Abstract
Fibroblast growth factor (FGF) 9 has oncogenic activity and plays an important role in the development of ovarian, lung, prostate, and gastric cancers. In the present study, with the aim of reducing the cost of utilizing growth factors in cancer research, a simple and efficient method for the preparation of recombinant human (rh)FGF9 in Escherichia coli was established. The rhFGF9 fusion protein (6 × His-TEV-rhFGF9) and the native protein released by tobacco etch virus (TEV) protease were obtained using a Ni-NTA system, with > 95% purity. Both purified forms of rhFGF9, with and without fusion tags, significantly stimulated the proliferation of NIH3T3 cells. The FGF9 subfamily, including FGF9, FGF16, and FGF20, in addition to rhFGF16, rhFGF9, and rhFGF20, were shown to stimulate the proliferation and migration of HuH7 human hepatocellular carcinoma (HCC) cells. Mechanistic studies revealed that the stimulation of HuH7 cell proliferation and migration with rhFGF9 and rhFGF20 were associated with the activation of the extracellular signal-regulated kinase (ERK) and nuclear factor κB (NF-κB) pathways and matrix metalloproteinase-26 (MMP26). Inhibition of the ERK and NF-κB pathways blocked cell migration, and NF-κB was demonstrated to be regulated by ERK. Therefore, the present study demonstrates a simple method for the preparation of biologically active rhFGF9 protein. Furthermore, the results indicate that exogenous rhFGF9- and rhFGF20-activated ERK/NF-κB signal transduction pathways play important roles in the regulation of HCC cell proliferation and migration, and this discovery helps to find the potential for new solutions of the treatment of liver cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is a common solid malignancy that is responsible for ~ 1 million mortalities each year (Di Bisceglie 2004). Typically, HCC is insensitive to chemotherapy and radiotherapy (Llovet et al. 2015) and, currently, the predominant method of clinical treatment is surgery (Bruix et al. 2005). However, most patients have a poor prognosis following surgery, and the incidence of cancer recurrence and metastasis remains high (Imamura et al. 2003). So far, studies have indicated that several signaling pathways, as well as the abnormal expression or activation of growth factors, are involved in the occurrence and development of HCC (Whittaker et al. 2010). Therefore, it is important to study the molecular mechanisms of HCC metastasis and explore novel therapeutic strategies to prevent it.
Fibroblast growth factors (FGFs) can initiate a series of biological responses, including regulating cell proliferation, migration, differentiation, and survival, by activating tyrosine kinase receptors (Ornitz and Itoh 2015). However, in various types of cancer tissues, aberrant FGF signaling can stimulate the excessive proliferation of cancer cells and promote their invasion and metastasis (Turner and Grose 2010). The FGF family consists of 22 members and is divided into seven subfamilies (Itoh and Ornitz 2008). FGF9, FGF16, and FGF20, which comprise one of the subfamilies, have high structural homology and similar receptor binding sites (Itoh and Ornitz 2008; Zhang et al. 2006), and thus are considered to share certain biological characteristics. In ovarian endometrioid adenocarcinomas, FGF9 and FGF20 have been found to be significantly overexpressed compared with normal ovarian tissues and are regulated by the classical Wnt signaling pathway (Chamorro et al. 2005; Hendrix et al. 2006). FGF16 is also closely associated with the development of human ovarian cancer (Basu et al. 2014). It has been shown that miR-140-5p can inhibit the proliferation, invasion, and metastasis of the human HCC cell line HCCLM3 by targeting transforming growth factor β receptor 1 (TGFβR1) and FGF9 (Yang et al. 2013). Concordantly, restoration of the expression of TGFβR1 and FGF9 can completely recover the biological functions of the hepatoma cells (Yang et al. 2013). However, the detailed mechanism underlying the role of FGF9 in HCC cell migration has not been established. Although the roles of FGF16 and FGF20 in the progression of HCC remain to be elucidated, FGF16 and FGF20 have been shown to be direct targets of the Wnt/β-catenin signaling pathway (Basu et al. 2014; Chamorro et al. 2005), which is closely related to the occurrence of HCC (Xu et al. 2012). Thus, in the present study, the specific roles of this FGF subfamily in the pathogenesis of HCC were investigated.
Matrix metalloproteinase (MMP)-26 is a newly identified member of the MMP family of matrix dissolution factors, which have important and unique functions in tumor invasion and angiogenesis (Uria and Lopez-Otin 2000). It has been established that MMP26 is closely related to the development and metastasis of various cancers, including those of the prostate (Riddick et al. 2005), breast (Zhao et al. 2004), esophageal (Ahokas et al. 2006), and lung (Marchenko et al. 2002). Recent reports have also revealed that HCC metastasis is associated with MMP26 expression (Wang et al. 2014; Yu et al. 2015).
Extracellular signal-regulated kinase (ERK) is an important member of the mitogen-activated protein kinase (MAPK) family, which regulates cell proliferation, differentiation, and survival (Roberts and Der 2007). ERK signaling may be stimulated by FGF, thus enabling it to mediate and amplify signals involved in tumor invasion and metastasis (Suyama et al. 2002). In addition, nuclear factor κB (NF-κB) is frequently activated in human hepatocellular malignant tumors (Luedde and Schwabe 2011). Therefore, whether these signaling proteins are regulated by the FGF9 subfamily in HCC, as well as the association between these signals, requires further study.
To study the role of the FGF9 subfamily in HCC, it is necessary to reduce the cost of obtaining the growth factor proteins for use in cancer research. The recombinant human (rh)FGF16 and rhFGF20 proteins have been successfully prepared in our laboratory previously. Although rhFGF9 has been successfully expressed in Escherichia coli and in Arabidopsis thaliana oil bodies, the preparation process of rhFGF9 was too complex (Koyama et al. 2001) and the expression cycle in plants was too long (Yi et al. 2016). Therefore, the present study used a 6 × His fusion strategy for the expression and purification of rhFGF9 protein. The target protein was released from the corresponding fusion protein by tobacco etch virus (TEV) protease, and high purity protein was obtained using Ni-NTA columns. Subsequent assays revealed strong biological activity of labeled and unlabeled rhFGF9 in NIH3T3. In addition, rhFGF9 and rhFGF20 could promote the proliferation and migration of HuH7 human HCC cells. Mechanistic studies indicated that rhFGF9 and rhFGF20 significantly stimulated ERK1/2 activity and could affect the protein levels of NF-κB and MMP26, thereby affecting cell proliferation and migration. In order to demonstrate the signal transduction cascade, specific inhibitors of ERK and NF-κB were applied to rhFGF9- and rhFGF20-stimulated HCC cells. The results revealed that the inhibition of ERK could affect the activation of NF-κB, whereas the expression of MMP26 was unaffected. Furthermore, the inhibition of NF-κB could affect the activation of MMP26. Therefore, we aimed to demonstrate the underlying mechanism of bioactive rhFGF9 and rhFGF20 on the proliferation and migration of HuH7 cells.
Materials and methods
Strains, plasmids, cell lines, and reagents
The E. coli strains DH5α and BL21(DE3)pLysS were purchased from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). The pETM-11-rhFGF9 plasmid was kindly provided by David G. Fernig (University of Liverpool, Liverpool, UK). DNA encoding hFGF9 (GenBank accession number NM_002010.2) was inserted into pETM-11 vector, which contains a 6 × His tag and a TEV cleavage site (ENLYFQ). Kanamycin, chloramphenicol, and isopropyl β-d-1-thiogalactopyranoside (IPTG) were purchased from Generay Biotech Co., Ltd. (Shanghai, China). TEV protease was purchased from Yeasen (Shanghai, China). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco (Thermo Fisher Scientific, Inc., Carlsbad, CA, USA). A bicinchoninic acid (BCA) kit and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Beyotime Institute of Biotechnology (Shanghai, China). Dimethyl sulfoxide (DMSO) and streptomycin were purchased from Sigma-Aldrich (St. Louis, MO, USA). HuH7 cells were kindly provided by Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China). NIH3T3 cells, rhFGF16, rhFGF20, and basic FGF (bFGF) proteins were stored in our laboratory. Primary antibodies against FGF9 and MMP26 were purchased from Abcam (Cambridge, MA, USA). The ERK inhibitor U0126 and primary antibodies against phospho-ERK1/2, ERK1/2, phospho-NF-κB p65, NF-κB p65, and β-actin were purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA). The NF-κB inhibitor BAY 11-7082 was purchased from Selleck Chemicals (Houston, TX, USA). Secondary antibodies and Western Blotting Luminol Reagent were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Expression and purification of fusion protein
The recombinant plasmid pETM-11-rhFGF9 was transformed into BL21(DE3)pLysS E. coli cells. After screening for strains with high-level plasmid expression, by selecting colonies that successfully grew on agar plates containing 50 μg/ml kanamycin and 34 μg/ml chloramphenicol, the recombinant E. coli cells were seeded with 1:100 (vol/vol) of 30 ml of lysogeny broth (LB) containing 50 μg/ml kanamycin and 34 μg/ml chloramphenicol and cultured at 37 °C with 180 rpm agitation overnight. The overnight medium was then inoculated into 350 ml fresh LB medium, and cells were incubated at 37 °C with 180 rpm agitation. When the OD600 reached 0.6–0.8, IPTG was added at a final concentration of 1 mM in order to induce protein expression. After incubation for another 4 h, cells were harvested by centrifugation at 9000 rpm for 10 min at 4 °C and the cell pellets were stored at − 80 °C.
The extracted frozen cells (10 g) were resuspended in 200 ml of lysate buffer (50 mM Tris buffer (pH 8.0) containing 0.3 M NaCl, 5% (vol/vol) glycerol and 1% (vol/vol) Triton X-100) and lysed on ice by sonication (repeated cycles of 5 s sonication followed by 5 s rest) for 15 min. After centrifugation at 48,000×g for 30 min at 4 °C, the supernatant was applied to a nickel chelating (Ni-NTA) column that had been treated with five column volumes of equilibration buffer (50 mM Tris, 0.3 M NaCl, pH 8.0). Bound fusion proteins (isoelectric point, 6.67) were eluted with 25, 50, 300, and 500 mM imidazole, respectively, and the eluted fraction was collected and analyzed by 12% (vol/vol) SDS-PAGE.
Target protein release and purification
Imidazole-eluted His-TEV-rhFGF9 was subjected to ultrafiltration via a microporous filter (3 kDa), and the BCA kit was used to measure the fusion protein concentration. Subsequently, TEV protease (5 U/μl) was added according to the restriction enzyme system (500 U TEV protease/1 mg protein) to release the target protein. The reaction solution was incubated at 4 °C with constant stirring overnight. The mixed product was then passed through a Ni-NTA column and the unbound portion, assumed to consist of rhFGF9 protein, was collected and analyzed by 12% (vol/vol) SDS-PAGE.
The purified His-TEV-rhFGF9 and rhFGF9 proteins were stored at − 80 °C for further use.
Cell proliferation analysis
E. coli-expressed rhFGF9 was used to stimulate NIH3T3 cells, and proliferation was measured by MTT assay. Standard basic FGF served as a positive control. PBS was used as negative control. In addition, the effects of rhFGF9, rhFGF16, and rhFGF20 on the proliferation of HuH7 hepatoma cells were analyzed. NIH3T3 and HuH7 cells were cultured in DMEM with low glucose (1.0 g/l) and high glucose (4.5 g/l), respectively, at 37 °C with 5% CO2. The complete medium contained 10% (vol/vol) FBS and 1% (vol/vol) streptomycin. NIH3T3 and HuH7 cells were seeded in 96-well plates at a density of 3000 cells/well for 24 h, then starved in DMEM containing 0.5% (vol/vol) FBS for 24 h. Subsequently, NIH3T3 cells were treated with various concentrations of His-TEV-hFGF9, rhFGF9, and bFGF, and HuH7 cells were treated with rhFGF9, rhFGF16, and rhFGF20 for 48 h prior to the addition of 20 μl 5 mg/ml MTT to each well followed by incubation for another 4 h. The original medium was removed by suctioning and 150 μl of DMSO solution was then added and shaken for 10 min. The absorbance of each well was measured at a wavelength of 490 nm on an enzyme-linked immunosorbent assay microplate reader (SpectraMax M2, Molecular Devices, Sunnyvale, CA, USA).
Scratch-healing experiment
HuH7 cells were inoculated in 6-well plates and incubated at 37 °C for 24 h until they had reached 100% confluence. Cell monolayers were scratched with a 10-μl pipette tip and rinsed twice with PBS solution. Subsequently, cells were co-cultured with various concentrations (0, 12.5, 25, and 50 ng/ml) of rhFGF9 or rhFGF20 for 48 h. In certain experiments, the cells were pretreated with 10 μM U0126 or 5 μM BAY 11-7082 for 1 h prior to wounding, and 50 ng/ml rhFGF9 or rhFGF20 were then added to each well and cultured for 48 h. Images of the wound areas were captured at 0 and 48 h under a microscope (IX70; Olympus, Tokyo, Japan), and migration distance was analyzed by Image J software (National Institutes of Health, Bethesda, MD, USA).
Western blotting
Total protein was extracted from cells with lysis buffer (Beyotime Institute of Biotechnology), and the protein concentration was determined BCA kit. Total protein (40 μg/lane) was separated by 12% (vol/vol) SDS-PAGE and transferred to 0.22-μm polyvinylidene fluoride membranes. The membranes were incubated in TBS containing 0.05% (vol/vol) Tween-20 (TBST) and 5% (wt/vol) skim milk for 1 h, and the following primary antibodies were applied overnight at 4 °C: anti-FGF9, anti-MMP26, anti-phospho-ERK1/2, anti-ERK1/2, anti-phospho-NF-κB p65, anti-NF-κB p65, and anti-β-actin. Following three washes in TBST, the membranes were incubated with donkey anti-mouse or goat anti-rabbit horseradish peroxidase-linked secondary antibodies at room temperature for 1 h. Following three further washes in TBST, the bands were detected with Western Blotting Luminol Reagent according to the manufacturer’s instructions. Quantification of relative expression of each targeting protein was performed by scanning densitometry using Image lab software (version 5.2).
Statistical analysis
Data processing was performed with Prism 5 software (GraphPad, San Diego, CA, USA). Individual groups were analyzed by a Student’s t test and one-way analysis of variance (ANOVA). P < 0.05 was considered to indicate statistical significance. All experimental data are expressed as the mean ± standard deviation.
Results
Expression and purification of rhFGF9 protein in E. coli
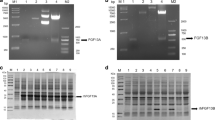
In the pETM-11-rhFGF9 plasmid, the target protein (FGF9) was fused to a 6 × His tag, which could be subsequently removed by TEV protease. The molecular weights of the fusion and native proteins were approximately 26.5 and 23.4 kDa, respectively (Fig. 1a). A high-expression E. coli strain was selected with IPTG at a final concentration of 1 mM and cultured for another 4 h. Following induction, expression of the target protein was verified by the presence of a 26.5 kDa band on 12% (vol/vol) SDS-PAGE (Fig. 1b). Western blotting revealed that the target protein was immunoreactive with rabbit anti-FGF9 antibody (Fig. 1c). The expressed product was observed in soluble form and the supernatant was subjected to a Ni-NTA column (Fig. 1d). After the 6 × His-TEV-rhFGF9 fusion protein bound to the column, the fusion protein was eluted with 300 mM imidazole. SDS-PAGE revealed that the proteins with weaker affinity for the Ni-NTA column could be eluted by 20 and 50 mM imidazole; the final purity of the fusion protein was > 95% (Fig. 1d). Furthermore, the target protein could be completely released by TEV protease (Fig. 1e). Subsequently, when the mixture was reapplied to the Ni-NTA column, the N-terminal 6 × His-tagged TEV enzyme and cleaved His tags bound to the Ni column, and the unbound native protein was present in the flow fraction (Fig. 1e).
Expression and purification of the fusion protein and target protein. a Schematic representation of expression vector pETM11-rhFGF9. b Expression of fusion protein induced by isopropyl β-d-1-thiogalactopyranoside in BL21(DE3) pLysS/pETM11-His-TEV-rhFGF9. Lane M, molecular-mass marker; lane 1, uninduced; lanes 2–5, after induction of the same clone for 1, 2, 3, or 4 h. c Expression of the His-TEV-rhFGF9 protein analyzed by western blotting. Lane 1, uninduced; lanes 2–5, after induction for 1, 2, 3, or 4 h. d SDS-PAGE analysis of the purification of His-TEV-rhFGF9. Lane 1, lysate supernatant; lane 2, flow-through fraction; lanes 3–5, eluted with 20, 50, or 300 mM imidazole, respectively. e Purification of rhFGF9 by removing the His tag with TEV protease. Lane 1, prior to digestion; lane 2, after digestion; lane 3, flow-through fraction
Mitogenic activity of fusion and native proteins on NIH3T3 cells
An MTT assay was use to detect the effect of purified rhFGF9 protein on the growth of NIH3T3 cells, and bFGF, which has strong mitogenic activity, was used as a positive control (Ornitz and Itoh 2015). Similar to bFGF, His-TEV-rhFGF9 and rhFGF9 showed strong, dose-dependent mitogenic activity in NIH3T3 fibroblasts (Fig. 2a). However, this effect was of a lower magnitude than that of bFGF, indicating that rhFGF9 is less potent than bFGF in NIH3T3 cells. In addition, the activity of the fusion protein was slightly lower than that of the target protein at concentrations of 16–64 ng/ml, but this difference was not significant. Therefore, the purified fusion protein and target protein obtained following TEV protease cleavage have strong mitogenic activity in NIH3T3 cells.
Mitogenic activity assay of purified rhFGF9 and bFGF on NIH3T3 cells and of the FGF9 subfamily on HuH7 cells. a NIH3T3 cells were cultured in 96-well plates for 24 h, starved in DMEM containing 0.5% (vol/vol) FBS for 24 h, and then treated with increasing concentrations (0, 0.5, 1, 2, 4, 8, 16, 32, 64, and 128 ng/ml) of bFGF, His-TEV-rhFGF9, or rhFGF9. PBS was used as a negative control. b HuH7 cells were seeded into 96-well plates for 24 h and starved for 24 h, then different concentrations (0, 3.125, 6.25, 12.5, 25, 50, and 100 ng/ml) of rhFGF9, rhFGF16, or rhFGF20 were added to each well for 48 h. PBS was used as negative control. Data are presented as the mean ± standard deviation (n = 3). *P < 0.05 vs. control
rhFGF9 and rhFGF20 promote the proliferation and migration of HuH7 cells
We further analyzed the effects of rhFGF9, rhFGF16, and rhFGF20 on the proliferation of HuH7 cells using an MTT assay. Cells were treated with rhFGF9, rhFGF16, and rhFGF20 at increasing concentrations (3.125, 6.25, 12.5, 25, 50, and 100 ng/ml) for 48 h. rhFGF9 and rhFGF20 each significantly stimulated the growth of HuH7 cells in a dose-dependent manner (P < 0.05) (Fig. 2b). However, there was no significant difference between the rhFGF16 and negative control groups. These results suggest that rhFGF9 and rhFGF20 serve important roles in the growth and survival of HCC cells. Subsequently, a wounding healing assay was used to detect the effects of rhFGF9 and rhFGF20 on the migration of HCC cells in vitro. The results showed that, following treatment with different concentrations (12.5, 25, and 50 ng/ml) of rhFGF9 or rhFGF20 for 48 h, the migration and healing ability of the cells was significantly higher than that of the control group (0 ng/ml) (P < 0.05) (Fig. 3a, b).
Effects of rhFGF9 and rhFGF20 on HuH7 cell migration. a A scratch-healing assay was performed following treatment of the cells with rhFGF9 (0, 12.5, 25, and 50 ng/ml) or rhFGF20 (0, 12.5, 25, and 50 ng/ml) for 48 h. Images of the wounded cell monolayers were captured after 48 h (white curve represents the wound borders). b Cell migration distance was measured according to the data shown in (a). c The levels of ERK, phospho-ERK, p65, phospho-p65, and MMP26 in the HuH7 cells after treatment with rhFGF9 (0, 12.5, 25, and 50 ng/ml) or rhFGF20 (0, 12.5, 25, and 50 ng/ml) for 1 h, which were assayed by western blotting. β-Actin was used as a loading control. d Quantification of the protein levels. Data are presented as the mean ± standard deviation (n = 3). *P < 0.05 vs. control
ERK, NF-κB signaling, and MMP26 activation are involved in the regulation of HuH7 cell proliferation and migration by rhFGF9 and rhFGF20
The aforementioned data suggest that rhFGF9 and rhFGF20 can significantly stimulate the proliferation of HuH7 cells. In subsequent experiments, HuH7 cells were incubated for 24 h in DMEM starvation medium, then stimulated with rhFGF9 or rhFGF20 (12.5, 25 and 50 ng/ml). After 1 h, ERK was quantitatively analyzed by western blotting. With increasing concentrations of rhFGF9 and rhFGF20, the level of ERK1/2 phosphorylation was significantly increased (P < 0.05) (Fig. 3c, d). A specific inhibitor of ERK, U0126 (10 μmol/l), was then used to inhibit the expression of ERK in HuH7 cells, thereby inhibiting the ERK/MAPK signal transduction cascade (Fig. 4a, b). The results indicated that U0126 could significantly attenuate the rhFGF9- or rhFGF20-induced increases in the proliferation and migration of HuH7 cells (P < 0.05) (Fig. 4c, d). These data suggest that ERK signaling is important in the proliferation and migration of rhFGF9- and rhFGF20-stimulated HuH7 cells.
Effects of NF-κB and ERK inhibitors on rhFGF9- and rhFGF20-induced HuH7 cell migration. a HuH7 cells were treated with various concentration of BAY 11-7082 (an NF-κB inhibitor; 0, 5, 10, and 20 μM) or U0126 (an ERK inhibitor; 0, 2.5, 5, and 10 μM) for 1 h. Cells were then collected and subjected to western blot analysis. b Quantification of the protein levels. c HuH7 cells were cultured on 96-well plates for 24 h and pretreated with U0126 (10 μM) for 1 h, then cultured with rhFGF9 (50 ng/ml) or rhFGF20 (50 ng/ml) for 48 h. Subsequently, MTT assay was used to access cell proliferation. d, e A wound-healing assay was performed following pretreatment of the cells with BAY 11-7082 (5 μM) or U0126 (10 μM) for 1 h and culture with rhFGF9 (50 ng/ml) or rhFGF20 (50 ng/ml) for 48 h. Cell migration distance was measured after 48 h (white curve represents the wound borders). Data are presented as the mean ± standard deviation (n = 3). *P < 0.05 vs. control; # P < 0.05 vs. FGF only
We then attempted to determine whether rhFGF9 and rhFGF20 could induce the expression of migration-related proteins in HCC cells, including MMP26 (Wang et al. 2014) and NF-κB (Li et al. 2011). The results showed that 25 ng/ml rhFGF9 and rhFGF20 induced a significant increase in MMP26 expression compared with the control group (P < 0.05) (Fig. 3c, d). In addition, the expression levels of NF-κB protein following rhFGF9 and rhFGF20 treatment were dose-dependent, showing the greatest increase at 50 ng/ml (Fig. 3c, d). When cells were pretreated with a selective inhibitor of NF-κB (5 μmol/l BAY 11-7082), the activation of NF-κB signal was successfully blocked (Fig. 4a, b). Following pretreatment of the cells with BAY 11-7082 for 1 h and incubation with 50 ng/ml rhFGF9 and rhFGF20 for 48 h, scratch-healing experiments showed significant inhibition of cell migration (P < 0.05) (Fig. 4d, e). Thus, the migration of rhFGF9- and rhFGF20-stimulated HuH7 cells was associated with the activation of MMP26 and NF-κB.
ERK/NF-κB signaling cascade is important in the expression of MMP26 induced by rhFGF9 and rhFGF20
To demonstrate whether ERK is involved in the production of NF-κB induced by rhFGF9 and rhFGF20, cells were pretreated with the ERK inhibitor U0126 for 1 h, followed by treatment with 50 ng/ml rhFGF9 or rhFGF20 for 1 h. The results showed that U0126 significantly attenuated the induction of NF-κB expression by rhFGF9 and rhFGF20 (P < 0.05), whereas MMP26 levels remained unaltered (Fig. 5a, b). Furthermore, to verify how rhFGF9- and rhFGF20-stimulated MMP26 expression was regulated by NF-κB, cells were pretreated with BAY 11-7082 for 1 h and then stimulated with 50 ng/ml rhFGF9 and rhFGF20. The results showed that the induction of MMP26 protein expression by FGF9 and FGF20 could be significantly inhibited by BAY 11-7082 (P < 0.05). However, phosphorylation of ERK1/2 could not be inhibited by BAY 11-7082 (Fig. 5c, d). Thus, NF-κB appears to be located downstream of the ERK signal. These results suggest that rhFGF9- and rhFGF20-stimulated ERK can affect NF-κB signaling via a cascade, and the activation of NF-κB can affect the expression of MMP26.
ERK and NF-κB were involved in rhFGF9- and rhFGF20-induced expression of MMP26. a, c HuH7 cells were pretreated with U0126 (10 μM) or BAY 11-7082 (5 μM) for 1 h, then cultured with rhFGF9 (50 ng/ml) or rhFGF20 (50 ng/ml) for 1 h. Western blot analysis was performed to examine the protein levels of ERK, phospho-ERK, p65, phospho-p65, and MMP26. b, d Quantification of the protein levels. Data are presented as the mean ± standard deviation (n = 3). *P < 0.05 vs. control; # P < 0.05 vs. FGF only
Discussion
The purification process of non-tagged proteins is complex, and it is therefore difficult to obtain a high level of purity. However, the expression of fusion proteins is able to provide affinity sites for protein purification to make the process simpler and more efficient. N-terminal His tagging is compatible with the transcriptional/translational mechanisms of bacteria and is favorable for protein expression. In addition, unlike the glutathione S-transferase (GST) tag, the His tag is less likely to form a dimer, and thus does not affect protein properties (Structural Genomics et al. 2008). In the present study, we selected the pETM11 vector containing a His tag and expressed the recombinant protein with IPTG at a final concentration of 1 mM. SDS-PAGE showed that the expression of recombinant FGF9 protein increased with time and reached a peak at ~ 4 h (Fig. 1b). A large quantity of soluble rhFGF9 protein was successfully expressed, and the N-terminal 6 × His tag of the fusion protein specifically bound to the Ni-NTA column. Lower concentrations of imidazole could better remove the weakly bound proteins, and the target protein could then be harvested at higher concentrations of imidazole (Fig. 1c). When the eluate was collected, dialysis was used to remove the imidazole. However, it was found that the rhFGF9 protein was prone to precipitate during dialysis. As imidazole has a strong buffering ability, especially at high concentrations, it is easy to alter the pH of the dialysis buffer, which may be the main cause of protein precipitation. In addition, when ultrafiltration technology was used to remove high concentrations of imidazole, the protein did not precipitate. A possible explanation for this effect could be that the experimental conditions required for ultrafiltration are milder (no phase change is required, in contrast with evaporation and freeze drying, and the temperature and pH remain unchanged), which can prevent the degeneration, inactivation or autolysis of biological macromolecules. Furthermore, the rhFGF9 protein has a strong affinity for heparin (Asada et al. 2009). Therefore, diluting the sample with a high concentration of imidazole prior to running through a heparin column may aid in purification. After the rhFGF9 protein binds to the heparin column, the buffer system can be replaced in order to remove the imidazole. Subsequent to this, TEV protease can specifically recognize the cleavage site (ENLYFQ) (Sun et al. 2015), and the fusion protein can be efficiently cleaved by TEV protease. Thus, binding of the His-tagged TEV protease and the soluble His tag to the Ni-NTA column can allow the effective separation of the native protein (Fig. 1d). In addition, limulus reagent was used to detect endotoxin produced by E. coli. Endotoxin levels from the purified protein were 25–50 EU/mg by gel semi-quantitative analysis. In the present study, however, protein samples used for cell experiments were diluted by at least 10,000 times with endotoxin-free media and endotoxin contents were almost negligible. Thus, the effect of residual endotoxin in cell experiments can be eliminated.
The biological activities of the purified fusion and non-fusion proteins were analyzed in vitro. Similar to bFGF, FGF9 was found to be capable of activating the FGF receptor (FGFR) tyrosine kinase, which activates a downstream signal to regulate DNA synthesis and proliferation in NIH3T3 cells (Ornitz and Itoh 2015; Yi et al. 2016). There was no significant difference in the mitogenic activities of the fusion protein and native protein (Fig. 2a), indicating that the His tag did not have a significant effect on the biological activity of the cells. However, the effect of rhFGF9 on NIH3T3 proliferation was significantly weaker than that of bFGF (Fig. 2b); this may be explained by differing ligand-receptor interaction specificities, which can affect FGF activity. The functional receptors in NIH3T3 cells are FGFR1 and FGFR2, which can be activated by bFGF (Li et al. 1994); however, FGF9 can specifically bind to FGFR2 and FGFR3 (Santos-Ocampo et al. 1996).
Aberrant FGF signaling can promote cancer development by influencing a series of downstream biological processes, and the incidence of cancer is often accompanied by excessive cell proliferation (Turner and Grose 2010). In the present study, the addition of exogenous rhFGF9 and rhFGF20 proteins could significantly stimulate the proliferation of HuH7 HCC cells (Fig. 2b). However, FGF16, as a member of the FGF9 subfamily, had no significant effect on the proliferation of HCC cells. Overexpression and gene amplification often produce excessive FGF signaling, leading to the occurrence and development of cancer (Ornitz and Itoh 2015). A previous study showed that FGF9 and FGF20 were expressed abnormally in HCC, whereas genetic alterations of FGF16 were not obvious (Tanner and Grose 2016). This also explains why FGF16 shows a weaker association with HCC, compared with the associations between HCC and FGF9 or FGF20.
In addition to increased cell proliferation, tumor cell migration is a key step in tumor infiltration and metastasis (Pathak and Kumar 2011). In the current study, wound-healing experiments showed that rhFGF9 and rhFGF20 could promote the migration of HuH7 cells (Fig. 3a). Previous studies have also shown that full-length recombinant FGF9 protein is capable of restoring HCCLM3 scratch healing following inhibition by miR-140-5p in vitro (Yang et al. 2013).
The Ras-MAPK/ERK signaling pathway plays a very important role in the promotion of tumorigenesis, invasion and metastasis (Roberts and Der 2007). The current results also demonstrated that rhFGF9 and rhFGF20 activated the activity of the MAPK/ERK signaling pathway and promoted the phosphorylation of ERK in HuH7 cells (Fig. 4a). NF-κB can be activated in human HCC, indicating that the factor plays an important role in processes related to cell malignancy (Luedde and Schwabe 2011). In addition, MMPs are closely associated with tumor invasion and metastasis (Pathak and Kumar 2011). The present study revealed that the addition of exogenous rhFGF9 and rhFGF20 could significantly increase the phosphorylation of NF-κB and the expression of MMP26 in HuH7 hepatoma cells (Fig. 4a). To investigate the association between ERK, NF-κB, and MMP26, specific inhibitors of ERK (U0126) and NF-κB (BAY 11-7082) were applied to the cells prior to FGF treatment. We found that U0126 suppressed the phosphorylation of NF-κB, while MMP26 levels were unaffected. Further experiments revealed that BAY 11-7082 could significantly inhibit the activation of NF-κB and MMP26, whereas the phosphorylation of ERK1/2 could not be attenuated, indicating that activation of NF-κB is regulated by ERK. This is consistent with the conclusion of a previous study, which showed that sorafenib enhanced the effect of suberoylanilide hydroxamic acid (an inhibitor of histone deacetylases) on HCC viability in HuH7 HCC cells by inducing exogenous and endogenous apoptosis via inhibition of the ERK/NF-κB pathway (Hsu et al. 2014).
In summary, the present study demonstrated the successful generation of a bioactive, soluble rhFGF9 fusion protein with > 95% purity using a simple and efficient purification strategy. The fusion protein could be cleaved by TEV protease to obtain the native protein. The purified fusion protein and non-fusion protein both showed strong biological activity. Furthermore, this study demonstrated that rhFGF9 and rhFGF20 serve important roles in the growth and migration of HuH7 cells. The induction of MMP26 expression by rhFGF9 and rhFGF20 is dependent on NF-κB rather than ERK, but NF-κB is directly regulated by ERK. Thus, the ERK/NF-κB transduction pathway is important in HCC cells, and this discovery laid the foundation for further study of liver cancer.
References
Ahokas K, Karjalainen-Lindsberg ML, Sihvo E, Isaka K, Salo J, Saarialho-Kere U (2006) Matrix metalloproteinases 21 and 26 are differentially expressed in esophageal squarnous cell cancer. Tumour Biol 27(3):133–141. https://doi.org/10.1159/000092774
Asada M, Shinomiya M, Suzuki M, Honda E, Sugimoto R, Ikekita M, Imamura T (2009) Glycosaminoglycan affinity of the complete fibroblast growth factor family. Biochim Biophys Acta 1790(1):40–48. https://doi.org/10.1016/j.bbagen.2008.09.001
Basu M, Mukhopadhyay S, Chatterjee U, Roy SS (2014) FGF16 Promotes invasive behavior of SKOV-3 ovarian cancer cells through activation of mitogen-activated protein kinase (MAPK) signaling pathway. J Biol Chem 289(3):1415–1428. https://doi.org/10.1074/jbc.M113.535427
Bruix J, Sherman M, Practice Guidelines Committee, American Association for the Study of Liver Diseases (2005) Management of hepatocellular carcinoma. Hepatology 42(5):1208–1236. https://doi.org/10.1002/hep.20933
Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE (2005) FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J 24(1):73–84. https://doi.org/10.1038/sj.emboj.7600460
Di Bisceglie AM (2004) Issues in screening and surveillance for hepatocellular carcinoma. Gastroenterology 127(5 Suppl 1):S104–S107
Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR (2006) Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res 66(3):1354–1362. https://doi.org/10.1158/0008-5472.CAN-05-3694
Hsu FT, Liu YC, Chiang IT, Liu RS, Wang HE, Lin WJ, Hwang JJ (2014) Sorafenib increases efficacy of vorinostat against human hepatocellular carcinoma through transduction inhibition of vorinostat-induced ERK/NF-kappa B signaling. Int J Oncol 45(1):177–188. https://doi.org/10.3892/ijo.2014.2423
Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, Sugawara Y, Minagawa M, Takayama T, Kawasaki S, Makuuchi M (2003) Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol 38(2):200–207
Itoh N, Ornitz DM (2008) Functional evolutionary history of the mouse Fgf gene family. Dev Dyn 237(1):18–27. https://doi.org/10.1002/dvdy.21388
Koyama N, Ohmae H, Tsuji S, Tanaka Y, Kurokawa T, Nishimura O (2001) Improved preparation and crystallization of 25 kDa human fibroblast growth factor-9. Biotechnol Appl Biochem 33(Pt 2):117–121
Li Y, Basilico C, Mansukhani A (1994) Cell transformation by fibroblast growth factors can be suppressed by truncated fibroblast growth factor receptors. Mol Cell Biol 14(11):7660–7669
Li J, Lau GK, Chen L, Dong SS, Lan HY, Huang XR, Li Y, Luk JM, Yuan YF, Guan XY (2011) Interleukin 17A promotes hepatocellular carcinoma metastasis via NF-kB induced matrix metalloproteinases 2 and 9 expression. PLoS One 6(7):e21816. https://doi.org/10.1371/journal.pone.0021816
Llovet JM, Villanueva A, Lachenmayer A, Finn RS (2015) Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 12(8):436. https://doi.org/10.1038/nrclinonc.2015.121
Luedde T, Schwabe RF (2011) NF-kappa B in the liver-linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 8(2):108–118. https://doi.org/10.1038/nrgastro.2010.213
Marchenko GN, Marchenko ND, Leng J, Strongin AY (2002) Promoter characterization of the novel human matrix metalloproteinase-26 gene: regulation by the T-cell factor-4 implies specific expression of the gene in cancer cells of epithelial origin. Biochem J 363(Pt2):253–262
Ornitz DM, Itoh N (2015) The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol 4(3):215–266. https://doi.org/10.1002/wdev.176
Pathak A, Kumar S (2011) Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol (Camb) 3(4):267–278. https://doi.org/10.1039/c0ib00095g
Riddick AC, Shukla CJ, Pennington CJ, Bass R, Nuttall RK, Hogan A, Sethia KK, Ellis V, Collins AT, Maitland NJ, Ball RY, Edwards DR (2005) Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. Br J Cancer 92(12):2171–2180. https://doi.org/10.1038/sj.bjc.6602630
Roberts PJ, Der CJ (2007) Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26(22):3291–3210. https://doi.org/10.1038/sj.onc.1210422
Santos-Ocampo S, Colvin JS, Chellaiah A, Ornitz DM (1996) Expression and biological activity of mouse fibroblast growth factor-9. J Biol Chem 271(3):1726–1731
Structural Genomics Consortium, China Structural Genomics Consortium, Northeast Structural Genomics Consortium, Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC (2008) Protein production and purification. Nat Methods 5(2):135–146. https://doi.org/10.1038/nmeth.f.202
Sun C, Li Y, Taylor SE, Mao XQ, Wilkinson MC, Fernig DG (2015) HaloTag is an effective expression and solubilisation fusion partner for a range of fibroblast growth factors. PeerJ 3:e1060. https://doi.org/10.7717/peerj.1060
Suyama K, Shapiro I, Guttman M, Hazan RB (2002) A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2(4):301–314. https://doi.org/10.1016/S1535-6108(02)00150-2
Tanner Y, Grose RP (2016) Dysregulated FGF signalling in neoplastic disorders. Semin Cell Dev Biol 53:126–135. https://doi.org/10.1016/j.semcdb.2015.10.012
Turner N, Grose R (2010) Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10(2):116–129. https://doi.org/10.1038/nrc2780
Uria JA, Lopez-Otin C (2000) Matrilysin-2, a new matrix metalloproteinase expressed in human tumors and showing the minimal domain organization required for secretion, latency, and activity. Cancer Res 60(17):4745–4751
Wang J, Su H, Han X, Xu K (2014) Inhibition of fibroblast growth factor receptor signaling impairs metastasis of hepatocellular carcinoma. Tumour Biol 35(11):11005–11011. https://doi.org/10.1007/s13277-014-2384-0
Whittaker S, Marais R, Zhu AX (2010) The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 29(36):4989–5005. https://doi.org/10.1038/onc.2010.236
Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang QF, Wu F (2012) MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/ss-catenin pathway. Liver Int 32(5):752–760. https://doi.org/10.1111/j.1478-3231.2011.02750.x
Yang H, Fang F, Chang R, Yang L (2013) MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology 58(1):205–217. https://doi.org/10.1002/hep.26315
Yi S, Yang J, Huang J, Guan L, Du L, Guo Y, Zhai F, Wang Y, Lu Z, Wang L, Li H, Li X, Jiang C (2016) Expression of bioactive recombinant human fibroblast growth factor 9 in oil bodies of Arabidopsis thaliana. Protein Expr Purif 116:127–132. https://doi.org/10.1016/j.pep.2015.08.006
Yu C, Wang Z, Xu X, Xiang W, Huang X (2015) Circulating hepatocellular carcinoma cells are characterized by CXCR4 and MMP26. Cell Physiol Biochem 36(6):2393–2402. https://doi.org/10.1159/000430201
Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM (2006) Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem 281(23):15694–15700. https://doi.org/10.1074/jbc.M601252200
Zhao YG, Xiao AZ, Park HI, Newcomer RG, Yan M, Man YG, Heffelfinger SC, Sang QXA (2004) Endometase/matrilysin-2 in human breast ductal carcinoma in situ and its inhibition by tissue inhibitors of metalloproteinases-2 and -4: a putative role in the initiation of breast cancer invasion. Cancer Res 64(2):590–598
Acknowledgments
This work was supported by grants of National Natural Science Foundation of China (No. 81471075) and granted by the Opening Project of Zhejiang Provincial TOP Key Discipline of Pharmaceutical Sciences (No.201720).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This article does not contain any studies with animals or human participants. All authors confirm that ethical principles have been followed in the experiments.
Conflicts of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Wang, S., Lin, H., Zhao, T. et al. Expression and purification of an FGF9 fusion protein in E. coli, and the effects of the FGF9 subfamily on human hepatocellular carcinoma cell proliferation and migration. Appl Microbiol Biotechnol 101, 7823–7835 (2017). https://doi.org/10.1007/s00253-017-8468-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8468-1