Abstract
Key message
PmSESY, a new wheat powdery mildew resistance gene was characterized and genetically mapped to the terminal region of chromosome 1RL of wild rye Secale sylvestre.
Abstract
The genus Secale is an important resource for wheat improvement. The Secale species are usually considered as non-adapted hosts of Blumeria graminis f. sp. tritici (Bgt) that causes wheat powdery mildew. However, as a wild species of cultivated rye, S. sylvestre is rarely studied. Here, we reported that 25 S. sylvestre accessions were susceptible to isolate BgtYZ01, whereas the other five confer effective resistance to all the tested isolates of Bgt. A population was then constructed by crossing the resistant accession SESY-01 with the susceptible accession SESY-11. Genetic analysis showed that the resistance in SESY-01 was controlled by a single dominant gene, temporarily designated as PmSESY. Subsequently, combining bulked segregant RNA-Seq (BSR-Seq) analysis with molecular analysis, PmSESY was mapped into a 1.88 cM genetic interval in the terminus of the long arm of 1R, which was closely flanked by markers Xss06 and Xss09 with genetic distances of 0.87 cM and 1.01 cM, respectively. Comparative mapping demonstrated that the corresponding physical region of the PmSESY locus was about 3.81 Mb in rye cv. Lo7 genome, where 30 disease resistance-related genes were annotated, including five NLR-type disease resistance genes, three kinase family protein genes, three leucine-rich repeat receptor-like protein kinase genes and so on. This study gives a new insight into S. sylvestre that shows divergence in response to Bgt and reports a new powdery mildew resistance gene that has potential to be used for resistance improvement in wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Modern cultivated wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is one of the most cultivated cereal crops that possesses abundant germplasm resources for improvement of its various desirable traits (Li et al. 2019a, b). Usually, wheat germplasm resources are classified into primary, secondary, and tertiary gene pools. The primary gene pool consists of hexaploid species containing AABBDD genome and their tetraploid and diploid progenitors, such as common wheat (AABBDD), T. spelta (AABBDD), T. dicoccoides (AABB), T. durum (AABB), T. urartu (AA), and T. tauschii (DD). The secondary gene pool includes the relatives sharing at least one homoloeogous genome with cultivated wheat, such as T. timopheevii (AAGG) and Aegilops species containing S genome related to B genome. Except the species from the primary and secondary gene pools, other distant species in Triticeae belong to the tertiary gene pool, such as Secale cereale (RR) and Dasypyrum villosum (VV) (Feuillet et al. 2007).
Blumeria graminis f. sp. tritici (Bgt) is a biotrophic fungal pathogen that causes powdery mildew in wheat and subsequent yield losses ranging from 5 to 40%. Exploring effective powdery mildew resistance genes and development of resistant wheat cultivars are important for controlling this disease (He et al. 2018; Li et al. 2020a). Up to now, 86 formally designated wheat powdery mildew resistance genes/alleles have been characterized from the primary (63), secondary (9), and tertiary (14) gene pools of wheat (McIntosh et al. 2017; Li et al. 2020b; He et al. 2020). Among the genes from the tertiary gene pool, Pm7, Pm8, Pm17, Pm20, and Pm56 are derived from S. cereale (Friebe et al. 1994; Singh et al. 2018; Hao et al. 2018), Pm21, Pm55, Pm62, and Pm67 from D. villosum (Chen et al. 1995; Zhang et al. 2016; Zhang et al. 2018a, b), Pm40 and Pm43 from Thinopyrum intermedium (Luo et al. 2009; He et al. 2009), Pm51 from Th. ponticum (Zhan et al. 2014), Pm2b from Agropryron cristatum (Ma et al. 2015), and Pm29 from Ae. ovata (Zeller et al. 2002).
The genus Secale (rye) consists of four species, including S. cereale, S. vavilovii, S. strictum (syn. S. montanum), and S. sylvestre (syn. S. fragile, Tibetan rye). Among them, S. sylvestre is an annual and autogamous wild species. It grows in sandy regions of rivers, shores, and steppe ecosystems, distributing from Hungary to Mongolia. Because morphological, cytogenetic, and molecular characters are obviously different from those of other rye species, S. sylvestre is considered to be highly divergent from other rye species (Tang et al. 2011). In wheat breeding, different species in the genus Secale are important germplasm resources; however, S. sylvestre is rarely concerned by breeders and researchers so far.
In the present study, we exploited the possibility of using S. sylvestre as a genetic resource for wheat improvement and identified five accessions conferring effective resistance against different isolates of Bgt. Using a population derived from the cross between the resistant accession SESY-01 and the susceptible accession SESY-11 of S. sylvestre, a new powdery mildew resistance gene, PmSESY, was mapped to the terminus of the long arm of chromosome 1R.
Materials and methods
Plant materials
S. sylvestre accessions were kindly provided by the National Centre for Plant Genetic Resources, Polish Genebank (NCPGR) (10), Genebank Information System of the IPK Gatersleben (GBIS-IPK) (12), and Germplasm Resources Information Network (GRIN) (8) (Table 1). S. strictum (PI 401405), S. vavilovii (PI 573649), S. cereale cv. Petkus (PI 428373), and cv. Kustro (PI 392065) were obtained from GRIN. The S. sylvestre accession SESY-01 (original accession number: 31356 in NCPGR) immune to isolate BgtYZ01 was crossed with the highly susceptible accession SESY-11 (original accession number: R 801 in GBIS-IPK), and the generated 345 F2 individuals and their corresponding F2:3 families were utilized to genetically map the powdery mildew resistance gene in SESY-01. All plants used in this study were grown under a daily cycle of 16 h of light and 8 h of darkness at 22 ± 2 °C in a greenhouse.
Evaluation of powdery mildew response to Bgt isolates
All plants of S. sylvestre accessions, F1 and F2 individuals, and ~ 50 seedlings of each F2:3 line generated from the cross SESY-01/SESY-11 were inoculated with Bgt isolate BgtYZ01 at one-leaf stage. The powdery mildew responses were evaluated at eight days after inoculation. The responses of the resistant accessions to another 15 Bgt isolates, collected from different regions of China, were also assessed, using the susceptible accession SESY-11 as control. Infection types (IT) were scored according to a 0 to 4 scale (Li et al. 2020b).
Non-denaturing fluorescence in situ hybridization (ND-FISH) analysis
S. sylvestre accessions SESY-01, SESY-11, S. strictum, S. vavilovii, S. cereale cv. Petkus, and cv. Kustro were used for ND-FISH assay. Root-tip metaphase chromosomes were prepared using the method described by Han et al. (2006). ND-FISH combined with oligonucleotide (oligo) probes Oligo-pSc119.2-1(Tang et al. 2014) and (AAC)6 was used to identify individual rye chromosomes. The ND-FISH procedure was carried out as described by Fu et al. (2015).
Molecular detection of rye species
The primers HAdh2e1 and HAdh8e1r were used for amplification of the partial Adh1 gene from S. sylvestre accessions (Petersen and Seberg 1998; Petersen et al. 2004). PCR amplification was carried out using the PrimeSTAR Max Premix (TaKaRa, Shiga, Japan). The obtained PCR products were extracted from agarose gel and then sequenced using the Sanger method. The phylogenetic tree was constructed by the neighbor-joining method in the MEGA7.0 software (Kumar et al. 2016).
Bulked segregant RNA-Seq (BSR-Seq)
The BSR-Seq method was conducted on the F2 individuals derived from the cross SESY-01/SESY-11. After estimating of the powdery mildew responses of F2 plants at one-leaf stage, equal size of the second leaves of 50 resistant and 50 susceptible plants was sampled as the R and S pools, respectively. Total RNA extraction, RNA-Seq analysis, quality control, and SNP/InDel calling were described in Wu et al. (2018) and He et al. (2020). SNP/InDel was called using the genome of rye cv. Lo7 (Martis et al. 2013; Rabanus-Wallace et al. 2019) as a reference and assessed by smoothed G (G’) value (Lott et al. 2009).
Development of molecular markers
In the target genomic region harboring powdery mildew resistance gene in SESY-01, rye genes (Martis et al. 2013; Rabanus-Wallace et al. 2019) containing the InDels and SNPs, revealed by BSR-Seq analysis, were used for developing molecular markers. Primers of InDel markers were designed according to the conserved sequences surrounding the polymorphism sites using Primer Premier 5.0. Primers of SNP markers were designed using the CAPS/dCAPS designer (Li et al. 2018). Polymorphic markers between the two parents SESY-01 and SESY-11 are listed in Table 2.
Marker analysis
Genomic DNA solution of each plant was prepared by the TE-boiling method and PCR amplified as described by He et al. (2017). PCR products of the InDel markers were directly separated in 10% non-denaturing polyacrylamide gel electrophoresis (PAGE), and those of the SNP markers were cut with the corresponding restriction enzymes (Takara, Shiga, Japan) (Table 2) prior to separation in 10% PAGE. DNA bands were visualized by silver staining.
Genetic analysis and comparative mapping
Genetic analysis of powdery mildew resistance gene in accession SESY-01 was conduct on an F2 population derived from the cross SESY-01/SESY-11. Chi-squared (χ2) test was used to determine the goodness-of-fit of the observed segregation ratio to the theoretical Mendelian ratio. Rye contigs corresponding to polymorphic markers were used to perform BLAST against the genomes of rye cv. Lo7 (Martis et al. 2013; Rabanus-Wallace et al. 2019) and wheat cv. Chinese Spring (IWGSC et al. 2018) to generate comparative genomics maps. Gene annotations of rye and wheat genomes were adopted to analyze the gene composition in the corresponding target region of S. sylvestre that was closely flanked by markers Xss06 and Xss09.
Results
Powdery mildew responses of different S. sylvestre accessions to Bgt isolates
Thirty S. sylvestre accessions obtained from different international germplasm resource institutions were tested against BgtYZ01, a virulent Bgt isolate prevailing in Yangzhou, Jiangsu province (China). The results demonstrated that five accessions (SESY-01, SESY-16, SESY-19, SESY-23, and SESY-28) were immune (IT 0), whereas the others were all complete susceptible (IT 4) (Fig. 1; Table 1). The resistance spectra of the above five resistant accessions were then assessed with another 15 Bgt isolates, collected from different regions of China. The five BgtYZ01-resistant accessions were still highly resistant, among which, accession SESY-01 conferred immunity to all isolates tested (Table 2).
Morphological, cytological and molecular characterization of S. sylvestre accessions
All plants of S. sylvestre accessions used in this study had slender culms and set slender seeds. These morphological characteristics were obviously distinguished from those of the other rye species (Tang et al. 2011). ND-FISH assay showed that the probe (AAC)6 produced signals on the satellites of 1RS arms of S. sylvestre accessions SESY-01 and SESY-11; however, these signals disappeared from the other four Secale accessions (Fig. 2a). This result was consistent with the findings of Cuadrado and Jouve (2002). In the phylogenetic tree based on the partial sequence of Adh1 gene, SESY-01, SESY-11, and S. sylvestre accession H4416 (AY294170) were clustered in the same clade (Fig. 2b). Taken together, accessions SESY-01 and SESY-11 belong to the species S. sylvestre.
Cytological and molecular detection of S. sylvestre accessions. a ND-FISH analysis of mitotic metaphase chromosomes of six Secale accessions using oligo probes Oligo-pSc119.2–1 (green) and (AAC)6 (red). Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm. b Phylogenetic tree based on the partial sequences of the Adh1 gene. GenBank accession numbers are shown in brackets
Genetic characteristics of the powdery mildew resistance in accession SESY-01
S. sylvestre accession SESY-01 immune to all tested Bgt isolates was crossed to the highly susceptible accession SESY-11 generating F1, F2, and F3 populations. After inoculation with isolate BgtYZ01, all the F1 plants displayed immunity. In the F2 population consisting of 345 individuals, 257 and 88 were resistant and susceptible, respectively, which fits to the ratio 3:1 (χ2 = 0.047, P = 0.828). In the F3 families, 90 were homozygous resistant, 167 were segregating, and 88 were homozygous susceptible, fitting to the ratio 1:2:1 (χ2 = 0.374, P = 0.829). Hence, it was concluded that the powdery mildew resistance in accession SESY-01 is governed by a single dominant gene, temporarily designated PmSESY.
BSR-Seq analysis of PmSESY
Using the RNA-Seq method, a total of 38,604,908 and 30,192,360 raw reads were obtained from the resistant and susceptible bulks, respectively. After quality control, 21,236,142 of 38,576,518 high-quality reads from the resistant pool and 17,521,940 of 30,156,107 high-quality reads from the susceptible pool were uniquely mapped to the genome of rye cv. Lo7, respectively. A total of 574,339 SNPs and InDels between the resistant and susceptible bulks were identified by variant calling, and 37,455 of them had a depth > 6. The results demonstrated that 157 SNPs and 15 InDels distributed in different chromosomes, of which, 137 SNPs and 13 InDels distributed on chromosome 1R. Further analysis revealed that most SNPs (84) and InDels (5) lied in 695–722 Mb on the long arm of chromosome 1R (1RL) (Fig. 3a, b), suggesting that this region provides powdery mildew resistance in SESY-01.
Genetic mapping of PmSESY
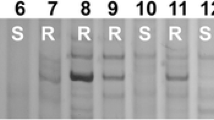
Based on the results obtained by the BSR-Seq analysis, rye genes located in 695–722 Mb of chromosome 1RL, which contained InDels and SNPs, were used for development of molecular markers. As a result, a total of 15 polymorphic markers between the two parents SESY-01 and SESY-11 were obtained, among which, 3 (Xss01, Xss02, and Xss15) were InDel markers and 12 (Xss03–Xss14) were SNP markers (Fig. 4; Table 3). These markers were then used to genotype 345 F2 individuals derived from the cross SESY-01/SESY-11. PmSESY was closely flanked by markers Xss06 and Xss09 with the corresponding genetic distances of 0.87 cM and 1.01 cM, respectively. In addition, two markers Xss07 and Xss08 were confirmed to co-segregate with PmSESY (Fig. 5a).
Polymorphic patterns of five representative markers (Xss02, Xss06, Xss09, Xss14, and Xss15). M, DL2000 DNA marker. 1–5, homozygous resistant F2 plants. 6–10, heterozygous resistant F2 plants. 11–15, homozygous susceptible F2 plants. 16, resistant accession SESY-01. 17, susceptible accession SESY-11. The polymorphic DNA bands are pointed by arrows
Genetic and comparative mapping of PmSESY. a Genetic map of PmSESY using the F2 population derived from the cross between the resistant accession SESY-01 and the susceptible accession SESY-11. b–d Comparative maps of the PmSESY locus corresponding to the orthologous regions on 1RL of rye cv. Lo7, and 1AL, 1BL and 1DL of wheat cv. Chinese Spring, respectively
Comparative mapping of PmSESY among the genomes of S. sylvestre, rye and wheat
Fifteen gene-derived markers, Xss01–Xss15, were used to carry out comparative genomics analysis among the genomes of S. sylvestre, rye cv. Lo7 and wheat cv. Chinese Spring. All the 15 markers were mapped to chromosome 1RL of the rye genome assembly. Furthermore, except the corresponding gene of marker Xss13, which could not be found on wheat 1AL, the corresponding genes of all the other markers could be well assigned to wheat chromosomes 1AL, 1BL, and 1DL (Fig. 5b–d). These results indicated that there is a good collinearity relationship among the tested genomic regions of S. sylvestre 1RL, rye 1RL, wheat 1AL, 1BL, and 1DL.
In the rye cv. Lo7 genome, the corresponding genes of flanking markers Xss06 and Xss09 were SECCE1Rv1G0061930 (Chr1R: 717,893,790–717,896,874) and SECCE1Rv1G0062920 (Chr1R: 721,703,553–721,707,206), respectively. Hence, PmSESY could be narrowed to a 3.81-Mb genomic region in the terminus of rye chromosome 1RL, where 98 genes (excluding SECCE1Rv1G0061930 and SECCE1Rv1G0062920) exist. Among them, 30 genes were involved in plant disease defense according to the annotation of rye genome, including five nucleotide-binding leucine-rich-repeat receptor (NLR)-type disease resistance genes, three kinase family protein genes, three leucine-rich repeat receptor-like protein kinase genes, three E3 ubiquitin-protein ligase genes, nine F-box protein genes, two zinc finger BED domain-containing protein genes, as well as one pathogen-related protein gene, lectin receptor kinase gene, calmodulin gene, calcium-binding protein gene, and flavin-containing monooxygenase gene each (Table 4). It was suggested that the PmSESY locus lies in a genomic region enriched with disease resistance-related genes.
Discussion
Blumeria graminis is a fungal pathogen that attacks species of the grass family, Poaceae. This pathogen is thought to be a single species but it can be classified into different forma speciales (f.sp.) according to host specialization. In general, one forma specialis infects only one specific host species (Troch et al. 2014; Menardo et al. 2017). For instance, B. graminis f. sp. tritici (B.g. tritici, Bgt) can infect wheat but not rye, whereas B. graminis f. sp. secalis (B.g. secalis, Bgs) can infect rye but not wheat. Recently, Menardo et al. (2016) reported that a forma specialis B.g. triticale can colonize on wheat, rye, and triticale, which originated from the hybridization between Bgt and Bgs in Europe where rye and triticale are widely planted. In China, both rye and triticale are small crops only planted in certain northern regions. As a result, B.g. triticale would not be prevailing in China. In this study, we found that S. sylvestre accession SESY-11 is highly susceptible to all the tested Bgt isolates collected from different wheat-producing regions of China. It was suggested that the susceptibility of SESY-11 is caused by losing resistance gene rather than by emergence of the new forma specialis B.g. triticale. We also found that most accessions of S. sylvestre examined are completely susceptible to Bgt isolate BgtYZ01. This is the first report that a species of the genus Secale can be colonized by wheat Bgt. Genetic analysis revealed that the resistance in S. sylvestre accession SESY-01 is controlled by a single dominant gene, PmSESY. Combining BSR-Seq analysis with genetic mapping, PmSESY was narrowed to a 1.88-cM genetic interval in the terminal region of the long arm of chromosome 1R (1RL), where no other powdery mildew resistance gene has been found before. Therefore, it was concluded that PmSESY is a novel gene conferring resistance against wheat powdery mildew.
The PmSESY locus corresponded to a 3.81-Mb region in rye cv. Lo7 genome, in which, 98 genes have been annotated (Rabanus-Wallace et al. 2019). Among them, 30 genes, such as NLR-type disease resistance genes and leucine-rich repeat receptor-like protein kinase genes, may serve as candidates involving in PmSESY resistance. Comparative analyses of the differences in sequences and transcriptional level of these genes between resistant SESY-01 and susceptible SESY-11 and then performing virus-induced gene silencing of differential genes in SESY-01 would allow to narrow down the PmSESY candidate(s). Further development of more high-density molecular markers according to precise reference genome of rye and larger population would contribute to fine genetic mapping of PmSESY. Moreover, using more markers in the target region carrying PmSESY to carry out association analysis on all resistant and susceptible accessions of S. sylvestre would also benefit to finding the PmSESY candidate(s).
The reference genome of rye cv. Lo7 may contribute to isolating PmSESY from S. sylvestre. However, it is not excluded that in the orthologous regions of the PmSESY locus, there may be different genomic structures and gene organizations between S. sylvestre and cultivated rye because the two species have diverged greatly during evolution (Tang et al. 2011). Recently, multiple cloning strategies have been used successfully to clone genes from wheat and barley. For example, through combination of mutagenesis with sequence capture, MutRenSeq has been adopted to identify wheat stem rust resistance genes Sr22 and Sr45 (Steuernagel et al. 2016). Based on mutagenesis, chromosome sorting, and next-generation sequencing, MutChromSeq has been used to clone the wheat powdery mildew resistance gene Pm2 (Sánchez-Martín et al. 2016) and barley leaf rust resistance gene Rph1 (Dracatos et al. 2019). These methods are independent of genetic analysis and positional cloning, even independent of reference genome. Therefore, creating susceptible mutants of S. sylvestre accession SESY-01 and using the above new methods may provide alternative ways to clone PmSESY.
The five S. sylvestre accessions were shown to be effectively resistant to all the tested Bgt isolates. It was suggested that PmSESY might possess broad-spectrum resistance to wheat powdery mildew and has great value for wheat breeding. Traditional method for utilization of genes originated from wild relatives is transferring them into common wheat through interspecific hybridization (Li et al. 2019b). In the past, S. sylvestre was crossed with Ae. tauschii and an amphiploid was obtained, suggesting that it is possible to transfer S. sylvestre genes to wheat (Yang et al. 2001). Since many wheat-rye addition lines and substitution lines involving chromosome 1R have been developed (Li et al. 2016), crossing S. sylvestre accession SESY-01 with such lines would allow the recombination between chromosomes 1R derived from different rye species, which may contribute to developing genetic stocks containing PmSESY. Recently, transgenic techniques for wheat have been fast developing and Agrobacterium tumefaciens-mediated transformation has been more stable and highly efficient (Zhang et al. 2018a, b), which will contribute to speeding up breeding application of PmSESY once it is cloned from S. sylvestre.
References
Cuadrado A, Jouve N (2002) Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. J Hered 93:339–345
Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ (1995) Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91:1125–1128
Dracatos PM, Bartoš J, Elmansour H, Singh D, Karafiátová M, Zhang P, Steuernagel B, Svačina R, Cobbin JCA, Clark B et al (2019) The coiled-coil NLR Rph1, confers leaf rust resistance in barley cultivar Sudan. Plant Physiol 179:1362–1372
Feuillet C, Langridge P, Waugh R (2007) Cereal breeding takes a walk on the wild side. Trends Genet 24:24–32
Friebe B, Heun M, Tuleen N, Zeller FJ, Gill BS (1994) Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci 34:621–625
Fu S, Chen L, Wang Y, Li M, Yang Z, Qiu L, Yan B, Ren Z, Tang Z (2015) Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep 5:10552
Han F, Lamb JC, Birchler A (2006) High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA 103:3238–3243
Hao M, Liu M, Luo J, Fan C, Yi Y, Zhang L, Yuan Z, Ning S, Zheng Y, Liu D (2018) Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat. Front Plant Sci 9:1040
He H, Ji Y, Zhu S, Li B, Zhao R, Jiang Z, Bie T (2017) Genetic, physical and comparative mapping of the powdery mildew resistance gene Pm21 originating from Dasypyrum villosum. Front Plant Sci 8:1914
He H, Liu R, Ma P, Du H, Zhang H, Wu Q, Yang L, Gong S, Liu T, Huo N et al (2020) Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor Appl Genet. https://doi.org/10.1007/s00122-020-03681-2
He H, Zhu S, Zhao R, Jiang Z, Ji Y, Ji J, Qiu D, Li H, Bie T (2018) Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol Plant 11:879–882
He R, Chang Z, Yang Z, Yuan Z, Zhan H, Zhang X, Liu J (2009) Inheritance and mapping of powdery mildew resistance gene Pm43 introgressed from Thinopyrum intermedium into wheat. Theor Appl Genet 118:1173–1180
International Wheat Genome Sequencing Consortium (IWGSC) (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Li H, Dong Z, Xia Q, Tian X, Sehgal S, Koo DH, Friebe B, Ma P, Liu W (2020) A spontaneous wheat-Aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor Appl Genet 133:1149–1159
Li H, Zhou Y, Xin W, Wei Y, Zhang J, Guo L (2019) Wheat breeding in northern China: achievements and technical advances. Crop J 7:718–729
Li J, Chen Q, Zhang P, Lang T, Hoxha S, Li G, Yang Z (2019) Comparative FISH and molecular identification of new stripe rust resistant wheat-Thinopyrum intermedium ssp. trichophorum introgression lines. Crop J 7:819–829
Li L, Liu J, Xue X, Li C, Yang Z, Li T (2018) CAPS/dCAPS Designer: a web-based high-throughput dCAPS marker design tool. Sci China Life Sci 61:992–995
Li Y, Shi X, Hu J, Wu P, Qiu D, Qu Y, Xie J, Wu Q, Zhang H, Yang L et al (2020) Identification of a recessive gene PmQ conferring resistance to powdery mildew in wheat landrace Qingxinmai using BSR-Seq analysis. Plant Dis 104:743–751
Li Z, Ren Z, Tan F, Tang Z, Ren T (2016) Molecular cytogenetic characterization of new wheat-rye 1R(1B) substitution and translocation lines from a Chinese Secale cereal L. Aigan with resistance to stripe rust. PLoS One 11:e0163642
Lott GK, Johnson BR, Bonow RH, Land BR, Hoy RR (2009) g-PRIME: a free, Windows based data acquisition and event analysis software package for physiology in classrooms and research labs. J Undergrad Neurosci Educ 8:A50–A54
Luo P, Luo H, Chang Z, Zhang H, Zhang M, Ren Z (2009) Characterization and chromosomal location of Pm40 in common wheat: a new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor Appl Genet 118:1059–1064
Ma P, Xu H, Xu Y, Li L, Qie Y, Luo Q, Zhang X, Li X, Zhou Y, An D (2015) Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor Appl Genet 128:613–622
Martis MM, Zhou R, Haseneyer G, Schmutzer T, Vrána J, Kubaláková M, König S, Kugler KG, Scholz U, Hackauf B et al (2013) Reticulate evolution of the rye genome. Plant Cell 25:3685–3698
McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Xia XC (2017) Catalogue of gene symbols for wheat: 2017 supplement (KOMUGI Wheat Genetic Resource Database). https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp
Menardo F, Praz CR, Wyder S, Ben-David R, Bourras S, Matsumae H, McNally KE, Parlange F, Riba A, Roffler S et al (2016) Hybridization of powdery mildew strains gives rise to pathogens on novel agricultural crop species. Nat Genet 48:201–205
Menardo F, Wicker T, Keller B (2017) Reconstructing the evolutionary history of powdery mildew lineages (Blumeria graminis) at different evolutionary time scales with NGS data. Genome Biol Evol 9:446–456
Petersen G, Seberg O (1998) Molecular characterization and sequence polymorphism of the alcohol dehydrogenase 1 gene in Hordeum vulgare L. Euphytica 102:57–63
Petersen G, Seberg O, Aagesen L, Frederiksen S (2004) An empirical test of the treatment of indels during optimization alignment based on the phylogeny of the genus Secale (Poaceae). Mol Phylogenet Evol 30:733–742
Rabanus-Wallace MT, Hackauf B, Mascher M, Lux T, Wicker T, Gundlach H, Báez M, Houben A, Mayer KFX, Guo L et al (2019) Chromosome-scale genome assembly provides insights into rye biology, evolution, and agronomic potential. bioRxiv. https://doi.org/10.1101/2019.12.11.869693
Sánchez-Martín J, Steuernagel B, Ghosh S, Herren G, Hurni S, Adamski N, Vrána J, Kubaláková M, Krattinger SG, Wicker T et al (2016) Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol 17:221
Singh SP, Hurni S, Ruinelli M, Brunner S, Sanchez-Martin J, Krukowski P, Peditto D, Buchmann G, Zbinden H, Keller B (2018) Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol Biol 98:249–260
Steuernagel B, Periyannan SK, Hernández-Pinzón I, Witek K, Rouse MN, Yu G, Hatta A, Ayliffe M, Bariana H, Jones JDG et al (2016) Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol 34:652–655
Tang Z, Yang Z, Fu S (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318
Tang ZX, Ross K, Ren ZL, Yang ZJ, Zhang HY, Chikmawati T, Miftahudin GJP (2011) Secale, Chapter 8. In: Kole C (ed) Wild crop relatives: genomic and breeding resources. Cereals, Springer, Berlin, pp 367–396
Troch V, Audenaert K, Wyand RA, Haesaert G, Höfte M, Brown JKM (2014) Formae speciales of cereal powdery mildew: close or distant relatives. Mol Plant Pathol 15:304–314
Wu P, Xie J, Hu J, Qiu D, Liu Z, Li T, Li M, Zhang H, Yang L, Liu H et al (2018) Development of molecular markers linked to powdery mildew resistance gene Pm4b by combining SNP discovery from transcriptome sequencing data with bulked segregant analysis (BSR-Seq) in wheat. Front Plant Sci 9:95
Yang Z, Li G, Jiang H, Ren Z (2001) Expression of nucleolus, endosperm storage proteins and disease resistance in an amphiploid between Aegilops tauschii and Secale silvestre. Euphytica 119:317–321
Zeller FJ, Kong L, Hartl L, Mohler V, Hsam SLK (2002) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 7. Gene Pm29 in line Pova. Euphytica 123:187–194
Zhan H, Li G, Zhang X, Li X, Guo H, Gong W, Jia J, Qiao L, Ren Y, Yang Z, Chang Z (2014) Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyru ponticum introgresion line. PLoS ONE 9:e113455
Zhang R, Fan Y, Kong L, Wang Z, Wu J, Xing L, Cao A, Feng Y (2018) Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor Appl Genet 131:2613–2620
Zhang R, Sun B, Chen J, Cao A, Xing L, Feng Y, Lan C, Chen P (2016) Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor Appl Genet 129:1975–1984
Zhang S, Zhang R, Song G, Gao J, Li W, Han X, Chen M, Li Y, Li G (2018) Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat. BMC Plant Biol 18:302
Acknowledgements
This study was supported by grants from Jiangsu Agricultural Science and Technology Innovation Fund [CX(19)2042], State Key Laboratory of Plant Cell and Chromosome Engineering (PCCE-KF-2019-04), Priority Academic Program Development of Jiangsu Higher Education Institutions, Jiangsu Education Department, National Natural Science Foundation of China (31872009), Leading Talents Plan of Hubei Academy of Agricultural Sciences (L2018013), Taishan Scholars Project (tsqn201812123) and State Key Laboratory of Crop Biology in Shandong Agricultural University (2020KF07). The authors are thankful to National Centre for Plant Genetic Resources, Polish Genebank (NCPGR), Genebank Information System of the IPK Gatersleben (GBIS-IPK) and Germplasm Resources Information Network (GRIN) for providing Secale sylvestre accessions. The authors are also grateful to Prof. Robert McIntosh (The University of Sydney, Australia) for constructive comments on this manuscript and Prof. Hongjie Li (Chinese Academy of Agricultural Sciences, China) for editing this manuscript.
Author information
Authors and Affiliations
Contributions
HH and SZ conceived and designed the experiments. HH, HD, RL, TL, LY, SG, ZT, HD, CL, RH, WS, and LW performed the experiments. HH and SZ analyzed the data and wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Standards
The authors state that they follow the Ethical Standards in research. We confirmed that all data in this submission have not been published previously
Additional information
Communicated by Beat Keller.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
He, H., Du, H., Liu, R. et al. Characterization of a new gene for resistance to wheat powdery mildew on chromosome 1RL of wild rye Secale sylvestre. Theor Appl Genet 134, 887–896 (2021). https://doi.org/10.1007/s00122-020-03739-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03739-1