Abstract
Key message
New powdery mildew resistance gene Pm68 was found in the terminal region of chromosome 2BS of Greek durum wheat TRI 1796. The co-segregated molecular markers could be used for MAS.
Abstract
Durum wheat (Triticum turgidum L. var. durum Desf.) is not only an important cereal crop for pasta making, but also a genetic resource for common wheat improvement. In the present study, a Greek durum wheat TRI 1796 was found to confer high resistance to all 22 tested isolates of Blumeria graminis f. sp. tritici (Bgt). Inheritance study on the F1 plants and the F2 population derived from the cross TRI 1796/PI 584832 revealed that the resistance in TRI 1796 was controlled by a single dominant gene, herein designated Pm68. Using the bulked segregant RNA-Seq (BSR-Seq) analysis combined with molecular analysis, Pm68 was mapped to the terminal part of the short arm of chromosome 2B and flanked by markers Xdw04 and Xdw12/Xdw13 with genetic distances of 0.22 cM each. According to the reference genome of durum wheat cv. Svevo, the corresponding physical region spanned the Pm68 locus was about 1.78-Mb, in which a number of disease resistance-related genes were annotated. This study reports the new powdery mildew resistance gene Pm68 that would be a valuable resource for improvement of both common wheat and durum wheat. The co-segregated markers (Xdw05–Xdw11) developed here would be useful tools for marker-assisted selection (MAS) in breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is one of the most widely cultivated cereal crops in the world, which provides about 18% of daily dietary calories consumed by humans (Tan et al. 2018; Li et al. 2019). Wheat production is greatly threatened by powdery mildew, caused by the fungal pathogen Blumeria graminis f. sp. tritici (Bgt) that has complex and variable virulence structures in natural populations (Wicker et al. 2013). Exploiting and utilizing broad-spectrum powdery mildew resistance (Pm) genes are important and urgent to effectively control the mildew disease. Although more than a hundred Pm genes/alleles have been documented, a few can provide resistance to most or all Bgt isolates (McIntosh et al. 2017; Chen et al. 2019). Commonly, Pm genes originated from distant relatives of wheat have resistance to more isolates, such as Pm12 from Aegilops speltoides Tausch (Song et al. 2009; Zhang et al. 2019) and Pm21 from Dasypyrum villosum L. Candagy (Chen et al. 1995; He et al. 2018). Some rare natural variations occurred in common wheat genes can also confer broad-spectrum resistance to powdery mildew, such as Pm24 (Lu et al. 2020), Pm38/Yr18/Lr34/Sr57 (Krattinger et al. 2009) and Pm46/Yr46/Lr67/Sr55 (Moore et al. 2015), among which the latter two genes also provide durable resistance to other diseases.
Durum wheat (Triticum turgidum L. var. durum Desf., simply T. durum, 2n = 4x = 28, AABB) is a tetraploid wheat species that is a relatively small cereal crop mainly used for pasta making. Durum wheat possesses good resistance to leaf rust, stem rust and stripe rust and has been used for wheat improvement (Miedaner et al. 2019). However, most durum wheat accessions are susceptible to powdery mildew. From this crop, only three powdery mildew resistance genes have been identified in the past decades, including Mld, Pm3h and PmDR147. Mld is a recessive gene reported on 4B that has been combined with other powdery mildew resistance gene, such as Pm2, and used for wheat breeding (Bennett 1984). Pm3h is a dominant resistance gene located on chromosome 1AS, probably originated from an Ethiopian durum wheat accession (Srichumpa et al. 2005). Pm3h has been cloned and confirmed to be identical to Pm3d at the nucleotide sequence level (Yahiaoui et al. 2006). PmDR147 is another dominant gene identified on 2AL of durum wheat accession DR147 (Zhu et al. 2004).
BgtYZ01, a super virulent isolate, was originally collected from Yangzhou, Jiangsu Province, China, where Bgt pathogen is prevailing (He et al. 2016, 2017). To effectively control this isolate, we attempted to screen new resistance resources from different relatives of wheat. From 100 durum wheat accessions, one resistant landrace TRI 1796 collected from Greece was obtained. TRI 1796 also conferred resistance to other 21 tested isolates of Bgt at the seedling stage and showed high resistance at the adult plant stage in fields of different regions. In the present study, genetic and comparative mapping of Pm68 in durum wheat TRI 1796 was carried out, which will facilitate its application in both common wheat breeding and durum wheat breeding.
Materials and methods
Plant materials
A collection of 100 accessions of T. durum were kindly provided by Genebank Information System of the IPK Gatersleben (GBIS-IPK) (82) and Germplasm Resources Information Network (GRIN) (18). The Greek T. durum accession TRI 1796 highly resistant to isolate BgtYZ01 was crossed with susceptible Canadian T. durum accession PI 584832, and the generated 224 F2 individuals and their corresponding F2:3 families were used to map the powdery mildew resistance gene in TRI 1796. The resistant wheat cv. Yangmai 18 carrying Pm21 and susceptible wheat cv. Yangmai 23 were kindly provided by Dr. Tongde Bie (Yangzhou Academy of Agricultural Sciences, China). The wheat lines carrying Pm26 and Pm42 were kindly provided by Prof. Zhiyong Liu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China).
Evaluation of powdery mildew response to isolate BgtYZ01
All T. durum accessions, F1 and F2 individuals as well as ~ 40 seedlings of each F2:3 line derived from the cross TRI 1796/PI 584832 at one-leaf stage were inoculated with isolate BgtYZ01 that is a highly virulent isolate collected from Yangzhou (He et al. 2016). The inoculated plants were grown under a daily cycle of 16 h of light and 8 h of darkness at 22 ± 2 °C in a greenhouse. The powdery mildew responses were assessed at 8 days after inoculation. The responses of parents TRI 1796 and PI 584832 to a set of 22 Bgt isolates collected from different regions of China were also detected. The resistance spectrum of TRI 1796 was compared with those of the wheat lines carrying Pm21, Pm26 and Pm42. Infection types (IT) were scored according to a 0–4 scale. ITs 0, 0, 1 and 2 were considered resistant, while those with an IT score of 3 and 4 were considered as susceptible (Li et al. 2020). Powdery mildew response of TRI 1796 at the adult plant stage in field was also estimated in Zhenjiang (Jiangsu Province), Yantai (Shandong Province) and Jinan (Shandong Province) in the 2018–2019 and 2019–2020 sowing seasons. Twenty seeds of each material were sown in a 1-m row. The flanking rows of TRI 1796 and PI 584832 were planted susceptible wheat cv. Yangmai 23 as the spreader. A mixture of Bgt isolates collected from the local site was inoculated on TRI 1796, PI 584832 and the spreader wheat at the jointing stage. At the milk stage, powdery mildew responses of the flag leaves were assessed on a 0–9 scale. Plants with scale 0, 1–2, 3–4, 5–6 and 7–9 were considered immune, highly resistant, moderately resistant, moderately susceptible and highly susceptible, respectively (Li et al. 2011).
Bulked segregant RNA-Seq (BSR-Seq)
The BSR-Seq method was conducted on the F2 individuals derived from the cross TRI 1796/PI 584832. Powdery mildew responses of the F2 individuals were assessed at one-leaf stage. Then, equal size of the second leaves of 60 resistant and 60 susceptible individuals were pooled separately. Total RNA of the two bulks of leaf samples was separately extracted using Illumina TruSeq RNA Sample Prep Kit (Illumina, Inc., San Diego, CA, United States) to be used for RNA-Seq analysis using the platform of Illumina HiSeq 4000 (Beijing Southern Genome Research Technology Co., Ltd., Beijing, China). The raw sequencing reads generated were quality controlled using software Trimmomatic v0.36 (Bolger et al. 2014) with default parameters. Using software STAR v2.5.1b (Dobin et al. 2013), the clean reads were aligned to the genome assembly sequences of durum wheat cv. Svevo (https://www.interomics.eu/durum-wheat-genome; Maccaferri et al. 2019) with the mismatch rate of less than 5%. The uniquely mapped read pairs were used in further analysis. The read alignments were masked for PCR duplications and split for reads spanning introns before they were used to call SNPs and InDels using module “HaplotypeCaller” of software GATK v3.6 (McKenna et al. 2010). The resulting SNPs and InDels with sequencing depth less than 6 were discarded, and the remaining ones were applied to BSA. Only variants with allele frequency difference (AFD) > 0.6 and P value of Fisher’s exact test on read count data < 1e−8 were classified as resistance-associated variants and used as templates for marker development. The genome of durum wheat cv. Svevo was further used as a reference to call SNPs and InDels.
Development of molecular markers
A total of 56 SSR markers reported to be located on chromosome 2BS (Somers et al. 2004; Hua et al. 2009; Liu et al. 2012) were used to screen polymorphisms between two parents TRI 1796 and PI 584832. The genes showed polymorphic SNP and their corresponding 3000-bp upstream and 3000-bp downstream sequences in the target region revealed by BSR-Seq were retrieved from the reference genome of durum wheat cv. Svevo (https://www.interomics.eu/durum-wheat-genome; Maccaferri et al. 2019) and used to perform BLAST against the genome of durum wheat cv. Kronos (https://opendata.earlham.ac.uk/opendata/data/Triticum_turgidum/EI/v1.1). The conserved sequences flanking the InDel regions between durum wheat cv. Svevo and cv. Kronos were then used to design primer pairs. A total of 171 gene-derived primer pairs were used to detect the polymorphisms between TRI 1796 and PI 584832.
Marker analysis
The crude genomic DNA solution was prepared using the TE-boiling method (He et al. 2017). PCR amplification was performed with polymorphic markers between the two parents TRI 1796 and PI 584832. Each reaction mixture (25 μl) contained 1 × PCR buffer (Mg2+ free), 2.7 mM of MgCl2, 0.2 mM of dNTP, 2 μM of each primer, 1 U of Taq DNA polymerase (Takara, Shiga, Japan) and 1 μl of DNA solution. DNA amplification was programmed at 94 °C for 3 min; 35 cycles of 94 °C for 20 s, 60 °C for 30 s, 72 °C for 1 min; at 72 °C for 5 min. PCR products of InDel markers were separated in 8% non-denaturing polyacrylamide gels, followed by silver staining. DNA fragment of TRITD2Bv1G010030 harboring a SNP site was obtained by PCR with marker Xdw04 and the polymorphism was then detected by Sanger sequencing.
Data analysis
Genetic analysis was performed on an F2 population derived from the cross TRI 1796/PI 584832. Chi-squared (χ2) test was used to determine the goodness-of-fit of the observed segregation ratio to theoretical Mendelian ratio.
Comparative genomics analysis
The corresponding genes of markers Xdw03 and Xdw16 linked to Pm68 were used to BLAST against the genomes of durum wheat cv. Svevo and common wheat cv. Chinese Spring (http://www.wheat-urgi.versailles.inra.fr; IWGSC et al. 2018). Gene annotations in the target intervals of the two genomes were provided by the online databases. For comparative mapping, the genomic information of T. dicoccoides (https://wheat.pw.usda.gov/GG3/wildemmer; Avni et al. 2017), T. urartu (http://202.194.139.32; Lin et al. 2018) and Aegilops tauschii (http://aegilops.wheat.ucdavis.edu/ATGSP/index.php; Luo et al. 2017) were also considered.
Results
Powdery mildew responses of different durum wheat accessions against isolate BgtYZ01
A collection of 100 accessions of durum wheat was used to assess powdery mildew responses to Bgt isolate BgtYZ01 at the seedling stage. The results showed that only one Greek landrace TRI 1796 was highly resistant (IT 0), whereas all the others were completely susceptible (IT 4). The resistance spectrum of TRI 1796 was further analyzed. The results demonstrated that TRI 1796 conferred effective resistance to 20 isolates (including BgtYZ01) at the level of IT 0; and 2 isolates at the level of IT 1. It was also found that isolates BgtYZ01, Bgt10 and Bgt16 completely infected the wheat line carrying Pm26 and isolates BgtYZ01, Bgt10, Bgt13 and Bgt19 completely infected the wheat line carrying Pm42 (Fig. 1a; Table 1). These results revealed that the resistance in TRI 1796 differs from those of Pm26 and Pm42. TRI 1796 and PI 584832 were also planted in fields of different regions during 2018–2020. Powdery mildew responses of their flag leaves were investigated at the milk stage. In each environment, TRI 1796 was immune (scale 0) to the mixture of Bgt isolates, whereas PI 584832 was highly susceptible (scale 7–9) (Fig. 1b). Hence, it was indicated that TRI 1796 could confer effective resistance against powdery mildew at the seedling and adult plant stages.
Powdery mildew responses of durum wheat accessions TRI 1796 and PI 584832. a Powdery mildew responses of TRI 1796 and PI 584832 to Bgt isolate BgtYZ01 at one-leaf stage, compared with wheat lines carrying Pm26, Pm42 and Pm21. b Powdery mildew responses of TRI 1796 and PI 584832 to Bgt natural population in field at the adult plant stage. The flag leaves shown here were collected from Yantai (Shandong Province) in the 2019–2020 sowing season
Genetic characteristics of the powdery mildew resistance in TRI 1796
The resistant Greek durum wheat TRI 1796 was then crossed with the susceptible Canada durum wheat PI 584832 (IT 4). When inoculated with isolate BgtYZ01, all the F1 plants showed resistance at the level of IT 0; (Fig. 1a) and the individuals of the F2 population showed resistance at the level of IT 0; or susceptibility at the level of IT 4. Among the 224 F2 plants, the segregation ratio of the resistant (166) and susceptible (58) individuals fits for 3:1, the theoretical Mendelian segregation ratio (χ2 = 0.10, P = 0.76). The F3 families segregated 56 homozygous resistant: 110 segregating: 58 homozygous susceptible, fitting for the ratio 1:2:1 (χ2 = 0.19, P = 0.91). Therefore, it was concluded that the powdery mildew resistance in TRI 1796 was controlled by a single dominant gene, which has been designated Pm68 based on the following analyses.
BSR-Seq analysis of Pm68 in durum wheat
Using RNA-Seq, a total of 28,722,281 and 27,111,764 raw reads were obtained from the resistant bulk and the susceptible bulk, respectively. After quality control, 23,969,694 of 28,703,103 high-quality reads from the resistant bulk and 21,673,784 of 27,095,223 high-quality reads from the susceptible bulk were uniquely mapped to the genome of durum wheat cv. Svevo, respectively. A total of 141,335 SNPs and InDels between the resistant and susceptible bulks were identified by variant calling, and 40,584 of them had a depth > 6. The result showed that 116 SNPs distributed in different chromosomes may be associated with the resistance provided by Pm68. Among them, 42 SNPs were enriched in an about 8.6-Mb region (16.2–24.8 Mb) on the short arm of chromosome 2B (2BS) (Fig. 2).
Genetic mapping of Pm68
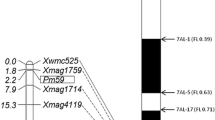
To screen polymorphic markers between two parents TRI 1796 and PI 584832, 56 SSR markers previously reported on 2BS were first used but none of them showed polymorphism. Then, 171 primer pairs were designed based on the InDels of the genes in the target regions of durum wheat cv. Svevo and cv. Kronos and 18 of them showed co-dominant polymorphisms between TRI 1796 and PI 584832. In addition, one SNP marker, Xdw04, corresponding to TRITD2Bv1G010030, was also developed (Fig. 3a, b; Table 2). Subsequently, these markers were used to genotype 224 F2 individuals derived from the cross between resistant TRI 1796 and susceptible PI 584832. Pm68 was closely flanked by markers Xdw04 and Xdw12/Xdw13, and both the genetic distances between Xdw04 and Pm68 and Xdw12/Xdw13 and Pm68 were 0.22 cM (Fig. 4a). Seven markers (Xdw05–Xdw11) were confirmed to co-segregate with Pm68. Among them, Xdw06, Xdw07 and Xdw08 were resistance gene analog (RGA) markers that were developed based on NBS-LRR-like genes TRITD2Bv1G010130, TRITD2Bv1G010230 and TRITD2Bv1G010240, respectively.
Polymorphic patterns of six representative markers. a PCR amplification patterns of five InDel markers (Xdw08, Xdw10, Xdw12, Xdw14, and Xdw15). M, DL2000 DNA marker. 1–5, homozygous resistant F2 plants. 6–10, heterozygous resistant F2 plants. 11–15, homozygous susceptible F2 plants. 16, resistant durum wheat TRI 1796. 17, susceptible durum wheat PI 584832. The polymorphic DNA bands are pointed by arrows. b DNA sequence chromatograms of PCR products obtained with SNP marker Xdw04. The polymorphic nucleotides marked by arrows are C, T and C/T in homozygous susceptible, homozygous resistant and heterozygous resistant individuals, respectively
Genetic map and comparative genomics map of chromosome 2BS carrying Pm68. a Genetic map of Pm68 which is closely flanked by markers Xdw04 and Xdw12/Xdw13. b–f Physical maps of homologous/orthologous regions of Pm68 on 2BS of durum wheat (T. durum) cv. Svevo, common wheat cv. Chinese Spring (T. aestivum) and wild emmer (T. dicoccoides), 2AS of T. urartu and 2DS of Ae. tauschii, respectively. Bar: 200 kb
Comparative mapping of Pm68 among related genomes
Fourteen gene-derived markers (Xdw04–Xdw17) were further used to perform comparative genomics analysis among the genomes of durum wheat cv. Svevo, common wheat cv. Chinese Spring, T. dicoccoides, T. urartu, and Ae. tauschii (Fig. 4b–f). The data demonstrated relatively good collinearity relationship between B genomes of durum wheat cv. Svevo and common wheat cv. Chinese Spring. In durum wheat, the corresponding genes of flanking markers Xdw04 and Xdw12 were TRITD2Bv1G010030 (Chr2B: 21587671–21591163) and TRITD2Bv1G010880 (Chr2B: 23374401–23375310), respectively. Therefore, Pm68 was narrowed to a 1.78-Mb genomic region on 2BS a durum wheat cv. Svevo. According to the gene annotation published, a total of 84 genes exist in the target physical region. Among them, 23 genes may be associated with plant disease defense, including 8 NBS-LRR-like resistance genes, 6 exocyst complex component genes, 2 LRR receptor-like protein kinase gene, one lectin receptor kinase gene, receptor protein kinase gene, calcineurin B-like gene, F-box/RNI-like superfamily gene, wall-associated kinase gene, chitinase gene and heat shock protein 90 gene each (Table 3).
We further analyzed the evolution of these disease resistance-related genes in the genomes of wheat species. The data showed that all the 23 durum wheat genes had the corresponding homologous genes or perfectly matched genomic sequences in wheat Chinese Spring and 19 of them maintained good collinearity relationship between durum wheat and common wheat. In particular, the 8 NBS-LRR-like resistance genes of durum wheat shared 99–100% identities with those of common wheat. Of the 23 genes of durum wheat, 22 had homologous genes/sequences in the genome of wild emmer and relatively good collinearity was also observed. However, the identities (75–100%) of these genes/sequences between durum wheat and wild emmer were relatively lower than those between durum wheat and common wheat. In T. urartu genome, 20 of 23 disease resistance-related genes could be found with 68–97% identities, but they did not keep the same order as shown in durum wheat genome. In addition, most of the 23 durum wheat genes could not match Ae. tauschii genes (Supplementary Table S1). In conclusion, most of the predicted candidate genes were highly conserved in the B- and A-subgenomes of wheat species.
Discussion
Three powdery mildew resistance gene, Mld (4B), PmDR147 (2AL) and Pm3h (1AS), have been documented in durum wheat (Bennett 1984; Zhu el al. 2004; Srichumpa et al. 2005). In the present study, a dominant powdery mildew resistance gene Pm68 was identified and mapped on the terminal part of 2BS in a Greek landrace durum wheat TRI 1796. In wheat crops, a total of six powdery mildew resistance genes have been reported on 2BS, including two dominant (Ml5323 and PmL962), three recessive (Pm26, Pm42 and PmWE99) and one incomplete dominant (MlIW170) genes. Among them, Pm26, Pm42 and MlIW170 are derived from T. dicocoides, PmWE99 and PmL962 from Thinopyrum intermedium, and Ml5323 from T. dicoccum (Rong et al. 2000; Hua et al. 2009; Piarulli et al. 2012; Liu et al. 2012; Liang et al. 2015; Shen et al. 2015; Ma et al. 2016). We comparatively analyzed the chromosomal locations of the six reported genes and found that Pm42, MlIW170, Ml5323 and PmL962 are all in the direction of proximal side of their common markers Xcau516/Xwg516/BF202540 (Fig. 5), and Pm26 is considered to be located in the same genomic region or be allelic to MlIW170 (Liu et al. 2012; Liang et al. 2015). Marker Xcau516/Xwg516/BF202540 responds to the durum wheat gene TRITD2Bv1G012960 (26.40 Mb). In this study, the closely linked marker Xgw12 of Pm68 responds to TRITD2Bv1G010880 (23.37 Mb). Both TRITD2Bv1G012960 and TRITD2Bv1G010880 lie in the proximal direction of Pm68. Therefore, the physical location of Pm68 is obviously different from those of the 5 reported genes Pm26, Pm42, MlIW170, Ml5323 and PmL962.
The sixth powdery mildew resistance gene is PmWE99 on 2BS. Although the relationship of PmWE99 and markers Xcau516/Xwg516/BF202540 are not clear, the genetic distance between PmWE99 and another marker Xgwm148 is tested to be 10.4 cM (Fig. 5; Ma et al. 2016). Xgwm148 can match at the position 101.24 Mb in the reference genome of durum wheat which lies between markers Xgw18 (94.79 Mb) and Xgw19 (105.45 Mb). In this study, the genetic distance between Pm68 and Xgw18 was 27.45 cM which is rather larger than 10.4 cM between PmWE99 and Xgwm148. Furthermore, Pm68 from durum wheat inherits as a dominant gene, whereas PmWE99 from Th. intermedium is a recessive gene. Hence, it was indicated that Pm68 also differs from PmWE99. Taken together, Pm68 reported here is a new powdery mildew resistance gene found in wheat crops.
Pm68 was mapped to a 0.44-cM genetic interval, flanked by markers Xdw04 and Xdw12, corresponding to the genes TRITD2Bv1G010030 and TRITD2Bv1G010880, respectively, suggesting the Pm68 locus responding to the physical region 21.59–23.37 Mb in the reference genome of durum wheat cv. Svevo. In the target region, a total of 84 genes have been annotated. Among them, 8 encode NBS-LRR-like resistance protein, 6 encode exocyst complex components and 9 encode other proteins, such as receptor-like kinase, wall-associated kinase, chitinase and heat shock protein 90. Comparative genomics analysis indicated that most of them were conserved in the B- and A-subgenomes of wheat species. Further comparative analyses of the sequences and the transcription patterns of these genes derived from resistant accession TRI 1796 and susceptible accession PI 584832 would allow to narrow the range of candidates of Pm68. However, there are many retrotransposons and transposons in the reference genome of durum wheat, accounting for 33.3%, which have potential impacts on the genome structure. Therefore, it is not excluded that the Pm68 locus in TRI 1796 has different gene organization from the reference. In addition, more saturated molecular markers and a larger population are required for fine mapping and cloning of Pm68.
Powdery mildew is a serious disease of durum wheat. Commonly, excellent resistance resource is lacking for controlling this disease in durum wheat (Bennett 1984). We investigated 100 durum wheat accessions hold by GBIS-IPK (82) and GRIN (18) and found only one accession resistant to virulent isolate BgtYZ01, suggesting that the resistant durum wheat is relatively rare resource. Resistance spectrum analysis demonstrated that Greek accession TRI 1796 carrying Pm68 possesses effective resistance to all 22 tested isolates of Bgt pathogen at the seedling stage, accompanied by hypersensitive response (HR). Interestingly, Pm68 conferred complete immunity to powdery mildew at the adult plant stage in fields of three regions of China in two sowing seasons (2018–2019 and 2019–2020). It is suggested that Pm68 resistance might be improved at the adult plant stage or there might be other gene(s) providing adult plant resistance in TRI 1796. Therefore, accession TRI 1796 would be a very useful resource for durum wheat breeding. When we mapped Pm68, seven co-segregated markers (Xdw05–Xdw11) were developed that would be used as powerful tools in marker-assisted selection (MAS). Furthermore, as one of the direct progenitors of common wheat, durum wheat is easy to cross with common wheat and generate offspring. Hence, Pm68 in durum wheat TRI 1796 also has potential to be used for common wheat resistance improvement.
References
Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale L, Mascher M, Spannagl M, Wiebe K et al (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357:93–97
Bennett FGA (1984) Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes. Plant Pathol 33:279–300
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ (1995) Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91:1125–1128
Chen F, Jia H, Zhang X, Qiao L, Li X, Zheng J, Guo H, Powers C, Yan L, Chang Z (2019) Positional cloning of PmCH1357 reveals the origin and allelic variation of the Pm2 gene for powdery mildew resistance in wheat. Crop J 7:771–783
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
He H, Zhu S, Jiang Z, Ji Y, Wang F, Zhao R, Bie T (2016) Comparative mapping of powdery mildew resistance gene Pm21 and functional characterization of resistance-related genes in wheat. Theor Appl Genet 129:819–829
He H, Ji Y, Zhu S, Li B, Zhao R, Jiang Z, Bie T (2017) Genetic, physical and comparative mapping of the powdery mildew resistance gene Pm21 originating from Dasypyrum villosum. Front Plant Sci 8:1914
He H, Zhu S, Zhao R, Jiang Z, Ji Y, Ji J, Qiu D, Li H, Bie T (2018) Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol Plant 11:879–882
Hua W, Liu Z, Zhu J, Xie C, Yang T, Zhou Y, Duan X, Sun Q, Liu Z (2009) Identification and genetic mapping of pm42, a new recessive wheat powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides). Theor Appl Genet 119:223–230
International Wheat Genome Sequencing Consortium (IWGSC), IWGSC RefSeq principal investigators, Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, IWGSC whole-genome assembly principal investigators, Pozniak CJ et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Li H, Wang X, Song F, Wu C, Wu X, Zhang N, Zhou Y, Zhang X (2011) Response to powdery mildew and detection of resistance genes in wheat cultivar from China. Acta Agron Sin 37:943–954
Li H, Zhou Y, Xin W, Wei Y, Zhang J, Guo L (2019) Wheat breeding in northern China: achievements and technical advances. Crop J 7:718–729
Li H, Dong Z, Xia Q, Tian X, Sehgal S, Koo DH, Friebe B, Ma P, Liu W (2020) A spontaneous wheat-Aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor Appl Genet 133:1149–1159
Liang Y, Zhang D, Ouyang S, Xie J, Wu Q, Wang Z, Cui Y, Lu P, Zhang D, Liu Z et al (2015) Dynamic evolution of resistance gene analogs in the orthologous genomic regions of powdery mildew resistance gene MlIW170 in Triticum dicoccoides and Aegilops tauschii. Theor Appl Genet 128:1617–1629
Lin H, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y et al (2018) Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557:424–428
Liu Z, Zhu J, Cui Y, Liang Y, Wu H, Song W, Liu Q, Yang T, Sun Q, Liu Z (2012) Identification and comparative mapping of a powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides) on chromosome 2BS. Theor Appl Genet 124:1041–1049
Lu P, Guo L, Wang Z, Li B, Li J, Li Y, Qiu D, Shi W, Yang L, Wang N et al (2020) A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat Commun 11:680
Luo M, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo N, Zhu T, Wang L, Wang Y et al (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551:498–502
Ma P, Xu H, Han G, Luo Q, Xu Y, Zhang X, An D, Li L, Sun Y (2016) Characterization of a segregation distortion locus with powdery mildew resistance in a wheat-Thinopyrum intermedium introgression line WE99. Plant Dis 100:1541–1547
Maccaferri M, Harris NS, Twardziok SO, Pasam RK, Gundlach H, Spannagl M, Ormanbekova D, Lux T, Prade VM, Milner SG et al (2019) Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet 51:885–895
McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Xia XC (2017) Catalogue of gene symbols for wheat: 2017 supplement (KOMUGI Wheat Genetic Resource Database). https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Miedaner T, Rapp M, Flath K, Longin CFH, Würschum T (2019) Genetic architecture of yellow and stem rust resistance in a durum wheat diversity panel. Euphytica 215:71
Moore JW, Herrera-Foessel S, Lan C, Schnippenkoetter W, Ayliffe M, Huerta-Espino J, Lillemo M, Viccars L, Milne R, Periyannan S et al (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet 47:1494–1498
Piarulli L, Gadaleta A, Mangini G, Signorile MA, Pasquini M, Blanco A, Simeone R (2012) Molecular identification of a new powdery mildew resistance gene on chromosome 2BS from Triticum turgidum ssp. dicoccum. Plant Sci 196:101–106
Rong JK, Millet E, Manisterski J, Feldman M (2000) A new powdery mildew resistance gene: introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 115:121–126
Shen XK, Ma LX, Zhong SF, Liu N, Zhang M, Chen WQ, Zhou YL, Li HJ, Chang ZJ, Li X, Bai GH, Zhang HY, Tan FQ, Ren ZL, Luo PG (2015) Identification and genetic mapping of the putative Thinopyrum intermedium-derived dominant powdery mildew resistance gene PmL962 on wheat chromosome arm 2BS. Theor Appl Genet 128:517–528
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song W, Xie C, Du J, Xie H, Liu Q, Ni Z, Yang T, Sun Q, Liu Z (2009) A “one-marker-for-two-genes” approach for efficient molecular discrimination of Pm12 and Pm21 conferring resistance to powdery mildew in wheat. Mol Breed 23:357–363
Srichumpa P, Brunner S, Keller B, Yahiaoui N (2005) Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 139:885–895
Tan C, Li G, Cowger C, Carver B, Xu X (2018) Characterization of Pm59, a novel powdery mildew resistance gene in Afghanistan wheat landrace PI 181356. Theor Appl Genet 131:1145–1152
Wicker T, Oberhaensli S, Parlange F, Buchmann JP, Shatalina M, Roffler S, Ben-David R, Doležel J, Šimková H, Schulze-Lefert P et al (2013) The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat Genet 45:1092–1096
Yahiaoui N, Brunner S, Keller B (2006) Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J 47:85–98
Zhang D, Zhu K, Dong L, Liang Y, Li G, Fang T, Guo G, Wu Q, Xie J, Chen Y et al (2019) Wheat powdery mildew resistance gene Pm64 derived from wild emmer (Triticum turgidum var. dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5. Crop J 7:761–770
Zhu Z, Kong X, Zhou R, Jia J (2004) Identification and microsatellite markers of a resistance gene to powdery mildew in common wheat introgressed from Triticum durum. Acta Botan Sin 46:867–872
Acknowledgements
This study was supported by grants from Jiangsu Agricultural Science and Technology Innovation Fund [CX(19)2042], Priority Academic Program Development of Jiangsu Higher Education Institutions, Jiangsu Education Department, Key Research and Development Program of Yantai City (2019YT06000470), National Natural Science Foundation of China (31872009), Leading Talents Plan of Hubei Academy of Agricultural Sciences (L2018013) and State Key Laboratory of Crop Biology in Shandong Agricultural University (2020KF07). The authors are thankful to Genebank Information System of the IPK Gatersleben (GBIS-IPK) and Germplasm Resources Information Network (GRIN) for providing durum wheat accessions, and Prof. Zhiyong Liu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China) for providing the wheat lines carrying Pm26 and Pm42. The authors are also grateful to Prof. Robert McIntosh (The University of Sydney, Australia) for constructive comments on this manuscript.
Author information
Authors and Affiliations
Contributions
HH, YQG and SZ conceived and designed the experiments. HH, RL, PM, HD, HZ, QW, LY, SG and TL performed the experiments. HH, RL, PM and NH analyzed the data. HH, RL, YQG and SZ wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Evans Lagudah.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table S1
Information of 23 disease resistance-related genes in different genomes of wheat species. - means no matched gene or sequence. (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
He, H., Liu, R., Ma, P. et al. Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor Appl Genet 134, 53–62 (2021). https://doi.org/10.1007/s00122-020-03681-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03681-2