Abstract
Cat, rabbit, rat, and squirrel species are very sensitive in food products because most of them are potential carriers of zoonotic diseases and rejected in most religions and cultures. Since cats and rats are abundant in most parts of the world and their meats do not carry any value in legal markets, these meats could be considered as potential adulterants in halal, kosher, and other food markets. Rabbit and squirrel meats are also susceptible to adulteration. Therefore, both health and economic interests in rat, rabbit, cat and squirrel species are significant. In this work, a novel tetraplex real-time PCR assay with TaqMan probes was described to discriminate and identify all four species (cat, rabbit, rat, and squirrel) in a single assay platform. Species-specific primers and probes were developed against ATP6, and cytochrome b genes to amplify 108, 123, 161 and 176 bp DNA fragments from rat, rabbit, squirrel and cat meat products under various states. A 141-bp internal amplification control (IAC) of 18S rRNA was used to avoid any false-negative results. Specificity was evaluated against 22 species but no cross-reactivity was found. Efficiency of PCR assay as well as target quantification were determined based on a standard curve that was generated using tenfold serially diluted mixed DNA extract (1:1:1:1) from squirrel, rat, rabbit and cat species. The assay was valid under pure, processed and admixed states with 10–0.1% (w/w) adulterant from each species. The limit of detection was 0.1% under admixed samples and 0.003 ng DNA under pure states from each species. Analyses of 18 model burgers (9 chicken and 9 beef) and 18 frankfurters (9 chicken and 9 beef) revealed 91–122% target recovery at 0.1–10% adulteration. Finally, 72 commercial burgers (36 chicken and 36 beef) and 72 frankfurters (36 chicken and 36 beef) were screened but no target species was detected except IAC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Authenticity issues in food products are intricately linked to public health, religious and cultural issues as well as fair-trade economic practices. Recently, rat meat was chemically modified to change physical appearances and sold as lamb [1]. This greatly aggravated the concerns and worries of general public and also health professionals because rats are carriers of several infectious and deadly diseases, such as listeriosis, yersiniosis, pasturellosis, meilioidosis and plague diseases [2]. The close cousins of rats are squirrels that are also a carrier of Creutzfeldt–Jakob disease (CJD) syndrome (neurodegenerative disease) [3] and Lyme disease [4]. On the other hand, rabbit and cat carcasses are almost identical to look at and so cats were sold as rabbits in several instances [5]. Recently, cat meat was sold as mutton in China [6] and India [7]. The issue is a grave concern because rabbit meat is a blooming industry but cat meat does not have any apparent market prices as cat consumption is a taboo in most of the societies and cultures [8]. While rabbit meat is appreciated because of its huge contents of essential minerals [9], and proteins but low content of saturated fats and cholesterol [10], cat meat is a carrier of hepatitis, severe acute respiratory syndromes, anthrax and some other deadly diseases [11]. Therefore, both health and economic interests in rat, rabbit, cat and squirrel species are huge and so there is a demand for a reliable, low cost and easily applicable authentication technique that could both detect and quantify any ingredients from these species under any matrices.
For meat and meat product authentication, recently, DNA-based PCR techniques have been evolved as the method of choice because protein- and lipid-based biomarkers are easily modified and so cannot offer so much reliability as is offered by robustly stable short-length polymorphic DNA sequences [12,13,14,15]. Several DNA-based methods such as species-specific PCR [16, 17], PCR-restriction fragment length polymorphism (PCR-RFLP) [18, 19] multiplex PCR [20, 21], randomly amplified polymorphic DNA (RAPD) [22], real-time PCR [23], and DNA barcoding systems [24] have been documented. However, among these methods, only real-time PCR approaches offer quantification, automation and high throughput [25, 26]. For the species targeted in this paper, a real-time PCR assay has been proposed which could detect red and gray squirrels [27]. On the other hand, several conventional PCR assays are available for rat [28, 29] and rabbit specification [30,31,32]. However, the main problems with these methods are that they are based on a single species target and long DNA amplicon (243–672 bp) that often breaks down under extensive food processing atmospheres, making them unreliable and quite expensive for forensic investigations [33]. To overcome the problems, recently, we have documented a short amplicon length (108–176 bp) multiplex PCR assay for rabbit, rat and squirrel detection in a conventional platform [5, 34]. However, this assay did not include cat species that is frequently adulterated in rabbit meat [5]. In this regard, multiplex real-time PCR (qPCR) assay with TaqMan probes are greatly advantageous because it provides data with high sensitivity and specificity at real time and ensures additional security through primers and specialized probes whose sequences are required to be perfectly matched with the selective sites of the target analytes for successful identification [35].
Recently, TaqMan-based multiplex qPCR assays have been reported for cattle, buffalo and pig [25], duck, pig and chicken [26], and deer [36], as well as pork, beef, turkey, sheep and chicken species [37]. However, such a featureful assay has not been reported for the cat, rabbit, rat and squirrel species. To address this knowledge gap, we documented here a short amplicon length (108–176 bp) tetraplex qPCR assay with TaqMan probes for the simultaneous detection of their DNA in processed food products for the first time. The method was validated with regard to specificity, limit of detection (LOD), accuracy, repeatability and robustness.
Materials and methods
Sample collection
Specimens of fresh muscle tissues were obtained from rabbit (Oryctolagus cuniculus), squirrel (Calloscious notatus), rat (Rattus rattus), cat (Felis catus), chicken (Gallus gallus), beef (Bos taurus), buffalo (Bubalus bubalis), sheep (Ovis aries), goat (Capra hiscus), pig (Sus scrofa), duck (Anas platyrhynchos), pigeon (Columba livia), crocodile (Crocodylus niloticus), amboina box turtle (Cuora amboinensis), Chinese edible frog (Hoplobatrachus rugulosus), deer (Cervus nippon yesoensis), dog (Canis lupus familiaris), tuna (Thunnus orientalis), salmon (Salmo salar) and plant species, namely, wheat (Triticum aestivum), cucumber (Cucumis sativus), onion (Allium cepa), and chili (Capsicum annuum). Commercially available meat, fish and plant samples were collected from local wet (Pudu Raya) and super markets (Aeon, Tesco and Giant) at Kuala Lumpur on three different days to increase the genetic diversity of the collected samples. Deer (Cervus nippon) meat was procured in triplicates from the Faculty of Veterinary Sciences at the University of Putra Malaysia, located at Serdang in Selangor. Stray dog (Canis lupus familiaris), cat (Felis catus) and rat (Rattus rattus) muscle tissues were donated by Kuala Lumpur City Hall (KLCH/DBKL) at Air Panas in Kuala Lumpur [38]. Monkey (Macaca fascicularis sp.) meat was a gift from the Department of Wildlife and National Park Malaysia (DWNPM/PERHILITAN) at Cheras in Kuala Lumpur. The commercial burger and frankfurter of chicken and beef origins of four different brands were purchased from Malaysian outlets. All samples were transported under ice-chilled conditions and were cut into the smallest possible pieces with surgical blades prior to storage at − 20 °C until further uses.
Preparation of admixtures and model meat products (burger and frankfurter)
Tertiary admixture (100 g) was prepared by mixing minced squirrel, rat, rabbit and cat at a ratio of 1:1:1:1 and was homogenized by blending. To prepare burgers and frankfurters, raw meat samples of squirrel, rat, rabbit and cat were minced and blended separately. Model burger and frankfurter of beef and chicken were made in the laboratory following Asing et al. [12] (Table 1). Certain amounts of beef and chicken were mixed with a balanced amount of squirrel, rat, rabbit and cat meat to make 10%, 1% and 0.1% adulteration for each target species.
DNA extraction
Total DNA was extracted from 30 mg of muscle tissue of each meat and fish species as well as their mixed meat products using a Yeastern Genomic DNA Mini Kit (Yeastern Biotech Co., Ltd. Taipei, Taiwan) [39]. Plant DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN GmgH, Hilden, Germany) [40]. DNA from commercial burger and frankfurter was extracted using NucleoSpin Food DNA kit (MACHEREY–NAGEL, GmbH & Co., Duren, Germany) [41]. The purity and concentration of all extracted DNA were determined using a UV–Vis spectrophotometer (Biochrom Libra S70, Biochrom Ltd, Cambridge, UK) based on absorbance at A260/A280 and the ratios were calculated [42,43,44]. All extracted DNAs were kept at − 20 °C until further uses.
Primer and probe design
The oligonucleotide primers and probes used in the present study were designed targeting mitochondrial cytb and ATP6 gene of squirrel, rat, rabbit and cat (Table 2). The 5′ and 3′ ends of each probe for squirrel, rat, rabbit and cat were labeled with ROX and TAO/3IAbRQSp; HEX and ZEN/3IABkFQ; Cy5 and TAO/3IAbRQSp; and TAMRA and TAO/3IAbRQSp, respectively. Eukaryotic 18S rRNA-specific primers and TaqMan probe (Table 2) were used as internal amplification control (IAC) [35]. The IAC probe was labeled with FAM at the 5′ end and ZAN/IOWA BLACK FQ at the 3′ end (Table 2). The designed primers and probes were supplied by Integrated DNA technologies (IDT), Singapore.
Conditions applied for tetraplex real-time PCR
Tetraplex real-time PCR assay of squirrel, rat, rabbit, cat and IAC was carried out using a Quant Studio 12 K flex real-time PCR system in a 20 µL reaction volume, comprising Prime time Gene Expression Master 2× Mix (IDT, Singapore), primer and probes, total DNA template of each target species and nuclease-free water. In the total volume of reaction mixture, the concentrations of each species DNA template and master mix were 30 ng and 1×, respectively. The primer and probe information is given in Table 2. The amplification was performed using initial denaturation step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 20 s, and annealing and extension at 58 °C for 60 s.
Specificity test
The specificity of the developed real-time tetraplex PCR assay was tested against the DNA templates of 22 non-target species including chicken, cow, goat, pig, pigeon, sheep, duck, buffalo, crocodile, turtle, donkey, deer, monkey, dog, turkey, Chinese frog, tuna, salmon, wheat, cucumber, onion and chili.
Limit of detection
To determine the limit of detection (LOD), the tetraplex qPCR assay was calibrated with a serially diluted DNA extract from a mixture of equal amounts of squirrel, rat, rabbit, and cat meat. A mixture was prepared with extracted DNA from the four target species (squirrel, rat, rabbit, and cat) consisting of 30 ng/μL of each species. After that, the DNA mixture was tenfold serially diluted [26] and the concentrations of the diluted DNA samples were 3, 0.3, 0.03, 0.003 ng/µL. In this assay, 1 µL of DNA mixture from each diluted sample was added to the 20 µL reaction mixture. As a result, the final volume of each reaction mixture contained the same amounts of DNA from all target species and the serially diluted five reaction mixtures contained the 30, 3, 0.3, 0.03, 0.003 ng of DNA, respectively. The tetraplex qPCR of each dilution was assayed in five replicates.
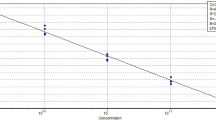
Generation of standard curve
Standard curve was constructed to determine qPCR efficiency and quantify PCR targets. To generate the standard curve of the tetraplex qPCR system for squirrel, rat, rabbit and cat, the DNA was extracted from the admixture (1:1:1:1) of squirrel, rat, rabbit and cat. The concentration of the extracted DNA was made 120 ng/µL consisting of 30 ng/µL DNA of each species. Then the admixture of DNA was tenfold serially diluted with nuclease-free water to make the concentration of DNA of each species of 3, 0.3, 0.03, 0.003 ng/µL. This resulted in mixtures containing 100–0.001% of DNA from each species. So, 1 µL of each diluted DNA was added to 20 µL of reaction mixture. After performing the tetraplex qPCR assay, the Ct values of each target species were plotted against the logarithmic concentration of DNA from each species [26, 45]. Subsequently, the standard curve was built up and the efficiency of the assay was calculated based on the slope of the curve according to the equation [36] as stated below:
The acceptance range of qPCR efficiency was between 90 and 110% that corresponded to a regression slope between − 3.1 and − 3.6 and an R2 value of ≥ 0.98 [45].
Sensitivity and validity
To evaluate the sensitivity and suitability of the tetraplex qPCR assay for food product analysis, two different types of model meat products, namely burger and frankfurter of beef and chicken origins were prepared in laboratory. Beef and chicken products were deliberately adulterated with 10, 1 and 0.1% (w/w) of squirrel, rat, rabbit and cat meat. The DNA was extracted from adulterated meat products and concentration was adjusted to 30 ng/µL with nuclease-free deionized water. The sensitivity and validity of the qPCR assay were determined based on respective Ct values according to the formula described in the generation of standard curve section.
Results and discussion
Assessment of DNA quality
Total genomic DNA was extracted from pure, admixed and commercial burger and frankfurter products at raw and post-processed states. Absorbance value at 260 nm and absorbance ratio at A260/A280 was used to measure concentration and purity of the extracted DNA, respectively. The concentration of extracted DNA was 110–245 ng/µL for animal and fish muscle tissues, 85–123 ng/µL for plant species and 20–45 ng/µL for meat products. The poor concentration of DNA in meat products was attributed to the complex nature of the food matrices that are composed of many ingredients. However, the absorbance ratio of all DNA samples at A260/A280 was 1.8–2.0 which reflected that good quality DNA was extracted from all specimens [13, 26].
Development of tetraplex qPCR model
The species-specific primers and probes were carefully evaluated for mismatch and melting temperature (Tm) because in a multiplex PCR system multiple primers and probes interact with several templates at the same or very closely related temperature [26]. In this assay, squirrel-, rat-, rabbit- and cat-specific primers and probes had very closely spaced Tm (59 ± 1 and 69 ± 1 °C) that ensured proper annealing of the primers and probes with their respective templates at a selective PCR condition [26]. The Tm values of the developed primers were 58.8–60.3 °C and so all the primers were annealed at 58 °C while Tms of the probes (68.5–70.70 °C) were 8–10 °C higher than that of the primers (Table 2). The higher Tm of the probes allowed preferential probes’ annealing before binding of the primers and it was the prerequisite for TaqMan chemistry [46]. These allowed the discrimination of four different amplicons in the same reaction tube through four different fluorescent reporter dyes tagged with the probes (Table 2). Initially, simplex qPCR system for each target species was optimized and then sequentially duplex, triplex and finally tetraplex qPCR was optimized. The Ct values of the tetraplex qPCR assay were 17.16 ± 0.05, 17.62 ± 0.07, 17.14 ± 0.04 and 16.92 ± 0.04 and they were very close to the corresponding Ct values of simplex qPCR for squirrel (Ct = 17.06 ± 0.05), rat (Ct = 17.46 ± 0.06), rabbit (Ct = 17.1 ± 0.03) and cat (Ct = 16.86 ± 0.06), respectively. Thus, the findings of the simplex and multiplex qPCR systems were very consistent, mutually validating each other.
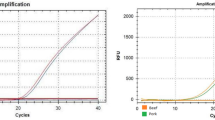
Specificity evaluation of the tetraplex qPCR system
The species specificity of the tetraplex qPCR system was critically evaluated because it is the foundation pillar of any PCR system. Following optimization, specificity was tested by cross-challenging the primers and probes against 22 non-target species on three different days in triplicates. To avoid false-negative detection, IAC was used as universal internal target to ensure that good quality DNA templates were present in all tubes (Fig. 1). Conversely, a blank or negative template control was made with everything but the template was replaced with equal volume of nuclease-free deionized water to eliminate the chances of any false-positive amplification. The obtained amplification profile clearly demonstrated that the species-specific amplification curves and background fluorescence were realized only for the relevant species in a 40 cycle PCR, confirming that no cross-amplifications took place in the tetraplex qPCR system (Fig. 1). While the amplification signal (Ct values) of the tetraplex qPCR assay for squirrel, rat, rabbit and cat were 17.143 ± 0.04, 17.48 ± 0.03,17.196 ± 0.05 and 16.8 ± 0.09, respectively, only IAC signal (Ct = 15.24–18.78) was obtained for the other 22 non-target species (Table 3).
Limit of detection
The limit of detection (LOD) of an assay determines the minimum amount of target analytes that could be detected in adulterated food stuff. So, it was ascertained by analyzing serially diluted DNA extracts mixed into equal proportion in a fixed amount of genomic DNA and subsequent PCR amplification. In this case, tenfold serially diluted mixed genomic DNA was used and consequently the concentration of the diluted DNA sample of each target species was 30, 3, 0.3, 0.03, 0.003 ng/µL. The amplification plots reflected detectable Ct from all concentrations, starting from 30 ng to 0.003 ng of DNA, suggesting the assay could detect and quantify minimum 0.003 ng of target DNA in a 20 μL reaction mixture. The resulting Ct values and the corresponding relative standard deviations (RSD) for all diluted DNA templates are listed in Table 4. It was found that the assay could detect and quantify the 0.003 ng/µL of DNA from each target species under mixed states with RSD values 0.07–0.56. Recently, Hossain et al. [25] reported an LOD of 0.003 ng/µL in a tetraplex qPCR assay for beef, buffalo and pork. On the other hand, Cheng et al. [26] detected 0.15 ng/µL DNA from blood curd samples of duck, pig and chicken. Similarly, 0.32 ng of DNA was determined by Koppel et al. [47] from boiled and raw sausages as well as fresh meat of beef, pork, chicken and turkey. These studies clearly revealed that LOD may vary from species to species and depends on many factors such as degree of decomposition, sample age, processing conditions and background matrices.
Target detection and qPCR efficiency
To determine the efficiency of the developed qPCR assay, four different standard curves were constructed for squirrel, rat, rabbit and cat by plotting the Ct values obtained from the tenfold serially diluted DNA (30, 3, 0.3, 0.03, 0.003 ng/µL) against the logarithmic concentration of DNA (Fig. 2). A good linear regression was found for all of the standard curves as reflected by the respective regression coefficient (R2), 0.9996, 0.9987, 0.9992 and 0.9988 for squirrel, rat, rabbit and cat, respectively. The corresponding slopes of each standard curve were − 3.1162, − 3.1671, − 3.174 and − 3.184, respectively. Thus, the calculated PCR efficiency was 109.36%, 106.87%, 106.53% and 106.06% for squirrel, rat, rabbit and cat, respectively. The obtained regression coefficient, corresponding slope and PCR efficiency were within the recommended values as described in published reports [35]. Previously Cheng et al. [26] found an mqPCR efficiency of 104.38, 91.75 and 97.46% for chicken, duck and pig species, respectively. On the other hand, Iwobi et al. [45] realized 101.1% and 91.6% efficiency for beef- and pork-specific multiplex qPCR system, respectively. Recently, Hossain et al. [25] got 108.73%, 107.82% and 94.68% efficiency for cow, buffalo, and pig, respectively. Thus, the developed method contained sufficient merit for the discriminatory detection of target species as the efficiency and R2 values of generated standard curves were found within recommended limits.
Sensitivity and validity of the tetraplex qPCR assay under commercial matrices
To evaluate the sensitivity of the tetraplex qPCR assay under complex matrices, two types of model meat products were made following Asing et al. [12]. The chicken and beef burger and frankfurter were spiked with 10%, 1% and 0.1% of squirrel, rat, rabbit and cat meat. The tetraplex qPCR assay was performed using the extracted DNA from adulterated meat products (beef and chicken burgers and frankfurters). For all the four target species the Ct values obtained for the lowest detectable quantity (0.1%) were 24.441 ± 0.266 to 25.83 ± 0.341 (Table 5) while that for IAC was constantly maintained mean between 14.534 ± 0.13 and 15.35 ± 0.19 for all levels of adulteration, indicating that any variation in adulteration level could not significantly change the endogenous target as all adulterants were from eukaryotic origins.
Commercial burger and frankfurter analysis
The commercial food products such as burger, meatball, frankfurter, hotdog, and nugget are very popular all over the world. These types of products are highly processed that results in total or partial annihilation or modification of morphological features and other physical properties. Therefore, manufacturers can easily mix an unexpected and lower priced meat in the final products for profit making purposes. To prevent or monitor these types of undesirable incidences, the accurate screening of commercial meat products can play a great role and build public confidence on health, religious, social and cultural perspectives. In this work, two types of commonly used commercial products, namely burger and frankfurter were evaluated using the developed tetraplex qPCR system. A total of 72 burgers (36 beef and 36 chickens) and 72 frankfurters (36 beef and 36 chickens) were purchased from different Malaysian outlets and were tested (Table 6). The experimental results revealed that no target species (cat, rabbit, rat, and squirrel) were present in the burger and frankfurter products but only IAC was amplified, reflecting the 100% accuracy of the qPCR assay. The meat of cat, rat, rabbit and squirrel species was not mixed with commercial burger and frankfurter, this might be due to the fact that halal products are under strict surveillance in Malaysia by Government agencies. Conversely, all model products were positively detected showing the efficiency of the assay (Table 6).
Conclusion
The developed tetraplex qPCR system was greatly promising in detecting cat, rabbit, rat and squirrel in a single assay platform even under any matrices and processing treatments given its species-specific primers, probes and short-length amplicon. The designed primers and probes bound only with complementary DNA at specific sites, offering a double-checking point for a well-protected secured target detection. The short amplicon size also increased the stability of the qPCR assay since short-length amplicon offers more stability over longer one for thermodynamic reasons. Additionally, the use of IAC effectively eliminated any false-negative results, enhancing the assay reliability. Above all, the designed primers, probes, shorter size of amplicon and IAC targets provided extraordinary specificity, stability and reliability of the developed tetraplex qPCR system. This assay detected 0.003 ng of DNA of the target species in pure and 0.1% of DNA in admixed states. Furthermore, it demonstrated high correlation coefficient (R2 = 0.999) between the actual values and reference values for 10–0.1% admixtures of the target species in burger and frankfurter formulations. The assay was also evaluated for the screening of commercial meat products. Overall the developed tetraplex qPCR assay for cat, rabbit, rat and squirrel was robust, specific, sensitive and cost effective for monitoring the target species in any forensic detection.
References
Ali ME, Razzak MA, Hamid SBA (2014) Multiplex PCR in species authentication: probability and prospects—a review. Food Anal Method 7:1933–1949
Anonymous (2017) Diseases carried by rats. http://www.macroevolution.net/ diseases-carried-by-rats.html. Accessed 5 Dec 2017
Joseph RB, Erick W, Beverly W (1997) Creutzfeldt-Jakob Disease and eating squirrel brains. http://www.mad-cow.org/~tom/victim23.html. Accessed 16 Dec 2017
Anonymous (2015) Grey squirrels can spread Lyme disease to humans. Retrieved from http://www.liverpoolecho.co.uk/news/uk-world-news/grey-squirrels-can-spread-lyme-9090382. Accessed 18 Dec 2017
Ahamad MNU, Ali ME, Hossain MM, Asing A, Sultana S, Jahurul M (2017) Multiplex PCR assay discriminates rabbit, rat and squirrel meat in food chain. Food Addit Contam A 34(12):2043–2057
Savadove B (2006) Cat meat dressed as mutton in street food. Retrieved from http://www.scmp.com/node/531745. Accessed 18 Dec 2017
Raj M (2015) Cat meat being sold as mutton in roadside eateries. THE TIMES OF INDIA. https://timesofindia.indiatimes.com/city/chennai/Cat-meat-being-sold-as-mutton-in-roadside-eateries/articleshow/46336721.cms. Accessed 13 Dec 2017
Ali ME, Al Amin M, Hamid SBA, Hossain MM, Mustafa S (2015) Lab-on-a-chip-based PCR-RFLP assay for the confirmed detection of short-length feline DNA in food. Food Addit Contam A 32:1373–1383
Dalle Zotte A, Szendrő Z (2011) The role of rabbit meat as functional food. Meat Sci 88:319–331
Hernandez P, Gondret F (2006) Rabbit meat quality. In: Maertens L, Coudert P (eds) Recent advances in rabbit sciences. Institute for Agricultural and Fisheries Research (ILVO), Animal Science Unit, Melle-Belgium, pp 269–290
Anitei S (2006) The origin of SARS epidemics found in civet cat meat consumption in Southern China. https://news.softpedia.com/news/The-Origin-of-SARS-Epidemics-Found-in-Civet-Cat-Meat-Consumption-in-Southern-China-40897.shtml. Accessed 20 Dec 2017
Asing Ali ME, Hamid SBA, Hossain MM, Ahamad MNU, Hossain SA, Zaidul I (2016) Duplex real-time PCR assay using SYBR Green to detect and quantify Malayan box turtle (Cuora amboinensis) materials in meatballs, burgers, frankfurters and traditional Chinese herbal jelly powder. Food Addit Contam A 33:1643–1659
Nejad FP, Tafvizi F, Ebrahimi MT, Hosseni SE (2014) Optimization of multiplex PCR for the identification of animal species using mitochondrial genes in sausages. Eur Food Res Technol 239:533–541
Focke F, Haase I, Fischer M (2010) DNA-based identification of spices: DNA isolation, whole genome amplification, and polymerase chain reaction. J Agr Food Chem 59:513–520
Lopez I, Pardo MA (2005) Application of relative quantification TaqMan real-time polymerase chain reaction technology for the identification and quantification of Thunnus alalunga and Thunnus albacares. J Agr Food Chem 53:4554–4560
Ali ME, Rashid NRA, Hamid SBA, Hossain SMA, Hossain MM, Zaidul ISM (2016) Development and validation of short-amplicon length PCR assay for macaques meat detection under complex matrices. Int J Food Prop 20(1):231–245
Hossain MM, Ali ME, Hamid SB, Mustafa S, Desa MN, Zaidul IS (2017) Targeting double genes in multiplex PCR for discriminating bovine, buffalo and porcine materials in food chain. Food Control 73:175–184
Pascoal A, Prado M, Castro J, Cepeda A, Barros-Velázquez J (2004) Survey of authenticity of meat species in food products subjected to different technological processes, by means of PCR-RFLP analysis. Eur Food Res Technol 218(3):306–312
Sultana S, Hossain MM, Naquiah NN, Ali ME (2018) Novel multiplex PCR-RFLP assay discriminates bovine, porcine and fish gelatin substitution in Asian pharmaceuticals capsule shells. Food Addit Contam A 35(9):1662–1673
Giusti A, Castigliego L, Rubino R, Gianfaldoni D, Guidi A, Armani A (2016) A conventional multiplex PCR assay for the detection of toxic Gemfish species (Ruvettus pretiosus and Lepidocybium flavobrunneum): a simple method to combat health frauds. J Agr Food Chem 64:960–968
Hossain MM, Ali ME, Hamid SB, Hossain SM, Asing Nizar NNA, Uddin MN, Ali L, Asaduzzaman M, Akanda MJH (2017) Tetraplex PCR assay involving double gene-sites discriminates beef and buffalo in Malaysian meat curry and burger products. Food Chem 224:97–104
Micheli MR, Bova R, Pascale E, D’Ambrosio E (1994) Reproducible DNA fingerprinting with the random amplified polymorphic DNA (RAPD) method. Nucleic Acids Res 22(10):1921
Moor D, Liniger M, Grohmann L, Felleisen R (2012) Real-time PCR method for the detection of figwort mosaic virus (FMV) to complement the FMV 34S promoter-specific PCR assay used for screening of genetically modified plants. Eur Food Res Technol 235(5):835–842
Abdullah A, Rehbein H (2014) Authentication of raw and processed tuna from Indonesian markets using DNA barcoding, nuclear gene and character-based approach. Eur Food Res Technol 239(4):695–706
Hossain MM, Ali ME, Sultana S, Bonny SQ, Kader MA, Rahman MA (2017) Quantitative tetraplex real-time polymerase chain reaction assay with TaqMan probes discriminates cattle, buffalo, and porcine materials in food chain. J Agrand Food Chem 65:3975–3985
Cheng X, He W, Huang F, Huang M, Zhou G (2014) Multiplex real-time PCR for the identification and quantification of DNA from duck, pig and chicken in Chinese blood curds. Food Res Int 60:30–37
O’Meara DB, Turner PD, Coffey L, O’Reilly C (2012) TaqMan assays for species identification of the red squirrel (Sciurus vulgaris) and the grey squirrel (Sciurus carolinensis). Conser Genet Res 4:603–604
Fang X, Zhang C (2016) Detection of adulterated murine components in meat products by TaqMan© real-time PCR. Food Chem 192:485–490
Rahmania H, Rohman A (2015) The employment of FTIR spectroscopy in combination with chemometrics for analysis of rat meat in meatball formulation. Meat Sci 100:301–305
Amaral JS, Santos CG, Melo VS, Oliveira MBP, Mafra I (2014) Authentication of a traditional game meat sausage (Alheira) by species-specific PCR assays to detect hare, rabbit, red deer, pork and cow meats. Food Res Int 60:140–145
Hanapi UK, Desa MNM, Ismail A, Mustafa S (2015) A higher sensitivity and efficiency of common primer multiplex PCR assay in identification of meat origin using NADH dehydrogenase subunit 4 gene. J Food Sci Tech 52:4166–4175
Rafayova A, Lieskovska Z, Trakovicka A, Kovacik A (2009) Detection of MSTN polymorphism in rabbit. Scientific Papers Anim Sci Biotechnol 42:637–641
Ali ME, Asing Hamid SBA, Razzak MA, Rashid NRA, Al Amin M, Mustafa S (2015) A suitable method to detect potential fraud of bringing Malayan box turtle (Cuora amboinensis) meat into the food chain. Food Addit Contam A 32:1223–1233
Ali ME, Ahamad MNU, Hossain MM, Sultana S (2018) Multiplex polymerase chain reaction-restriction fragment length polymorphism assay discriminates of rabbit, rat and squirrel meat in frankfurter products. Food Control 84:148–158
Ali ME, Hashim U, Dhahi TS, Mustafa S, Man YBC, Latif MA (2012) Analysis of pork adulteration in commercial burgers targeting porcine-specific mitochondrial cytochrome B gene by TaqMan probe real-time polymerase chain reaction. Food Anal Method 5:784–794
Druml B, Mayer W, Cichna-Markl M, Hochegger R (2015) Development and validation of a TaqMan real-time PCR assay for the identification and quantification of roe deer (Capreolus capreolus) in food to detect food adulteration. Food Chem 178:319–326
Cammà C, Di Domenico M, Monaco F (2012) Development and validation of fast real-time PCR assays for species identification in raw and cooked meat mixtures. Food Control 23:400–404
Amin MA, Hamid SBA, Ali ME (2016) A method for the detection of potential fraud of bringing feline meat in food chain. Int J Food Prop 19(7):1645–1658
Rashid NRA, Ali ME, Hamid SBA, Rahman MM, Razzak MA, Asing Amin MA (2015) A suitable method for the detection of a potential fraud of bringing macaque monkey meat into the food chain. Food Addit Contam A 32:1013–1022
Datukishvili N, Gabriadze I, Kutateladze T, Karseladze M, Vishnepolsky B (2010) Comparative evaluation of DNA extraction methods for food crops. Int J Food Sci Tech 45(6):1316–1320
Hird H, Lloyd J, Goodier R, Brown J, Reece P (2003) Detection of peanut using real-time polymerase chain reaction. Eur Food Res Technol 217:265–268
Ahmad Nizar NN, Ali ME, Hossain MM, Sultana S, Ahamad MN (2018) Double gene targeting PCR assay for the detection of Crocodylus porosus in commercial products. Food Addit Contam A 35(6):1038–1051
Hossain MM, Ali ME, Abd Hamid SB, Mustafa S, Mohd Desa MN, Zaidul I (2016) Double gene targeting multiplex polymerase chain reaction-restriction fragment length polymorphism assay discriminates beef, buffalo, and pork substitution in frankfurter products. J Agr Food Chem 64:6343–6354
Ahmad Nizar NN, Sultana S, Hossain MM, Johan MR, Ali ME (2018) Double gene targeting multiplex PCR-RFLP detects Crocodylus porosus in chicken meatball and traditional medicine. Int J Food Prop 21(1):2037–2051
Iwobi A, Sebah D, Kraemer I, Losher C, Fischer G, Busch U, Huber I (2015) A multiplex real-time PCR method for the quantification of beef and pork fractions in minced meat. Food Chem 169:305–313
Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HR (2005) Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn 5(2):209–219
Köppel R, Ruf J, Zimmerli F, Breitenmoser A (2008) Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, chicken and turkey. Eur Food Res Technol 227:1199–1203
Acknowledgements
This study was supported by the University of Malaya Grant no. FP054-2016 to M.E. Ali. The authors would like to thank to Dewan Bandaraya, Kuala Lumpur, and Wildlife and National Parks, Malaysia, for providing cat, dog, rat and monkey meat samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have contributed to this article and they do not have any conflict of interest to publish it in journal.
Compliance with ethics requirements
Ethical clearance of Ref. no.: NANOCAT/23/07/2013/A(R) was obtained from the Institutional Animal Care and Use Committee, University of Malaya (UM IACUC), and all experiments were conducted following the national and institutional guidelines while handling animal meats used in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ahamad, M.N.U., Hossain, M.A.M., Uddin, S.M.K. et al. Tetraplex real-time PCR with TaqMan probes for discriminatory detection of cat, rabbit, rat and squirrel DNA in food products. Eur Food Res Technol 245, 2183–2194 (2019). https://doi.org/10.1007/s00217-019-03326-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-019-03326-9