of SAC-TRAIL against SK-OV-3 and MCF-7 cells were 0.46 nM and 44.33 nM, respectively. This indicated that different orders of fusion units obviously affect the antitumor activity of the fusion protein. These differences are likely due to the smaller spatial site hindrance of the independent domains in SAC-TRAIL, which allows for more effective exertion of their respective functions.
Previous studies have indicated that the length and amino acid sequence of the linker are important factors affecting the biological activity of fusion proteins (Yu et al. 2015). The antitumor activity of the recombinant protein SAC-TRAIL with different linkers was evaluated and the fusion proteins with a flexible linker (G4S)3 were found to have a relatively higher proliferation inhibition effect on tumor cells. As fusion proteins with different linkers have various antitumor activities, we analyzed the physicochemical properties of each protein using Antheprot software. This analysis showed that the flexible linker has low solvent accessibility and high hydrophobicity. The optimal antitumor activity of fusion protein with linker (G4S)3 may, therefore, be caused by its high hydrophobicity, which brings the SAC and TRAIL active centers spatially closer together to facilitate binding of recombinant proteins to receptors on the tumor cell surface.
Circular dichroism results also indicated that different properties of the linker could affect the composition of the secondary structure of the recombinant protein. CDNN analysis demonstrated that the flexible linker (G4S)3 had a lower proportion of α-helix (55.3%) and higher random coiling (20.2%). The higher random coiling conferred greater specificity of biological activity while fewer α-helices reduced the rigidity of the recombinant protein, thus facilitating the specific binding to respective receptors.
Flow cytometry assays showed that SAC-(G4S)3-TRAIL induced the apoptosis of SK-OV-3 and MCF-7 cells. In comparison to TRAIL, SAC-(G4S)3-TRAIL demonstrated a remarkably enhanced apoptosis-inducing ability. Immunofluorescence assay revealed that the amount of recombinant protein SAC-(G4S)3-TRAIL bound to the surface of SK-OV-3 cells was significantly greater in comparison to MCF-7 cells at the same concentration. Most importantly, no fluorescence was observed on the surface of normal cells HEK-293. Overall, TRAIL exhibited stronger targeted cytotoxicity to cancer cells after fusion with SAC.
In conclusion, the novel fusion protein SAC-(G4S)3-TRAIL exhibits superior anticancer effects toward tumor cells without harming normal cells. Optimized strategies for fusion expression and linker selection will provide a useful platform for the treatment of drug-resistant cancers. These results indicate that SAC-(G4S)3-TRAIL may be a promising candidate for treating GRP78-overexpressed tumors.
Data availability
The authors can confirm that all relevant data are included in the article and supplementary information files.
References
Abdollahi T (2004) Potential for TRAIL as a therapeutic agent in ovarian cancer. Vitam Horm 67:347–364
Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T (2001) Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng 14:529–532
Baeshen MN, Al-Hejin AM, Bora RS, Ahmed MM, Ramadan HA, Saini KS, Baeshen NA, Redwan EM (2015) Production of biopharmaceuticals in E. coli: current scenario and future perspectives. J Microbiol Biotechnol 25:953–962
Boehrer S, Nowak D, Puccetti E, Ruthardt M, Sattler N, Trepohl B, Schneider B, Hoelzer D, Mitrou PS, Chow KU (2006) Prostate-apoptosis-response-gene-4 increases sensitivity to TRAIL-induced apoptosis. Leuk Res 30:597–605
Chaudhry P, Singh M, Parent S, Asselin E (2012) Prostate apoptosis response 4 (Par-4), a novel substrate of caspase-3 during apoptosis activation. Mol Cell Biol 32:826–839
Chu L, Gu J, He Z, Xiao T, Liu X (2006) Adenoviral vector expressing CYLD augments antitumor activity of TRAIL by suppression of NF-kappaB survival signaling in hepatocellular carcinoma. Cancer Biol Ther 5:615–622
Cousin FJ, Jouan-Lanhouet S, Theret N, Brenner C, Jouan E, Le Moigne-Muller G, Dimanche-Boitrel MT, Jan G (2016) The probiotic Propionibacterium freudenreichii as a new adjuvant for TRAIL-based therapy in colorectal cancer. Oncotarget 7:7161–7178
Dubuisson A, Micheau O (2017) Antibodies and derivatives targeting DR4 and DR5 for cancer therapy. Antibodies 6:16
El-Guendy N, Zhao Y, Gurumurthy S, Burikhanov R, Rangnekar VM (2003) Identification of a unique core domain of par-4 sufficient for selective apoptosis induction in cancer cells. Mol Cell Biol 23:5516–5525
Han MA, Min KJ, Woo SM, Seo BR, Kwon TK (2016) Eupafolin enhances TRAIL-mediated apoptosis through cathepsin S-induced down-regulation of Mcl-1 expression and AMPK-mediated Bim up-regulation in renal carcinoma Caki cells. Oncotarget 7:65707–65720
Hassanzadeh A, Hagh MF, Alivand MR, Akbari AAM, Asenjan KS, Saraei R, Solali S (2018) Down-regulation of intracellular anti-apoptotic proteins, particularly c-FLIP by therapeutic agents; the novel view to overcome resistance to TRAIL. J Ce Physiol 233:6470–6485
Hebbar N, Wang C, Rangnekar VM (2012) Mechanisms of apoptosis by the tumor suppressor Par-4. J Cell Physiol 227:3715–3721
Huang Z, Zhang C, Chen S, Ye F, Xing XH (2013) Active inclusion bodies of acid phosphatase PhoC: aggregation induced by GFP fusion and activities modulated by linker flexibility. Microb Cell Fact 12:25
Hymowitz SG, O’Connell MP, Ultsch MH, Hurst A, Totpal K, Ashkenazi A, de Vos AM, Kelley RF (2000) A unique zinc-binding site revealed by a high-resolution X-ray structure of homotrimeric Apo2L/TRAIL. Biochemistry 39:633–640
Idriss HT, Naismith JH (2000) TNF alpha and the TNF receptor superfamily: structure-function relationship(s). Microsc Res Tech 50:184–195
Jagtap JC, Dawood P, Shah RD, Chandrika G, Natesh K, Shiras A, Hegde AS, Ranade D, Shastry P (2014) Expression and regulation of prostate apoptosis response-4 (Par-4) in human glioma stem cells in drug-induced apoptosis. PLoS One 9:e88505
Jiao P, Zhang J, Dong Y, Wei D, Ren Y (2018) Construction and characterization of the recombinant immunotoxin RTA-4D5-KDEL targeting HER2/neu-positive cancer cells and locating the endoplasmic reticulum. Appl Microbiol Biotechnol 102:9585–9594
Johnstone RW, Frew AJ, Smyth MJ (2008) The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer 8:782–798
Ki MR, Pack SP (2020) Fusion tags to enhance heterologous protein expression. Appl Microbiol Biotechnol 104:2411–2425
Lebendiker M, Danieli T (2014) Production of prone-to-aggregate proteins. FEBS Lett 588:236–246
Lee TJ, Jang JH, Noh HJ, Park EJ, Choi KS, Kwon TK (2010) Overexpression of Par-4 sensitizes TRAIL-induced apoptosis via inactivation of NF-kappaB and Akt signaling pathways in renal cancer cells. J Cell Biochem 109:885–895
Lin CH, Pan YC, Liu FW, Chen CY (2017) Prokaryotic expression and action mechanism of antimicrobial LsGRP1C recombinant protein containing a fusion partner of small ubiquitin-like modifier. Appl Microbiol Biotechnol 101:8129–8138
Meynier S, Kramer M, Ribaux P, Tille JC, Delie F, Petignat P, Cohen M (2015) Role of PAR-4 in ovarian cancer. Oncotarget 6:22641–22652
Mun EJ, Babiker HM, Weinberg U, Kirson ED, Von Hoff DD (2018) Tumor-treating fields: a fourth modality in cancer treatment. Clin Cancer Res 24:266–275
Rozanov D, Spellman P, Savinov A, Strongin A (2015) A humanized leucine zipper-TRAIL hybrid induces apoptosis of tumors both in vitro and in vivo. PLoS One 10:e0122980
Sachdev D, Chirgwin JM (1998) Order of fusions between bacterial and mammalian proteins can determine solubility in Escherichia coli. Biochem Biophys Res Commun 244:933–937
Sanchez-Garcia L, Martin L, Mangues R, Ferrer-Miralles N, Vazquez E, Villaverde A (2016) Recombinant pharmaceuticals from microbial cells: a 2015 update. Microb Cell Fact 15:33
Sanlioglu AD, Dirice E, Aydin C, Erin N, Koksoy S, Sanlioglu S (2005) Surface TRAIL decoy receptor-4 expression is correlated with TRAIL resistance in MCF7 breast cancer cells. BMC Cancer 5:54
Sells SF, Wood DP Jr, Joshi-Barve SS, Muthukumar S, Jacob RJ, Crist SA, Humphreys S, Rangnekar VM (1994) Commonality of the gene programs induced by effectors of apoptosis in androgen-dependent and -independent prostate cells. Cell Growth Differ 5:457–466
Seo OW, Kim JH, Lee KS, Lee KS, Kim JH, Won MH, Ha KS, Kwon YG, Kim YH (2012) Kurarinone promotes TRAIL-induced apoptosis by inhibiting NF-kappa B-dependent cFLIP expression in HeLa cells. Exp Mol Med 44:653–664
Shrestha-Bhattarai T, Rangnekar VM (2010) Cancer-selective apoptotic effects of extracellular and intracellular Par-4. Oncogene 29:3873–3880
Snajdauf M, Havlova K, Vachtenheim J, Ozaniak A, Lischke R, Bartunkova J, Smrz D, Strizova Z (2021) The TRAIL in the treatment of human cancer: an update on clinical trials. Front Mol Biosci 8:628332
Sudhakar A (2009) History of cancer, ancient and modern treatment methods. J Cancer Sci Ther 1:1–4
Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA, Smith CA, Goodwin RG, Rauch CT (1997) TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J 16:5386–5397
Wang S, Lin H, Zhao T, Huang S, Fernig DG, Xu N, Wu F, Zhou M, Jiang C, Tian H (2017) Expression and purification of an FGF9 fusion protein in E. coli, and the effects of the FGF9 subfamily on human hepatocellular carcinoma cell proliferation and migration. Appl Microbiol Biotechnol 101:7823–7835
Wilding KM, Hunt JP, Wilkerson JW, Funk PJ, Swensen RL, Carver WC, Christian ML, Bundy BC (2019) Endotoxin-free E coli-Based cell-free protein synthesis: pre-expression endotoxin removal approaches for on-demand cancer therapeutic production. Biotechnol J 14:e1800271
Yu K, Liu C, Kim BG, Lee DY (2015) Synthetic fusion protein design and applications. Biotechnol Adv 33:155–164
Zhang J, Sun A, Dong Y, Wei D (2018) Recombinant production and characterization of SAC, the core domain of Par-4, by SUMO fusion system. Appl Biochem Biotechnol 184:1155–1167
Zhao E, Zhou M, Fu C, Shen B, Zhang Q, Lian L (1995) Effects of TNF alone or in combination with chemotherapeutic agents on human ovarian cancers in vitro and in nude mice. Chin Med J (engl) 108:571–575
Acknowledgements
We thank Yuting Li for providing language help.
Funding
This study was supported by the grants from the National Natural Science Foundation of China (No. 21706072/B060806).
Author information
Authors and Affiliations
Contributions
JZ conducted experiments and wrote the original draft. WD conceived and designed research. DW validated and analyzed data. YR contributed new methods. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, J., Dong, W., Ren, Y. et al. SAC-TRAIL, a novel anticancer fusion protein: expression, purification, and functional characterization. Appl Microbiol Biotechnol 106, 1511–1520 (2022). https://doi.org/10.1007/s00253-022-11807-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11807-3