Abstract
The continuous development of fast and simple new methods to identify animal-derived ingredients is very important for the authentication of meat products. This study intended to develop a multiplex PCR method using new species-specific nuclear DNA (nDNA) sequences for the detection of ingredients derived from sheep/goat, bovine, chicken, duck and pig in meat products. Sequence alignment analysis in 53 species showed high specificity of species-specific nDNA. Species-specific primers were designed on the conservative region of each species-specific nDNA sequence. The specificity and conservation of the sequences and primers were verified by PCR reaction and sequencing with the limit of detection down to 0.5 ng. Then, a species-specific multiplex PCR method was developed and optimized to simultaneously detect sheep/goat (237 bp), bovine (223 bp), chicken (192 bp), duck (168 bp) and pig (154 bp) in one reaction. Various processed meat products containing one or more animal-derived ingredients were detected by the developed multiplex PCR method, and the results were consistent with their labeled meat species. Our study provides a fast and simple detection method for regulating labeling of animal-derived ingredients in meat products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The authenticity of meat products has attracted increasing attention since the European horse meat case in 2013 [1]. Food labeling regulations require that the species of meat in food products must be accurately declared to consumers. However, high-priced meats, such as beef and lamb, are often substituted or adulterated with low-priced meats (e.g. pork, chicken and duck meats) for a huge profit. Adulterations of one or multiple animal ingredients have been widely reported in commercial meat products [2]. An investigation performed on 100 meat products highlighted that 22.0% of cases contained undeclared species, primarily with poultry substitution in beef [3]. The analysis of 224 Halal meat products in Iran showed that 7.58% of the total samples contained Haram meat [4]. Food products containing undeclared meat may cause economic problems, health risks and violation of ethical/religious principles [5]. Therefore, it is a necessary task to establish efficient and reliable species identification methods to monitor animal-derived ingredients in food for comprehensive risk analysis.
Beef, lamb, pork, chicken and duck meats are the main meat products for humans in daily life. According to a report published by the Food and Agriculture Organization of the United Nations (FAO), meat production in 2013 reached 308.2 million tons, and this number is increasing significantly every year [6]. Unfortunately, commercial meat products (ham, kebab and meat rolls, etc.) are often hard to recognize their components owing to the deep processing. To ensure correct declaration, many DNA-based polymerase chain reaction (PCR) approaches for species identification have been developed [7], mainly including PCR-restriction fragment length polymorphism (PCR–RFLP) [8, 9], species-specific PCR [10, 11], real-time PCR [12] and droplet digital PCR [13]. In these methods, species-specific DNA sequences for species identification were mostly based on mitochondrial DNA (mtDNA), due to its heat stability and multi-copy number [14]. PCR methods based on nucleotide sequence variations in the mitochondrial genes, such as 12S rRNA, Cytb, COX1 and D-loop, have been widely used for species identification in animal foods [6, 15, 16], even in the analysis of processed products [17, 18]. Although these methods can solve some problems of meat adulteration in food, PCR methods based on mtDNA sequences are hard to effectively distinguish closely related species, due to little variation of the mtDNA sequence exists among them. And the mutability of mtDNA sequence may lead to inaccurate results in the detection of different breeds or individuals of target species if the mutation occurs in the primer binding region [19]. In contrast, nuclear DNA (nDNA) does not have the above problems and can provide more suitable species-specific sequences for species identification. Some methods have been developed to find species-specific nDNA for species identification in recent years [20,21,22,23], but still rare. Limitations in the number of species-specific sequences have become the bottleneck in species identification using PCR techniques. In addition, to achieve rapid, simple and reliable species identification, multiplex PCR based on species-specific primers is the preferred choice. That is not only because it can simultaneously detect multiple animal species in one reaction, but also is easy to use and cost-saving. Thus, the development of the multiplex PCR method using new species-specific nDNA sequences and primers to simultaneously identify various animal-derived ingredients in meat products are urgently required.
In this study, five species-specific nDNA sequences were independently screened and used to design species-specific primers for accurate species identification in meat products. After that, the inter-species specificity of the assay was assessed in 19 animal species. Then we examined the sequence variability in different breeds and individuals to validate the intra-species conservation and evaluated the sensitivity with serially diluted DNA. Furthermore, a multiplex PCR method was developed to simultaneously detect sheep/goat, bovine (cattle and buffalo), chicken, duck and pig, and applied to the detection of commercial meat products in the market.
Materials and methods
Animal species and genomic DNA preparation
Nineteen muscle samples from different animal species were obtained and employed as experimental materials [24]. Bubalus bubalis (buffalo), Ovis aries (sheep), Capra hircus (goat), Sus scrofa (pig), Canis lupus familiaris (dog), Oryctolagus cuniculus (rabbit), Mus musculus (mouse), Rattus norvegicus (rat), Cricetulus griseus (hamster), Cavia porcellus (guinea pig), Gallus gallus (chicken), Anas platyrhynchos (duck) and Anser domestica (goose) were collected from Huazhong Agricultural University. Vulpes vulpes (fox), Mustela putorius furo (mink) and Nyctereutes procyohoides Grag (racoon dog) were collected from a farm in Hebei province of China. Bos Taurus (cattle) was provided by the Chinese Academy of Agricultural Sciences. Equus caballus (horse) was provided by Xinjiang Academy of Animal Sciences. Equus asinus (donkey) was provided by Shandong Dong-e E-jiao Co., Ltd.. All samples were cut into small pieces using a sterile scalpel, kept in a sterile plastic bag, and stored at − 20 ℃. Moreover, forty-four individuals from different breeds of each species were collected to check for the possible intra-species polymorphism of the species-specific sequences (Supplementary Table 1). To assess the application of our established method, seventeen processed meat products of lamb, beef, pork, chicken and duck were also obtained from nearby markets or online shopping, including a homemade meatball with an equal proportion of cattle, sheep, pig, chicken and duck meat produced and cooked in high-pressure (100 kPa) and high-temperature (121 ℃) for 20 min.
The genomic DNA isolation of samples was carried out in accordance with the proteinase K-SDS-phenol/chloroform protocol described by Sambrook et al. [25]. The concentration and purity of genomic DNA extracts were measured by using Nanodrop 2000 (Thermo Scientific, USA). The genomic DNA of all species were diluted to 50 ng/μL with ddH2O for subsequent use.
Species-specific sequences and species-specific primers
DNA sequences of sheep, cattle, chicken, duck and pig were aligned with those of other species available in GenBank of NCBI using Local-BLAST. Only these sequences with “E-value” < 10–5, “Identity” < 90%, “Query cover length” > 200 bp and located on the autosomes were retained and kept for further filtering. In particular, the sequences of cattle and sheep with “E-value” < 10–5, “Query cover length” > 200 bp and “Identity” > 95%, compared with buffalo and goat, respectively, were screened out as specific sequences for subfamily Bovinae and Caprinae.
To further ensure specificity of the candidate sequences of each species for species identification, these sequences were verified through BLAST online and multiple sequence alignment by ClustalW against the reference genome of 53 species (cattle, zebu, yak, bison, buffalo, horse, przewalskii horse, donkey, sheep, goat, deer, camel, pig, rabbit, fox, dog, wolf, dingo, cat, bear, ferret, monkey, human, mouse, rat, jerboa, hamster, guinea pig, squirrel, beaver, pika, dolphin, whale, elephant, bat, hedgehog, chicken, turkey, duck, goose, ostrich, anole, frog, zebrafish, salmon, fruit fly, soybean, maize, rice, wheat, yeast, Escherichia coli and Salmonella). Meanwhile, the single nucleotide polymorphism (SNP) of each sequence was also analyzed as a screening factor using SNP database of NCBI, Ensembl and UCSC, as well as from the results of PCR sequencing using mixed DNA of different breeds and individuals. Species-specific primers were designed in conserved regions based on the comparison of target species sequences of the database recorded. Degenerate bases were used when different nucleotides exist in the primer binding regions of the target species. Primer design was used Primer 5.0 (PREMIER Biosoft International, Palo Alto, USA). The primers that achieved a high score of rating without hairpin structures or dimers were selected. The specificity of them was preliminarily verified in the 19 species mentioned above by NCBI Primer-BLAST. Then, the primers (Table 1) were synthesized by Beijing Tsingke Company (Beijing, China).
Single PCR
For single PCR amplification, the reaction mixture consisted of 1 × Taq buffer, 0.2 mM dNTPs, 2.5 units of Taq DNA polymerase (TaKaRa Biotechnology Co. Ltd., China), 0.6 µM forward/reverse primer, 50 ng DNA template and sterile distilled H2O up to a final volume of 20 μL. The PCR program consists of the following steps: initial denaturation step at 95 ℃ for 5 min; 35 cycles of denaturation at 95 ℃ for 30 s, annealing at 60 ℃ for 40 s, extension at 72 ℃ for 40 s; final extension at 72 ℃ for 5 min; and a temperature reduction at 12 ℃ for 3 min. The PCR products were electrophoresed on a 2% agarose gel stained with GelRed (Biotium, USA) in 1 × Tris-acetate-EDTA (TAE) buffer.
Specificity evaluation of the primers for species-specific PCR
The DNA samples extracted from 19 animal species were used as templates for validating the specificity of the designed primers. Sterile distilled H2O was added into PCR mixture instead of DNA as blank control. These species-specific primers, which could amplify target species with expected length and melting temperature (Tm), were used for species identification in 19 animal.
Evaluation of intra-species conservation and sequencing
To verify the broad applicability of each pair of species-specific primers in different breeds and individuals, more than 44 individuals were amplified and sequenced. One μL of non-target species DNA mixture and sterile distilled H2O were added into PCR mixture as negative control and blank control, respectively. Additionally, DNA mixtures from several breeds and individuals were used as templates for PCR and amplified products were sent to Beijing Tsingke Company for sequencing.
Sensitivity
In the determination of the limit of detection (LOD), a series of target species DNA (10, 5, 1, 0.5, 0.1, 0.05 and 0.01 ng/μL) were diluted with ddH2O. Each dilution was added separately to the reaction mixtures. Single PCR was performed in 20 μL reaction mixtures with 1 μL DNA for each species, respectively. Ultimately, 5 μL amplified products were electrophoresed on agarose (2%) gels and stained with GelRed™ after which they were visualized by gel documentation system.
Multiplex PCR
For multiplex PCR amplification, 0.4, 0.25, 0.25, 0.2 and 0.3 μΜ of five primer pairs (sheep/goat, bovine, chicken, duck and pig, respectively) were mixed together. And multiplex PCR was applied the same as single PCR in a T100™ thermal cycler (Bio-Rad Laboratories, Inc., USA). The PCR products were analyzed by 2% agarose gel electrophoresis and visualized by ethidium bromide staining in 0.5 × Tris-borate-EDTA (TBE) with GelRed staining.
Application of detection method in processed meat products
Lamb, beef, chicken, duck, pork and mixed meat products were purchased from nearby markets or online shopping. These processed meat products were minced and homogenized separately and then stored at − 20 ℃. After DNA extraction, species-specific multiplex PCR amplification was carried out to detect the animal-derived ingredients in triplicate according to the optimized system and conditions.
Results and discussion
Species-specific sequences and primers were selected for species identification by sequence alignment analysis
Suitable species-specific sequence for species identification should contain two characters: inter-species specificity and intra-species conservation. Inter-species specificity means the low-level DNA homology among different species, while intra-species conservation means the high conservation of gene structures and sequences among different breeds of the target species. Based on this rule, five species-specific nDNA sequences from the conservative nuclear genome regions (NC_019479.2, AC224113.1, NC_006091.5, NC_040071.1 and AK349170.1) were selected as species-specific sequences for detection of sheep/goat, bovine, chicken, duck and pig, respectively. These species-specific sequences performed BLAST against reference genomes of 53 species, which include representative species of 4 kingdoms, 10 classes, 21 orders, 36 families and 47 genera. Then, the sequences similar to the target species were downloaded and analyzed by ClustalW multiple sequence alignment software. As illustrated in Fig. 1, only a few homologous sequences from the species closely related to our target species were observed. And there are still obvious differences for these homologous sequences, especially in the primer binding regions. For instance, the homology of all non-bovine species sequences to bovine-specific sequence was less than 90%, with significant base differences in primer binding regions. Our study showed that species-specific sequences were only homologous in the genomes of closely related species. Therefore, it is essential to include at least one closely species in the selection of species-specific sequences. On the contrary, it is highly conservative on the subfamily level of Bovinae (genus Bos and Bubalus) or Caprinae (genus Ovis and Capra) (Fig. 1). For example, we were fortunate to obtain a highly conserved bovine-specific sequence for different species of subfamily Bovinae (cattle, zebu, yak, bison, buffalo).
Then, we analyzed the conservation of each sequence from SNP databases and sequencing. As shown in Supplementary Fig. 1 , SNPs in the sequences were marked by different colors, accounting for less than 5% of the total length of each sequence. This implies that the similarity of almost all the individual sequences within each target species is more than 95%. It has been reported that key mismatches in primer binding region interfere with the efficiency of PCR and may even lead to false-negative detection [26]. In the sequences of this study, there are few SNPs in primer binding regions, especially at the 3′end, which ensured the reliability of the detection results. When there were intra-species polymorphisms in the primer binding regions of the target species, degenerate bases were used to eliminate the differences in amplification efficiency among different individuals.
Verification of specificity, conservation and sensitivity of species-specific sequences and primers
Specificity evaluation in 19 animal species
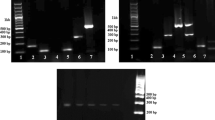
To check the specificity of species-specific primers for species identification, we first evaluated the selected species-specific primers against 19 animal species using Primer-BLAST program to make sure no amplification in non-target species. Meanwhile, single PCR was performed using 50 ng of genomic DNA extracted from different species of domestic and model animals including the representative breeds of corresponding species (Fig. 2). The expected size bands of 237 bp, 223 bp, 192 bp, 168 bp and 154 bp were amplified from sheep/goat, bovine (cattle and buffalo), chicken, duck and pig, respectively, without any amplification produced from non-target species. The results revealed that our species-specific primers possessed high specificity and could be utilized for species identification. It is worthwhile to mention that same size amplification products were found in the representative animals of genus Ovis (sheep) and genus Capra (goat) for sheep/goat-specific primers. Similar results were observed in the amplification of genus Bos (cattle) and genus Bubalus (buffalo) using bovine-specific primers.
Specificity evaluation of PCR in 19 species. a specific amplification results of sheep/goat-specific primers; b species-specific amplification results of bovine-specific primers; c species-specific amplification results of chicken-specific primers; d species-specific amplification results of duck-specific primers; e species-specific amplification results of pig-specific primers; f patterns of species-specific amplification
Conservation detection among different breeds and individuals
Sequence conservation within target species is a necessary factor to ensure accurate species identification and avoid false-negative results. In this study, single PCR was performed on the target species genomes of different breeds and individuals, as well as a negative control (mixture DNA of non-target species) and blank control (water). As shown in Supplementary Fig. 2 , all samples (100%) were identified without false-positive or false-negative results for each pair of species-specific primers. This implies that the selected species-specific primers can accurately identify all target species in meat. Then, amplified products were mixed and sequenced. The sequencing results of the PCR products also confirmed over 95% similarity within different individuals of each target species, which were consistent with the results of sequence analysis (Fig. 1, Supplementary Fig. 1). Previous studies have generally evaluated their methods in terms of specificity and sensitivity, but rarely analyzed the sequence conservation [10, 27, 28]. Considering that sequence mismatch is likely to lead to false-negative results, conservation analysis of sequences, especially in primer binding regions, is indispensable in species identification. In our study, the conservation of the sequence was strictly evaluated in three aspects: (1) exonic sequences were used as target sequences for species identification, because of its more stable inheritance and fewer mutations in different breeds relative to intronic sequences and intergenic regions; (2) alignment analysis of sequences within multiple genera of the target species was carried out, and primers were designed in conservative regions of sequences (Fig. 1, Supplementary Fig. 1). The effect of base differences in the primer binding regions of the target species on PCR amplification efficiency was eliminated by degenerate bases; (3) the conservation was validated in the DNA of 44 individuals of different breeds, and the accuracy and reliability of our method in detecting target species from different geographical origin were further confirmed by sequencing and SNP databases.
Sensitivity of the species-specific PCR
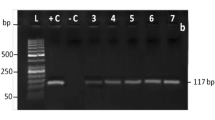
Sensitivity also plays an important role in species identification. Five species-specific primers were used to amplify seven serially diluted DNA of their respective species according to the optimized system and conditions. Each pair of species-specific primers can amplify the target band of expected size from at least 0.5 ng of DNA by single PCR. The sheep/goat-specific primers successfully produced amplification from 0.05 ng of template DNA (Fig. 3). It has been documented that when meat is heated to 100 ℃, the DNA is degraded to about 1100–300 bp [29, 30]. To improve the sensitivity of species-specific PCR for processed meat products, short DNA fragments (< 300 bp) were amplified to ensure that they can still be detected under various processing conditions [31]. Selection of small DNA fragments for amplification enables our method to detect effectively even in processed meat products.
Development of multiplex PCR system of sheep/goat, bovine, chicken, duck and pig
Considering that lamb and beef are often adulterated by cheaper meats including pork, chicken and duck meats, we developed a multiplex PCR to detect meat simply and quickly. By optimizing the ratio of primers in PCR, the direct multiplex assay for the simultaneous detection of sheep/goat, bovine, chicken, duck and pig was successfully developed. As shown in Fig. 4, clear and sharp bands of the expected sizes of 237, 223, 192, 168 and 154 bp for sheep/goat, bovine, chicken, duck and pig, respectively, were observed on an agarose gel electrophoresis. There was no apparent primer cross-reaction with species-specific primers in DNA samples, which indicated the specificity of primers for each species. Obviously, the number and size of the electrophoretic bands were in accordance with the number and species of the added DNA mixture. In addition, the multiplex PCR system with 50 ng total DNA (10 ng DNA for each species) can simultaneously and accurately identify five animal-derived ingredients (Lane 21 of Fig. 4). This means that as low as 10 ng of DNA for each species can be detected in the multiplex PCR. This is enough to meet the sensitivity requirements for the detection of animal-derived ingredients in meat products in real life.
Multiplex PCR results for method validation. Lane 1, blank control (sterile distilled H2O); the DNA templates used in Lanes 2–6 were sheep, cattle, chicken, duck, and pig DNA, respectively; Lanes 7–12, sheep/chicken, sheep/duck, sheep/pig, cattle/chicken, cattle/duck, cattle/pig DNA mixtures; Lanes 13–18, sheep/chicken/duck, sheep/chicken/pig, sheep/duck/pig, cattle/chicken/duck, cattle/chicken/pig, cattle/duck/pig DNA mixtures; Lanes 19–20, sheep/chicken/duck/pig, cattle/chicken/duck/pig DNA mixtures; Lane 21, sheep/cattle/chicken/duck/pig DNA mixture
Accurate identification of various DNA mixtures indicated that the five species could be recognized successfully and simultaneously. Traditionally, accurate analytical methods are essential for species identification in meat products, which requires simple and fast detection procedures. The multiplex PCR method we employed is an effective method for simultaneously identifying five species from mixed samples. Compared with the traditional single PCR, this method saves the experimental time and cost, and can improve the efficiency of detecting animal-derived ingredients in food [32]. The multiplex PCR can be applied to determine whether meat contains one or more of five animal-derived ingredients according to the number and size of bands on agarose gel electrophoresis. The presence of any of these species-specific bands of unlabeled ingredients in food would indicate adulteration.
Detection of processed meat products in the market by multiplex PCR
To verify the detection ability of our method for processed meat products, species identification experiments were carried out for 17 commercial meat products by multiplex PCR. Whether these meats were dried, fried, stewed, or boiled, our method could successfully identify the species they represented from processed meat products, even the mixed processed meat products with multiple ingredients (Table 2, Supplementary Fig. 3). This result provided the potential application prospects for our method in the detection and monitoring of commercial meat products in the meat market. Actually, the detection ability to processed meat products determines whether the developed methods can be widely applied in the meat market. Food processing can break up and degrade a lot of DNA. It has been reported that 99% of DNA degraded into small fragments after 20 min of high-pressure cooking [33]. In this study, short species-specific nDNA sequences (< 300 bp) for sheep/goat, bovine, chicken, duck and pig were screened, and showed good specificity, conservation and sensitivity in the detection of meat products. Our method provides a highly specific, simple and reliable species identification method for conventional livestock and poultry meat products.
Conclusion
Five new species-specific nDNA sequences were selected for the detection of sheep/goat, bovine, chicken, duck and pig. Species-specific PCR was developed to detect lamb, beef, chicken, duck and pork in raw and processed meat products. The assay utilized very short nDNA sequences with high inter-species specificity as detection targets and thus very sensitive and specific for various processed foods. Then, a multiplex PCR system was developed for the simultaneous identification of five species and used to detect various processed meat products. The detection results in several processed meat products demonstrated the potential of our method for the detection of meat products in the market. We believe that our method would find widely application in the food industry for the authentication of lamb, beef, chicken, duck and pork derived ingredients in raw and produced foods.
References
Hsieh YH, Ofori JA (2014) Detection of horse meat contamination in raw and heat-processed meat products. J Agric Food Chem 62(52):12536
Hsieh YHP, Chen FC, Sheu SC (1997) AAES research developing simple, inexpensive tests for meat products. Highlights Agric Res 44(2):19–20
Ayaz Y, Ayaz ND, Erol I (2010) Detection of species in meat and meat products using enzyme-linked immunosorbent assay. J Muscle Foods 17(2):214–220
Doosti A, Ghasemi Dehkordi P, Rahimi E (2014) Molecular assay to fraud identification of meat products. J Food Sci Technol 51(1):148–152
Di Pinto A, Bottaro M, Bonerba E, Bozzo G, Ceci E, Marchetti P et al (2015) Occurrence of mislabeling in meat products using DNA-based assay. J Food Sci Technol 52:2479–2484
Izadpanah M, Mohebali N, Elyasi Gorji Z, Farzaneh P, Vakhshiteh F, Shahzadeh Fazeli SA (2018) Simple and fast multiplex PCR method for detection of species origin in meat products. J Food Sci Technol 55(2):698–703
Sheikha AFE, Mokhtar NFK, Amie C, Lamasudin DU, Isa NM, Mustafa S (2017) Authentication technologies using DNA-based approaches for meats and halal meats determination. Food Biotechnol 31(4):281–315
Guan F, Jin Y-T, Zhao J, Xu A-C, Luo Y-Y (2018) A PCR method that can be further developed into PCR-RFLP assay for eight animal species identification. J Anal Methods Chem. https://doi.org/10.1155/2018/5890140
Teixeira LV, Teixeira CS, Oliveira DAA (2015) Species-specific identification of red-meat and meat derivative products with the PCR-RFLP technique. Arquivo Brasileiro De Medicina Veterinária E Zootecnia 67:309–314
Amaral JS, Santos CG, Melo VS, Costa J, Oliveira MBPP, Mafra I (2015) Identification of duck, partridge, pheasant, quail, chicken and turkey meats by species-specific PCR assays to assess the authenticity of traditional game meat Alheira sausages. Food Control 47:190–195
Prusakova OV, Glukhova XA, Afanas'eva GV, Trizna YA, Nazarova LF, Beletsky IP (2018) A simple and sensitive two-tube multiplex PCR assay for simultaneous detection of ten meat species. Meat Sci 137:34–40
Ahamad MNU, Hossain MM, Uddin SMK, Sultana S, Nizar NNA, Bonny SQ et al (2019) Tetraplex real-time PCR with TaqMan probes for discriminatory detection of cat, rabbit, rat and squirrel DNA in food products. Eur Food Res Technol 245:2183–2194
Köppel R, Ganeshan A, Weber S, Pietsch K, Graf C, Hochegger R et al (2019) Duplex digital PCR for the determination of meat proportions of sausages containing meat from chicken, turkey, horse, cow, pig and sheep. Eur Food Res Technol 245:853–862
Kumar A, Kumar RR, Sharma BD, Gokulakrishnan P, Mendiratta SK, Sharma D (2014) Identification of species origin of meat and meat products on the dna basis: a review. CRC Crit Rev Food Technol. https://doi.org/10.1080/10408398.2012.693978
Hassan B, El-Garhy HAS, Moustafa MMA (2014) Detection of pork adulteration in processed meat by species-specific PCR-QIAxcel procedure based on D-loop and cytb genes. Appl Microbiol Biotechnol 98(23):9805–9816
Hou B, Meng X, Zhang L, Guo J, Li S, Jin H (2015) Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Sci 101:90–94
Qin P, Qu W, Xu J, Qiao D, Yao L, Xue F, Chen W (2019) A sensitive multiplex PCR protocol for simultaneous detection of chicken, duck, and pork in beef samples. J Food Sci Technol 56(3):1266–1274
Yasemin D, Pelin U, Senyuva HZ (2012) Detection of porcine DNA in gelatine and gelatine-containing processed food products-Halal/Kosher authentication. Meat Sci 90(3):686–689
Ballin NZ, Vogensen FK, Karlsson AH (2009) Species determination Can we detect and quantify meat adulteration? Meat Sci 83(2):165–174
Floren C, Wiedemann I, Brenig B, Schutz E, Beck J (2015) Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chem 173:1054–1058
Köppel R, Ruf J, Zimmerli F, Breitenmoser A (2008) Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, chicken and turkey. Eur Food Res Technol 227(4):1199–1203
Laube I, Zagon J, Spiegelberg A, Butschke A, Kroh LW, Broll H (2007) Development and design of a ‘ready-to-use’reaction plate for a PCR-based simultaneous detection of animal species used in foods. Int J Food Sci Technol 42(1):9–17
Xiang W, Shang Y, Wang Q, Xu Y, Zhu P, Huang K, Xu W (2017) Identification of a chicken (Gallus gallus) endogenous reference gene (Actb) and its application in meat adulteration. Food Chem 234:472
Wang W, Liu J, Zhang Q, Zhou X, Liu B (2019) Multiplex PCR assay for identification and quantification of bovine and equine in minced meats using novel specific nuclear DNA sequences. Food Control 105:29–37
Reid GA (1991) Molecular cloning: a laboratory manual. In: Sambrook J, Fritsch EF, Maniatis T (eds) Trends in biotechnology. Cold Spring Harbor Laboratory Press, New York
Kirsten H, Teupser D, Weissfuss J, Wolfram G, Emmrich F, Ahnert P (2007) Robustness of single-base extension against mismatches at the site of primer attachment in a clinical assay. J Mol Med (Berl) 85(4):361–369
Amaral JS, Santos CG, Melo VS, Oliveira MBPP, Mafra I (2014) Authentication of a traditional game meat sausage (Alheira) by species-specific PCR assays to detect hare, rabbit, red deer, pork and cow meats. Food Res Int 60:140–145
Ha J, Kim S, Lee J, Lee S, Lee H, Choi Y, Oh H, Yoon Y (2017) Identification of pork adulteration in processed meat products using the developed mitochondrial DNA-based primers. Korean J Food Sci Anim 37(3):464–468
Tartaglia M, Saulle E, Pestalozza S, Morelli L, Antonucci G, Battaglia PA (1998) Detection of bovine mitochondrial DNA in ruminant feeds: a molecular approach to test for the presence of bovine-derived materials. J Food Prot 61(5):513–518
Xu X, Gullberg A, Arnason U (1996) The complete mitochondrial DNA (mtDNA) of the donkey and mtDNA comparisons among four closely related mammalian species-pairs. J Mol Evol 43(5):438–446
Ali ME, Rahman MM, Hamid SBA, Mustafa S, Bhassu S, Hashim U (2013) Canine-specific pcr assay targeting cytochrome b gene for the detection of dog meat adulteration in commercial frankfurters. Food Anal Methods 7(1):234–241
Wu Q, Xiang S, Wang W, Zhao J, Xia J, Zhen Y et al (2017) Species Identification of Fox-, Mink-, Dog-, and Rabbit-Derived Ingredients by Multiplex PCR and Real-Time PCR Assay. Appl Biochem Biotechnol 185:1–12
Ebbehøj KF, Thomsen PD (1991) Species differentiation of heated meat products by DNA hybridization. Meat Sci 30(3):221
Acknowledgement
This work was supported by the National Science and Technology Major Project of China (2018ZX08012-001-010).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Wenjun Wang declares that he has no conflict of interest. Xiaokang Wang declares that he has no conflict of interest. Qingde Zhang declares that he has no conflict of interest. Zuhong Liu declares that he has no conflict of interest. Xiang Zhou declares that he has no conflict of interest. Bang Liu declares that he has no conflict of interest. We have filed patents for primers to detect bovine and pig specific nuclear DNA sequences.
Compliance with ethics requirements
This article does not contain any studies involving human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
217_2020_3494_MOESM1_ESM.pdf
Supplementary file1 Supplementary Fig. 1. All SNPs in the species-specific sequences of target species collected from NCBI, Ensembl and UCSC databases, as well as from the PCR sequencing results of different breeds and individuals. The underlined sequences were primer binding regions; degenerate bases M=A/C, S=G/C, Y=C/T, R=A/G, K=G/T. (PDF 119 kb)
217_2020_3494_MOESM2_ESM.pdf
Supplementary file2 Supplementary Fig. 2. Conservation analysis of PCR for sheep/goat (A), bovine (B), chicken (C), duck (D), and pig (E). In the agarose gel, M, BM2000 DNA Marker, B, blank control, N, negative control, 1-44, samples of different breeds or individuals. (PDF 299 kb)
217_2020_3494_MOESM3_ESM.pdf
Supplementary file3 Supplementary Fig. 3. Detection results in lamb, beef, chicken, duck, pork and mixed meat products. M, BM2000 DNA Marker; B, blank control; N, negative control; P1, positive control of sheep/goat; P2, positive control of bovine; P3, positive control of chicken; P4, positive control of duck; P5, positive control of pig; RLK, roasted lamb kebab; BLR, boiled lamb roll; BH, beef ham; BJ, beef jerky; DCC, deep-fried chicken chop; GCK, grilled chicken kebab; SDN, stewed duck neck; RD, roasted duck; PS, pork sausage; CBS, cooked bacon slices; (a), mixed meat rolls; (b), beef dumplings; (c), beef ham slices; (d), western ham; (e), pork ham slices; (f), sausage; (g), homemade meatball. (PDF 250 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Wang, X., Zhang, Q. et al. A multiplex PCR method for detection of five animal species in processed meat products using novel species-specific nuclear DNA sequences. Eur Food Res Technol 246, 1351–1360 (2020). https://doi.org/10.1007/s00217-020-03494-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-020-03494-z