Abstract
Monoamine oxidases (MAOs) are flavoenzymes that metabolize biogenic amines, dietary amines, and catecholamines in the brain and peripheral tissues. While MAOs are known to contribute to psychiatric and neurodegenerative (Parkinson’s and Alzheimer’s) diseases for a long time, recent studies have established their role in heart diseases as these enzymes potently generate reactive oxygen species (ROS) in cardiomyocytes via oxidative deamination of mainly norepinephrine and serotonin. Indeed, MAOs have emerged as important regulators of mitochondrial/endothelial/cardiac dysfunction, essential hypertension, ventricular hypertrophy, myocardial infarction, cardiomyocyte apoptosis, postischemic cardiac damage, and heart failure. Transcriptional and posttranscriptional regulation of MAOs (via certain transcription factors or microRNAs) may emerge as new therapeutic strategies for treatment of cardiovascular pathological conditions. The next-generation MAO inhibitors (that do not cause irreversible inhibition of MAOs) may also be useful for management of cardiovascular disease states involving dysregulated expression/activity of MAOs.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Monoamine oxidases (MAOs) (EC 1.4.3.4) are flavin adenine dinucleotide (FAD)-dependent enzymes which metabolize biogenic amines, dietary amines, and catecholamines (viz., epinephrine, norepinephrine, and dopamine) in the brain and peripheral tissues. MAOs oxidatively deaminate these amines into corresponding aldehydes and generate hydrogen peroxide (H2O2) and ammonia (NH3) during this reaction. Aldehydes generated in these reactions are further metabolized into corresponding organic acids by aldehyde dehydrogenases [1]. MAOs are expressed as integral proteins in the outer membrane of mitochondria. Based on the differences observed in substrate/inhibitor specificity and cell-/tissue-specific expression, MAOs are classified into two types, namely, MAOA and MAOB [2]. In brief, epinephrine, norepinephrine, and serotonin are preferentially metabolized by MAOA, while phenylalanine and benzylamine are mainly metabolized by MAOB. Dopamine, tyramine, and tryptamine are common substrates for both the MAOs [3]. Selective MAOA and MAOB inhibitors are clinically used to treat depression and Parkinson’s disease [4].
Apart from sharing ~70% identity between their amino acid sequences, both MAOs have a conserved pentapeptide sequence (viz., Ser-Gly-Gly-Cys-Tyr), which serves as the FAD binding domain [1]. In several mammalian species including human, mouse, and rat, MAOs are mapped to the p arm of the X chromosome; these two genes are located next to each other in a tail-to-tail fashion. Identical exon-intron organization, equal number of exons, and high sequence similarity suggest that MAOA and MAOB are derived from a common ancestral gene (Fig. 6.1). Both MAOs are ubiquitously expressed in all cell types except red blood cells in a tissue-specific manner [5]. The human heart contains high levels of both isozymes; in the rat heart, MAOA is abundant, while MAOB is almost absent and the reverse is true for the mouse heart [6, 7]. MAOA expression is regulated by several transcription factors including circadian-clock components (via E-box elements), GATA2 (GATA binding protein-2), Krüppel-like factor-11 (Klf11), R1, sirtuin 1, Sp1 (specificity protein 1), SRY (sex-determining region gene on the Y chromosome), and TBP (TATA-binding protein), while MAOB expression is reported to be regulated by c-Jun, Egr1 (early growth response protein1), Klf-11, and Sp1 [8,9,10,11,12]. Interestingly, MAOs are also regulated by molecules of cardiovascular relevance such as androgen, glucocorticoid, retinoic acid (RA), forskolin, and tumor necrosis factor-α (TNF-α) [8, 9].
Schematic representation of human MAOA and MAOB genes and their protein products. The human MAOA (panel A) and MAOB (panel B) genes consist of 15 exons seperated by 14 introns (UCSC Genome Browser refGenes NM_000240 and NM_000898). The lengths of UTRs, exons, and introns are mentioned. FAD flavin adenine dinucleotide, UTR untranslated region, Ex exon, Int intron, bp base pair. MAOA protein consists of 527 amino acids and the amino acids 403–407 serve as the FAD binding site. MAOB protein consists of 520 amino acids and the amino acids 394–398 serve as the FAD binding site
Besides the well-studied functions of MAOs in neuronal/behavioral disorders, cancer metastasis, and embryonic development [13,14,15,16], a lot of research has been performed in recent years to explore their possible roles in mitochondrial/endothelial/cardiac dysfunction, essential hypertension, ventricular hypertrophy, myocardial infarction, cardiomyocyte apoptosis, postischemic cardiac damage, and heart failure as discussed below [17,18,19,20,21,22,23,24]. This chapter aims to summarize our current understanding on the role of these enzymes in heart diseases.
2 Role of MAOs in Cardiac Cell Death and Chronic Ventricular Dysfunction
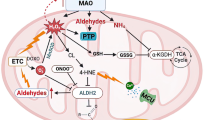
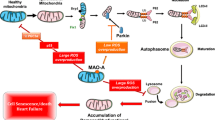
MAOs are potent generators of reactive oxygen species (ROS) or oxidative stress due to oxidative deamination of mainly norepinephrine and serotonin in cardiac tissues [25,26,27,28]. Depending on the type of available substrate and ROS generated by MAOs, different signal transduction mechanisms lead to distinct phenotypes including cell proliferation/hypertrophy, basilar artery contraction, or apoptosis/necrosis [17, 18, 27,28,29,30]. For example, transgenic mice with cardiac-specific MAOA overexpression displays oxidative stress-induced p53 activation, which leads to downregulation of peroxisome proliferator-activated receptor gamma co-activator 1α (PGC1α) (a crucial regulator of mitochondrial biogenesis/function) that in turn causes mitochondrial dysfunction, cardiomyocyte necrosis, and chronic ventricular dysfunction [18] (Fig. 6.2). Moreover, ROS generated via MAOA can also block autophagic flux of lysosomes by reducing the lysosomal acidification and by preventing the nuclear translocation of transcription factor-EB (TF-EB) (that acts as a master regulator of lysosomal biogenesis and autophagy) [29] (Fig. 6.2).
Plausible mechanisms of MAOA-mediated apoptosis, necrosis, endothelial dysfunction, and ventricular dysfunction. MAOA-generated oxidative stress causes p53 activation and consequently downregulates PGC1α. Oxidative stress also impairs lysosome’s function by blocking the nuclear translocation of TF-EB, which in turn leads to blockade of autophagic flux. p53- and TF-EB-mediated pathways lead to necrosis/chronic ventricular dysfunction. MAO-generated oxidative stress can also promote apoptosis via ceramide accumulation and downregulation of S1P in cardiomyocytes. Blunt-headed arrow indicates “inhibition” of nuclear translocation of TF-EB. S1P sphingosine 1-phosphate, PGC1α peroxisome proliferator-activated receptor-coactivator 1α, P phosphorylation
3 Role of MAOs in Cardiac Hypertrophy and Heart Failure
In contrast to cardiac cell death via apoptosis or necrosis, MAOs can lead to cardiac hypertrophy via different signaling pathways. In biomechanically stretched cardiomyocytes, MAOA has been reported to be upregulated (by ~four-fold) leading to cardiac hypertrophy and consequent heart failure [31]. These cellular changes are due to oxidative stress generated during oxidative deamination of serotonin and norepinephrine.
Serotonin (5-hydroxytyramine [5-HT]), a MAOA-specific substrate and a potent vasoactive amine, induces cardiomyocyte hypertrophy in a MAOA-dependent manner via activation of extracellular-regulated kinases (ERK1/2 that are essential signaling molecules for cell growth) [28] (Fig. 6.3). This cardiac hypertrophy is partly 5-HT2B receptor dependent as reflected by cellular response following treatment with amine transporter inhibitors (imipramine) and MAO inhibitor (pargyline) [28]. In corroboration, MAOA contributes to oxidative stress in human heart valves following exposure to serotonin and dopamine [25]. In the circulatory system, the major source of 5-HT is platelets. Upon aggregation/activation, a large amount of 5-HT is released from the platelets into the circulation causing either vasorelaxation via endothelial cells or vasoconstriction via vascular smooth muscle cells [24]. In addition, 5-HT-dependent MAOA-mediated ROS also leads to basilar artery contraction in rats [26].
Signaling pathways underlying development of cardiomyocyte hypertrophy via MAOA-mediated catabolism of serotonin and norepinephrine. Serotonin and norepinephrine are released from the activated platelets and intracardiac nerves, respectively. Following interaction with their respective receptors and signaling, they are sequestered into the cytoplasm via respective transporters present in the membrane. Serotonin and norepinephrine are degraded by MAOA-generating hydrogen peroxide/oxidative stress that, in turn, contributes to basilar artery contraction or cardiomyocyte hypertrophy. ERK1/2, extracellular signal-regulated kinase 1/2
Norepinephrine stimulates the MAOA enzyme activity in neonatal and adult cardiomyocytes in vitro that leads to ROS production and maladaptive hypertrophy [27]. These in vitro changes may involve the transcription factor NFAT (nuclear factors of activated T cells) that contributes to maladaptive hypertrophic signaling (Fig. 6.3). In line with this finding, pharmacological or genetic inhibition of MAOA prevents the occurrence of heart failure in mice subjected to pressure overload [27]. Corroboratively, transcriptomic and proteomic analyses reveal that MAOA is one of the most upregulated proteins in a well-defined rat model of chronic heart failure (which has volume overload due to surgically created aorto-caval fistula) [32]. Similarly, enzyme activity and expression of both MAOs are significantly elevated in left and right ventricles of end-stage ischemic failing hearts in human [33].
In addition, MAOB knockout mice show compensated cardiac hypertrophy following pressure overload induced by transverse aortic constriction. They are also found to be resistant to adverse left ventricular (LV) dilation and dysfunction upon pressure overload. Thus, MAOB activity also contributes to oxidative stress and structural and functional derangements in the heart [19]. Moreover, oxidative stress also diminishes the activity of aldehyde dehydrogenase which may, in turn, cause the accumulation of toxic aldehydes. These accumulated aldehydes may induce mitochondrial dysfunction contributing to myocardial damage [19].
4 Role of MAOs in Blood Pressure Homeostasis
Essential hypertension (EH), a common, multifactorial/polygenic health problem, is the chief risk factor for cardiovascular/renal diseases (viz., myocardial infarction, heart failure, stroke, and end-stage renal disease) [34]. Catecholamines have been implicated to play an important role in the pathogenesis of EH. For example, dopamine modulates blood pressure via generation of ROS, interaction with the renin-angiotensin-aldosterone system (RAAS), regulation of epithelial sodium transport, and vascular smooth muscle contractility [35, 36] (Fig. 6.4). Therefore, MAOs are logical candidate genes for blood pressure regulation. Of note, there are three blood pressure QTLs (Bp65, Bp64, and Bp56) (source: Rat Genome Database) in the X chromosome of rat; both MAOA and MAOB are localized in the Bp65 and Bp64 QTLs (with LOD scores of 5.8 and 5.2, respectively) in line with their plausible contributions to blood pressure modulation (Fig. 6.5).
Plausible molecular mechanisms of blood pressure regulation by catecholamines. Catecholamines alter the blood pressure homeostasis either through adrenergic/dopaminergic receptors or by increasing the release of renin from the adrenal cortex. Higher level of renin produces more angiotensin II which leads to vasoconstriction via angiotensin receptor 1, increasing endothelin-1, aldosterone secretion, and ROS generation (via enhancing the expression/activity of MAOs and NADPH oxidase)
Graphical representation of the blood pressure QTLs on the X chromosome of rat. Blood pressure QTLs (Bp65, Bp64, and Bp56) on the rat X chromosome and their respective LOD scores obtained from Rat Genome Database. Two of these three BP QTLs harbor the MAOA and MAOB genes, suggesting their important roles in BP regulation. The genomic positions of MAOA and MAOB genes in the BP QTLs are indicated
Several studies reported higher level of catecholamines in hypertensive individuals and in rodent models of hypertension compared to their respective normotensive controls [37,38,39]. This difference may, at least partly, be attributed to altered expression or enzyme activity of catecholamine catabolizing enzymes (e.g., MAOs and catecholamine-o-methyltransferase). Notably, two independent microarray studies on adrenal gland and kidney tissues of mouse models of human essential hypertension (viz., BPH (blood pressure high) and BPL (blood pressure low) mice) showed that MAOA expression was elevated by ~1.3- and ~3.3-fold, respectively, in BPL mice [40, 41]. Based on these observations, we speculate that higher MAOA levels in BPL may contribute to lower catecholamine levels, which in turn could lead to low blood pressure phenotype. MAOs also inhibit nitric oxide synthase (NOS2) expression and consequently reduce the levels of the vasodilator nitric oxide (NO) (Fig. 6.4) [42]. Consistently, MAOA enzyme activity was ~1.4-fold higher in the kidneys of normotensive Wistar-Kyoto (WKY) rat than the spontaneously hypertensive rat (SHR) [43]. Surprisingly, some studies reported higher MAOA enzyme activity in the heart, aorta, femoral arteries, isolated cardiomyocytes, and brain of SHR compared to WKY rats [20, 42, 43]. SHR rats have also been reported to have higher MAOA protein level in their basilar arteries compared to WKY rats [26]. Similarly, MAOB enzyme activity was reported to be ~2.8-fold higher in isolated cardiomyocytes of SHR compared to WKY rats [21]. However, comparative microarray analysis showed that SHR adrenal gland tissues exhibited ~0.52-fold underexpression of MAOB than that of WKY [44]. The mechanism of such differential expression/activity pattern of MAOs across different tissues of SHR and WKY remains unclear. Of note, a recent study reported that in vivo administration of lipopolysaccharide and angiotensin II augments the vascular expression of both the MAOs leading to increased generation of H2O2 and subsequent endothelial dysfunction [22] (Fig. 6.4).
MAOB-specific substrates (phenylethylamine, tyramine, and tryptamine) are bioactive endogenous amines present in mammalian peripheral as well as central nervous system in low concentration (less than 1% of biogenic amines); therefore, they are called trace amines (TAs) [45]. These amines lack the catechol nucleus but are similar to biogenic amines in terms of structure and metabolism; these are described as “false neurotransmitters” or “sympathomimetic amines.” TAs are present in food products like cheese, red wine, chocolates, etc. MAO inhibitor-treated patients consuming a TA-rich diet may develop complications such as tachycardia and hypertension. This hypertensive crisis is described as “cheese effect” irrespective of the nature of TA-rich food [46, 47]. The molecular mechanism of TA-induced hypertension is based on the fact that tyramine and phenylethylamine are structurally similar to norepinephrine. Therefore, these molecules enter sympathetic neurons by the same monoamine membrane transporter and displace norepinephrine. Consequently, norepinephrine is diffused from the cytoplasm into the synaptic cleft, leading to α-adrenoceptor-mediated vasoconstriction and the sudden rise in blood pressure [46, 48]. Thus, various studies support the role of MAOs in modulating blood pressure under pathophysiological conditions.
5 Mechanisms of Transcriptional Regulation of MAOs
5.1 Transcriptional Regulation of MAOA
Because transcription factors play crucial roles in gene regulation, a potential strategy for developing novel therapeutics against disease conditions can be attained by modulating the expression and/or activity of a specific transcription factor [49,50,51]. Regulatory mechanisms for both MAOs have been studied extensively. For example, previous reports showed that Sp1 and SRY synergistically enhanced the human MAOA (hMAOA) promoter activity in a dose-dependent manner. Of note, SRY plays a very important role in blood pressure homeostasis [52]. Indeed, apart from MAOA, promoters of several other key cardiovascular-regulatory genes including tyrosine hydroxylase (the rate-limiting enzyme in the catecholamine biosynthesis pathway) [53], chromogranin B [54], and genes in RAAS pathway [55] are responsive to Sry and influence blood pressure.
Sp/Klf family, Sp3, Sp4, and KLF11 are the other transcription factors which have also been reported to regulate hMAOA promoter. KLF11 and Sp4 are known to trans-activate the hMAOA gene expression; on the other hand, Sp3 and a novel transcription factor known as R1 (RAM2/CDCA7L/JPO2) repress hMAOA gene expression as they compete for the same binding site with Sp1 [9, 56]. KLF transcription factors, in general, interact with histone acetyltransferases (HATs), including p300, for gene regulation. Consistently, co-transfection of p300 and KLF11 expression plasmids with hMAOA promoter luciferase construct showed that activation of hMAOA by KLF11 was further augmented in the presence of p300 [56]. The mouse MAOA (mMAOA) promoter is also well-characterized; mMAOA gene expression is regulated by GATA2, Sp1, and TBP in a coordinated manner [8]. Of note, not only mMAOA, these transcription factors also enhance the MAOA protein levels in humans [8]. It is interesting to note that GATA2 may also hamper the inflammatory state in atherosclerosis and obesity [57], indicating a possible role of MAOA in these disease conditions. In addition, circadian-clock components (via E-box elements) and NAD-dependent deacetylase sirtuin 1 (SIRT1) have also been reported to regulate mMAOA gene expression [11, 12]. SIRT1-/GATA2-mediated MAOA gene regulation is critical because single-nucleotide polymorphisms (SNPs) present in both of these upstream regulators of MAOA are associated with cardiovascular/cardiometabolic disorders or their risk traits [58,59,60,61].
Dopamine, a common substrate for both the MAOs, regulates the expression and enzymatic activity of MAOA via D-2-like receptors in mesangial renal cells although such regulation has not been observed in proximal tubule renal cells [62]. Dexamethasone, a synthetic glucocorticoid hormone, has also been shown to augment MAOA gene expression in human skeletal myocytes via glucocorticoid receptor and Sp1. These results provide molecular mechanism for the pathogenesis of glucocorticoid-induced myopathy [63]. In addition, forskolin-mediated cAMP-PKA (protein kinase A) pathway and TNF-α also increase MAOA gene expression via Sp1 [8]. This observation has therapeutic implications since forskolin (a diterpene isolated from root of Coleus forskohlii) was reported to have beneficial effects in cardiovascular diseases including congestive heart failure and hypertension [64,65,66,67]. It may also be noted that a recent study established the role of GATA2, Sp1, and TBP in regulating MAOA gene expression under ischemia-like pathophysiological conditions [8].
5.2 Transcriptional Regulation of MAOB
Several studies reported the characterization of the human MAOB (hMAOB) promoter. Unlike the hMAOA promoter, the core hMAOB promoter contains a TATA box; it also harbors two Sp1 binding domains, which are separated by a CACCC element [68]. Egr1 also regulates hMAOB expression by binding to the distal Sp1 domain [69, 70]. Another transcription factor called Sp4 trans-activates hMAOB promoter activity via direct interaction with the Sp1 sites; this activation has been reported to be repressed by Sp3 and Krüppel-like zinc-finger transcription factor KLF5 (also called BTEB2) as Sp3/KLF5 compete for the Sp1-binding sites [9, 56]. Site-directed mutagenesis revealed that CACCC sequence (present between the two Sp1-binding sites) is a repressor element. It is important to note that the transforming growth factor-β-inducible early gene TIEG2 (also called KLF11) and Sp3 exhibit dual functions for the regulation of hMAOB. TIEG2 acts as a repressor at the CACCC element whereas it acts as an activator at the distal Sp1 site of hMAOB promoter. However, due to its higher affinity for the Sp1 site than the CACCC element, the overall effect of TIEG2 is activation of the hMAOB gene expression [68]. Egr1 and c-Jun can also regulate hMAOB gene expression by interacting with the overlapping Sp1/Egr-1/Sp1 sites [9]. Interestingly, phorbol 12-myristate 13-acetate enhances hMAOB gene expression by increasing the Egr1/c-Jun gene expression via activation of PKC (protein kinase C) and MAPK (mitogen-activated protein kinase) signaling pathways [9]. Our recent studies suggest that MAOB gene expression may also be regulated by cyclic AMP/PKA/CREB (cAMP response element binding protein) pathway (unpublished observation).
The roles of a number of hormones, such as androgen, glucocorticoid, estrogen, and RA, have been demonstrated in hMAOB gene regulation [9]. Of note, RA enhances MAOB expression through retinoic acid receptor α (RARα) and retinoid X receptor α (RXRα) transcription factors. RARα physically interacts with Sp1 to form a transcriptional regulatory complex and recruited to Sp1-binding sites at hMAOB promoter [9]. Of note, RAR/RXR have a crucial role in cardiovascular pathophysiology as evident from the fact that knockout of RAR/RXR in mice leads to the development of heart defects such as defects in the conduction system, heart malformations, and heart failure. On the other hand, elevated level of RAR or RXR leads to dilated cardiomyopathy and congestive heart failure [71, 72].
Similar to MAOA gene regulation, dexamethasone has been reported to stimulate hMAOB promoter activity via glucocorticoid response element (GRE) and Sp1-binding sites in vitro and in vivo. The molecular mechanism of this activation involves activation of glucocorticoid receptor by dexamethasone, which then translocates into the nucleus and binds to GRE [9, 56, 73]. Interestingly, glucocorticoids and their receptors have direct effects on the heart, blood vessels, and cardiometabolic risk factors which are discussed in detail elsewhere [74, 75]. Dopamine may also activate MAOB expression similar to the case of MAOA; the dopamine-mediated upregulation of MAOB seems to be modulated by cyclic AMP response element (CRE) in the proximal MAOB promoter (unpublished observation).
5.3 Potential Therapeutic Application of the Transcriptional Regulators of MAOs
As detailed above, some of the transcription factors (viz., Sp1, KLF11, possibly Egr1, and CREB) are common regulators of MAOA and MAOB. Regulation of these molecules as a new therapeutic strategy for management of cardiovascular diseases may be worth studying. Of note, mithramycin A, an antibiotic produced by Streptomyces argillaceus, is used to treat various diseases including testicular carcinoma and chronic myeloid leukemia by virtue of its ability to diminish binding of Sp1 and Egr1 to regulatory promoter elements (and thereby modulating gene expression) [76, 77]. Mithramycin A has also been reported to diminish the binding of Sp1 and Egr1 to the MAOB promoter, thereby offering neuroprotection in a mouse model of Parkinson’s disease [78, 79]. Moreover, in endothelial cells, mithramycin A prevented the TNF-α-mediated fractalkine (a chemokine) expression suggesting that it could function as an anti-inflammatory agent [80]. In view of these reports, it will be interesting to evaluate therapeutic potential of mithramycin A and other agents that may regulate the expression of MAOs via interactions with the key transcription factors in the context of cardiovascular diseases.
6 Posttranscriptional Regulation of MAOs: Potential Role for Several microRNAs
MicroRNAs (miRNAs) are small (~22 nucleotides), noncoding RNAs which have emerged as important posttranscriptional regulators of gene expression either by inhibiting translation or by degrading mRNA [81]. They are involved in regulating various physiological processes including development, metabolism, and maintaining homeostasis [82, 83]. Dysregulated expression of miRNAs is associated with various complications including cardiovascular diseases. In addition to this, circulating miRNAs serve as excellent noninvasive biomarkers for diagnosis and prognosis of diseases [84]. Some miRNAs are also being evaluated for their therapeutic applications in various disease states. For example, miravirsen (a miR-122 inhibitor) is under clinical trials for the treatment of chronic hepatitis C infection [85]. Similarly, a few miRNAs are at various preclinical/clinical stages for the plausible treatment of various pathological conditions [86].
MiRNA-142 is reported to diminish MAOA expression in neuronal cells by downregulating SIRT1 [87]. Computational analysis of the MAOA and MAOB 3’-UTRs using ten miRNA prediction tools (DIANA-microT [88], miRanda [89], miRDB [90], miRWalk [91], RNAhybrid [92], PICTAR4, PICTAR5 [93], PITA [94], RNA22 [95], and Targetscan [93]) revealed putative binding sites for 641 and 297 miRNAs, respectively. MiRNAs predicted by at least three tools and based on the thermodynamic scores obtained using PITA (ΔΔG < -10) and RNAhybrid (ΔG < -20 kcal/mole) are presented in Table 6.1. Interestingly, miR-608 and miR-125a-3p harbor putative binding sites in the 3’-UTRs of both the MAOs representing these miRNAs as candidates for further studies. However, experimental validations of interactions between miR-608/miR-125a-3p and MAOA/MAOB are required for confirmation of their roles in regulating MAOA/MAOB expression.
An early increase in plasma levels of miRNA-133a and miR-133b in myocardial infarction (MI) and coronary artery disease is well-documented indicating that these miRNAs could serve as novel diagnostic markers for these diseases [96, 97]. Interestingly, in silico analysis using PITA and RNAhybrid revealed putative binding sites for miR-133a and miR-133b in the 3’-UTR of both the MAOs. Both MAOA and MAOB are also predicted by miRwalk (version 3.0) as putative targets of miR-1224. Besides this, a recent study reported the increase in miR-1224 levels in human hepatocytes and serum under acute liver failure. The levels of miR-1224 were also augmented in mice subjected to ischemia/reperfusion compared to control [98]. Furthermore, in mouse, lipopolysaccharide (LPS)-induced miR-1224 was shown to downregulate the expression of Sp1 [99]; this finding suggests that miR-1224 may also indirectly regulate MAOs, since Sp1 governs the expression of both the MAOs. miR-1224 may also regulate MAOs via modulation of the expression of CREB, a key regulator of catecholamine biosynthetic genes, since CREB is a target of miR-1224 [100] and forskolin/cAMP augments MAOA [8]/MAOB expression/activity [101]. Our in vitro experiments also provided evidence for regulation of MAOB by miR-1224 (unpublished observation). Of note, the expression of MAOs is augmented by LPS and angiotensin II (AngII) in murine aortic rings, which is mediated by phosphatidylinositol kinase and nuclear factor-κB [22]. Taken together, this increase in miR-1224 could be a compensatory mechanism to block MAOA/MAOB gene expression by binding to their 3′-UTRs and by targeting both Sp1 and CREB (Fig. 6.6). This is further substantiated by the evidence that MAO inhibitors are protective against oxidative stress [102]. All these observations indicate a complex interplay between miR-1224, Sp1 and CREB in regulating MAOA/MAOB expression which warrants further investigation.
Possible interplay of miR-1224, Sp1, and CREB in governing MAO gene regulation. The transcription factors Sp1 and CREB regulate MAOA/MAOB gene expression, which in turn may contribute to oxidative stress during ischemia/reperfusion (I/R) injury. The levels of miR-1224 are augmented under I/R condition which could be a compensatory mechanism to block Sp1, CREB, and MAOA/MAOB gene expressions. Upward arrows indicate “increase” and blunt-headed arrows indicate “inhibition” of function of the corresponding molecules. Sp1 specificity protein 1, CREB cAMP response element binding protein, miR-1224 microRNA-1224
7 Cardiovascular Implications of Systemic Ablation of MAOA/MAOB in Mouse
Generation of MAOA or MAOB knockout mice was carried out by inserting interferon β transgene or neomycin resistance gene into exon 2 and 6 of MAOA and MAOB, respectively [103]. As expected, MAOA knockout mice displayed higher levels of its substrates (catecholamines and serotonin) in the brain along with various neurochemical and physiological changes in comparison with the wild-type animals [103]. Similarly, adult MAOB knockout mice showed ~8.0-fold higher level of phenylethylamine in the brain while no statistically significant increase in serotonin, norepinephrine, and dopamine, due to the substrate specificity of MAOB. The most striking cardiovascular characteristic of MAOA/MAOB knockout mice was their hypotensive nature and reduced heart rate in the resting, restrained state [103]. This finding is in corroboration with the resting hypotension in Norrie disease patients who have deletions in MAOA gene [104, 105]. But this is in contrast to the fact that higher level of catecholamine is mostly associated with hypertension. This phenotype can be explained by the fact that high level of catecholamine may be a cause or effect, which can lead to higher/lower blood pressure. Most probably, these knockout mice developed some compensatory mechanism which leads to lowered blood pressure than that of wild-type mice. Expectedly, MAOA/MAOB knockout mice were found to have increased baroreceptor activity that serves to regulate blood pressure and leads to hypotensive state [106].
8 Human Genetic Studies Link MAOs and Their Upstream Regulators with Cardiovascular and Cardiometabolic Risk Factors
Genome-wide linkage analysis in human hypertensive population revealed several blood pressure quantitative trait loci (QTLs); among them, the blood pressure QTL on the X chromosome (Xp11.4-Xq11) harbors several genes of cardiovascular relevance including MAOA and MAOB [107]. This observation is in line with the identification of blood pressure QTLs that harbor these genes in the X chromosome in rats (Fig. 6.5). Some of the well-characterized polymorphisms (VNTR (variable number of tandem repeats) and EcoRV polymorphism) present in MAOA gene are also associated with cardiovascular or cardiometabolic risk factors including body mass index, lipid levels, and obesity [108,109,110,111]. In brief, the most widely studied polymorphism in hMAOA gene is a VNTR (30-bp repeat sequence present in 3, 3.5, 4, or 5 copies) present at ~1.2 kb upstream of the coding region in hMAOA. Several studies in the last few decades reported the functional role of this VNTR in the context of neuronal/behavioral traits. According to those studies, alleles with 3.5 or 4 copies of the repeat sequence displayed substantially higher (two- to tenfold) transcriptional activity when compared to alleles with 3 or 5 copies of the VNTR [112,113,114]. Another polymorphism present in MAOA gene, i.e., EcoRV polymorphism or T/C polymorphism (rs1137070) located within exon 14, has been associated with altered MAOA enzyme activity [115]. Briefly, MAOA gene with allele T harbors an EcoRV site and higher MAOA activity than that of MAOA gene with allele C and no EcoRV site. Interestingly, this T/C polymorphism causes a nucleotide substitution at the third position of a codon and does not affect the amino acid sequence (Asp to Asp). Perhaps, the polymorphism is in linkage disequilibrium with another genetic variation to regulate MAOA enzyme activity [115], or the rate of translation of the mRNA transcript could be altered due to this synonymous T/C polymorphism [116]. Moreover, this SNP was also associated with gout and hyperuricemia (another risk factor for cardiovascular disorders) [52, 117].
Several studies have associated SNPs in the upstream regions of the crucial transcriptional regulators of MAOA including SIRT1 and GATA2 with weight/body mass index/systolic blood pressure/diastolic blood pressure/hypertension/hyperglycemia in different populations across the world [58,59,60,61, 118, 119]. Sirtuin proteins (SIRT1–SIRT7) are nicotinamide adenine dinucleotide (NAD)-dependent deacetylases. The most conserved member of the sirtuin family, SIRT1, regulates the PGC1α activity via deacetylation, thereby protecting the cells against oxidative stress. In addition, SIRT1 deacetylates many other crucial transcription factors and cofactors including p53 [120], forkhead box class O (FOXO) proteins [121], and nuclear factor-κB [122]. Of note, MAOA upregulation leads to necrosis or chronic ventricular dysfunction via p53-PGC1α-mediated pathway as shown in Fig. 6.2. It is evident from human genetic studies that the SIRT1 SNP rs2273773 (C/T in exon 5, a silent mutation) is associated with seasonal variation in weight and diastolic blood pressure/hypertension in Finnish nationwide population [58]. Another study has also probed the association of SIRT1 SNPs (rs7895833 (A/G in the promoter region), rs7069102 (C/G in intron 4), and rs2273773 (C/T in the coding region)) with cardiovascular/cardiometabolic risk factors. For example, the mutant alleles for rs7069102 and rs2273773 were detected at significantly higher frequencies in cardiovascular disease patients compared to control subjects, increasing the disease risk by 2.4- and 1.9-fold, respectively, in mutant allele carriers than in wild-type allele carriers. In contrast, the allele frequency for rs7895833 did not differ between both groups [61]. Another study in a Japanese population showed the association of rs7895833, rs7069102, and rs2273773 with different cardiovascular/cardiometabolic phenotypes including fasting glucose/hyperglycemia/body fat ratio/systolic blood pressure/diastolic blood pressure/hypertension [60]. Thus, SIRT1 emerged as a potential therapeutic target for metabolic syndrome [123,124,125]. In addition to SIRT1, human genetic studies have identified GATA2 as a novel susceptibility gene for coronary artery disease by showing the association of GATA2 SNPs with cardiovascular/cardiometabolic risk traits [118, 119].
9 Conclusions and Perspectives
A growing body of research suggests that dysregulation of MAOs plays an important role in several cardiovascular pathophysiological conditions (including essential hypertension, LV remodeling, heart failure, cardiomyocyte hypertrophy, and I/R injury) possibly due to ROS generated by MAOs. Therefore, regulation of MAOs (perhaps, by tissue-specific regulation of some transcription factors) may emerge as a new therapeutic strategy for treatment of cardiovascular pathologies. Although, so far, conclusive studies on the applicability of MAO inhibitors with heart disease patients are lacking, general MAO inhibitors were previously used as therapeutics for cardiovascular diseases and have been reported to reduce blood pressure and intensity/frequency of anginal pain [126]. However, the main concern for the use of these irreversible MAO inhibitors is a phenomenon called “cheese effect” which, subsequently, causes hypertensive crisis. The efficiency of the next-generation reversible MAO inhibitors that are devoid of these harmful effects remains to be evaluated in cardiovascular pathologies. It is also important to note that although MAOB is highly abundant in the human myocardium, most of the studies focused on MAOA; therefore, future research should be designed to understand the contribution of MAOB to these complications. Systematic studies identifying posttranscriptional regulators (certain microRNAs or their inhibitors) of MAOs may also lead to identification of novel cardiovascular therapeutics. Based on human genetic studies, computational predictions, and regulatory mechanisms, certain common molecular factors may also emerge as potential therapeutic agents for dysregulated MAO expression/activity.
References
Nagatsu T (2004) Progress in monoamine oxidase (MAO) research in relation to genetic engineering. Neurotoxicology 25:11–20
Grimsby J, Chen K, Wang LJ, Lan NC, Shih JC (1991) Human monoamine oxidase A and B genes exhibit identical exon-intron organization. Proc Natl Acad Sci U S A 88:3637–3641
Shih JC, Chen K, Ridd MJ (1999) Monoamine oxidase: from genes to behavior. Annu Rev Neurosci 22:197–217
Finberg J (2014) Update on the pharmacology of selective inhibitors of MAO-A and MAO-B: focus on modulation of CNS monoamine neurotransmitter release. Pharmacol Ther 143:133–152
Grimsby J, Lan NC, Neve R, Chen K, Shih JC (1990) Tissue distribution of human monoamine oxidase A and B mRNA. J Neurochem 55:1166–1169
Saura J, Richards JG, Mahy N (1994) Age-related changes on MAO in Bl/C57 mouse tissues: a quantitative radioautographic study. J Neural Transm Suppl 41:89–94
Strolin Benedetti M, Thomassin J, Tocchetti P, Dostert P, Kettler R, Da Prada M (1994) Species differences in changes of heart monoamine oxidase activities with age. J Neural Transm Suppl 41:83–87
Gupta V, Khan AA, Sasi BK, Mahapatra NR (2015) Molecular mechanism of monoamine oxidase A gene regulation under inflammation and ischemia-like conditions: key roles of the transcription factors GATA2, Sp1 and TBP. J Neurochem 134:21–38
Shih JC, Wu JB, Chen K (2011) Transcriptional regulation and multiple functions of MAO genes. J Neural Transm (Vienna) 118:979–986
Wong W, Ou X, Chen K, Shih J (2002) Activation of human monoamine oxidase B gene expression by a protein kinase C MAPK signal transduction pathway involves c-Jun and Egr-1. J Biol Chem 277:22222–22230
Libert S, Pointer K, Bell EL, Das A, Cohen DE, Asara J, Kapur K, Bergmann S, Preisig M, Otowa T et al (2011) SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell 147:1459–1472
Hampp G, Ripperger J, Houben T, Schmutz I, Blex C, Perreau-Lenz S, Brunk I, Spanagel R, Ahnert-Hilger G, Meijer J et al (2008) Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol 18:678–683
Bortolato M, Chen K, Shih J (2008) Monoamine oxidase inactivation: from pathophysiology to therapeutics. Adv Drug Deliv Rev 60:1527–1533
Wang C, Billett E, Borchert A, Kuhn H, Ufer C (2013) Monoamine oxidases in development. Cell Mol Life Sci 70:599–630
Wu J, Shao C, Li X, Li Q, Hu P, Shi C, Li Y, Chen Y, Yin F, Liao C et al (2014) Monoamine oxidase A mediates prostate tumorigenesis and cancer metastasis. J Clin Invest 124:2891–2908
Nicotra A, Pierucci F, Parvez H, Senatori O (2004) Monoamine oxidase expression during development and aging. Neurotoxicology 25:155–165
Pchejetski D, Kunduzova O, Dayon A, Calise D, Seguelas M, Leducq N, Seif I, Parini A, Cuvillier O (2007) Oxidative stress-dependent sphingosine kinase-1 inhibition mediates monoamine oxidase A-associated cardiac cell apoptosis. Circ Res 100:41–49
Villeneuve C, Guilbeau-Frugier C, Sicard P, Lairez O, Ordener C, Duparc T, De Paulis D, Couderc B, Spreux-Varoquaux O, Tortosa F et al (2013) p53-PGC-1α pathway mediates oxidative mitochondrial damage and cardiomyocyte necrosis induced by monoamine oxidase-A upregulation: role in chronic left ventricular dysfunction in mice. Antioxid Redox Signal 18:5–18
Kaludercic N, Carpi A, Nagayama T, Sivakumaran V, Zhu G, Lai E, Bedja D, De Mario A, Chen K, Gabrielson KL et al (2014) Monoamine oxidase B prompts mitochondrial and cardiac dysfunction in pressure overloaded hearts. Antioxid Redox Signal 20:267–280
Kaludercic N, Carpi A, Menabò R, Di Lisa F, Paolocci N (2011) Monoamine oxidases (MAO) in the pathogenesis of heart failure and ischemia/reperfusion injury. Biochim Biophys Acta 1813:1323–1332
Pino R, Failli P, Mazzetti L, Buffoni F (1997) Monoamine oxidase and semicarbazide-sensitive amine oxidase activities in isolated cardiomyocytes of spontaneously hypertensive rats. Biochem Mol Med 62:188–196
Sturza A, Leisegang M, Babelova A, Schröder K, Benkhoff S, Loot A, Fleming I, Schulz R, Muntean D, Brandes R (2013) Monoamine oxidases are mediators of endothelial dysfunction in the mouse aorta. Hypertension 62:140–146
Sturza A, Duicu O, Vaduva A, Dănilă M, Noveanu L, Varró A, Muntean D (2015) Monoamine oxidases are novel sources of cardiovascular oxidative stress in experimental diabetes. Can J Physiol Pharmacol 93:555–561
Kaludercic N, Mialet-Perez J, Paolocci N, Parini A, Di Lisa F (2014) Monoamine oxidases as sources of oxidants in the heart. J Mol Cell Cardiol 73:34–42
Peña-Silva R, Miller J, Chu Y, Heistad D (2009) Serotonin produces monoamine oxidase-dependent oxidative stress in human heart valves. Am J Physiol Heart Circ Physiol 297:H1354–H1360
Poon C, Seto S, Au A, Zhang Q, Li R, Lee WYW, Leung GPH, Kong SK, Yeung JHK, Ngai SM et al (2010) Mitochondrial monoamine oxidase-A-mediated hydrogen peroxide generation enhances 5-hydroxytryptamine-induced contraction of rat basilar artery. Br J Pharmacol 161:1086–1098
Kaludercic N, Takimoto E, Nagayama T, Feng N, Lai EW, Bedja D, Chen K, Gabrielson KL, Blakely RD, Shih JC et al (2010) Monoamine oxidase A-mediated enhanced catabolism of norepinephrine contributes to adverse remodeling and pump failure in hearts with pressure overload. Circ Res 106:193–202
Bianchi P, Pimentel DR, Murphy MP, Colucci WS, Parini A (2005) A new hypertrophic mechanism of serotonin in cardiac myocytes: receptor-independent ROS generation. FASEB J 19:641–643
Santin Y, Sicard P, Vigneron F, Guilbeau-Frugier C, Dutaur M, Lairez O, Couderc B, Manni D, Korolchuk VI, Lezoualc’h F et al (2016) Oxidative stress by monoamine oxidase-A impairs transcription factor EB activation and Autophagosome clearance, leading to Cardiomyocyte necrosis and heart failure. Antioxid Redox Signal 25:10–27
Maceyka M, Payne SG, Milstien S, Spiegel S (2002) Sphingosine kinase, sphingosine-1-phosphate, and apoptosis. Biochim Biophys Acta 1585:193–201
Frank D, Kuhn C, Brors B, Hanselmann C, Lüdde M, Katus HA, Frey N (2008) Gene expression pattern in biomechanically stretched cardiomyocytes: evidence for a stretch-specific gene program. Hypertension 51:309–318
Petrak J, Pospisilova J, Sedinova M, Jedelsky P, Lorkova L, Vit O, Kolar M, Strnad H, Benes J, Sedmera D et al (2011) Proteomic and transcriptomic analysis of heart failure due to volume overload in a rat aorto-caval fistula model provides support for new potential therapeutic targets – monoamine oxidase A and transglutaminase 2. Proteome Sci 9:69
Manni ME, Rigacci S, Borchi E, Bargelli V, Miceli C, Giordano C, Raimondi L, Nediani C (2016) Monoamine oxidase is Overactivated in left and right ventricles from ischemic hearts: an intriguing therapeutic target. Oxidative Med Cell Longev 2016:4375418
Deng AY (2007) Genetic basis of polygenic hypertension. Hum Mol Genet 16(Spec No. 2):R195–R202
Missale C, Nash SR, Robinson SW, Jaber M, Caron MG (1998) Dopamine receptors: from structure to function. Physiol Rev 78:189–225
Zeng C, Zhang M, Asico LD, Eisner GM, Jose PA (2007) The dopaminergic system in hypertension. Clin Sci 112:583–597
Andrew R, Best SA, Watson DG, Midgley JM, Reid JL, Squire IB (1993) Analysis of biogenic amines in plasma of hypertensive patients and a control group. Neurochem Res 18:1179–1182
Goldstein DS (1983) Plasma catecholamines and essential hypertension. An analytical review. Hypertension 5:86–99
Grobecker G, Roizen MF, Weise V, Saavedra JM, Kopin IJ (1975) Sympathoadrenal medullary activity in young, spontaneously hypertensive rats. Nature 258:267–268
Fries RS, Mahboubi P, Mahapatra NR, Mahata SK, Schork NJ, Schmid-Schoenbein GW, O’Connor DT (2004) Neuroendocrine transcriptome in genetic hypertension: multiple changes in diverse adrenal physiological systems. Hypertension 43:1301–1311
Puig O, Wang I-M, Cheng P, Zhou P, Roy S, Cully D, Peters M, Benita Y, Thompson J, Cai T-Q (2010) Transcriptome profiling and network analysis of genetically hypertensive mice identifies potential pharmacological targets of hypertension. Physiol Genomics 42A:24–32
Vega A, Chacón P, Monteseirín J, El Bekay R, Alvarez M, Alba G, Conde J, Martín-Nieto J, Bedoya FJ, Pintado E et al (2004) A new role for monoamine oxidases in the modulation of macrophage-inducible nitric oxide synthase gene expression. J Leukoc Biol 75:1093–1101
Yasuhara H, Tonooka M, Wada I, Oguchi K, Sakamoto K, Kamijo K (1983) Hemodynamics and monoamine oxidase activity in spontaneously hypertensive rats (SHR). Jpn J Pharmacol 33:1057–1064
Friese RS, Mahboubi P, Mahapatra NR, Mahata SK, Schork NJ, Schmid-Schönbein GW, O’Connor DT (2005) Common genetic mechanisms of blood pressure elevation in two independent rodent models of human essential hypertension. Am J Hypertens 18:633–652
Berry MD (2004) Mammalian central nervous system trace amines. Pharmacologic amphetamines, physiologic neuromodulators. J Neurochem 90:257–271
Knoll J, Miklya I, Knoll B, Markó R, Rácz D (1996) Phenylethylamine and tyramine are mixed-acting sympathomimetic amines in the brain. Life Sci 58:2101–2114
Blackwell B, Marley E, Ryle A (1964) Hypertensive crisis associated with monoamine-oxidase inhibitors. Lancet 1:722–723
Raiteri M, Del Carmine R, Bertollini A, Levi G (1977) Effect of sympathomimetic amines on the synaptosomal transport of noradrenaline, dopamine and 5-hydroxytryptamine. Eur J Pharmacol 41:133–143
Neef DW, Jaeger AM, Thiele DJ (2011) Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov 10:930–944
Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H (2007) RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov 6:793–810
Evans WE, Guy RK (2004) Gene expression as a drug discovery tool. Nat Genet 36:214–215
Ely D, Underwood A, Dunphy G, Boehme S, Turner M, Milsted A (2010) Review of the Y chromosome, Sry and hypertension. Steroids 75:747–753
Milsted A, Serova L, Sabban EL, Dunphy G, Turner ME, Ely DL (2004) Regulation of tyrosine hydroxylase gene transcription by Sry. Neurosci Lett 369:203–207
Zhang K, Rao F, Wang L, Rana BK, Ghosh S, Mahata M, Salem RM, Rodriguez-Flores JL, Fung MM, Waalen J et al (2010) Common functional genetic variants in catecholamine storage vesicle protein promoter motifs interact to trigger systemic hypertension. J Am Coll Cardiol 55:1463–1475
Milsted A, Underwood AC, Dunmire J, DelPuerto HL, Martins AS, Ely DL, Turner ME (2010) Regulation of multiple renin-angiotensin system genes by Sry. J Hypertens 28:59–64
Duncan J, Johnson S, Ou X-M (2012) Monoamine oxidases in major depressive disorder and alcoholism. Drug Discov Ther 6:112–122
Menghini R, Marchetti V, Cardellini M, Hribal ML, Mauriello A, Lauro D, Sbraccia P, Lauro R, Federici M (2005) Phosphorylation of GATA2 by Akt increases adipose tissue differentiation and reduces adipose tissue-related inflammation: a novel pathway linking obesity to atherosclerosis. Circulation 111:1946–1953
Kovanen L, Donner K, Partonen T (2015) SIRT1 polymorphisms associate with seasonal weight variation, depressive disorders, and diastolic blood pressure in the general population. PLoS One 10:e0141001
Shimoyama Y, Suzuki K, Hamajima N, Niwa T (2011) Sirtuin 1 gene polymorphisms are associated with body fat and blood pressure in Japanese. Transl Res 157:339–347
Shimoyama Y, Mitsuda Y, Tsuruta Y, Suzuki K, Hamajima N, Niwa T (2012) SIRTUIN 1 gene polymorphisms are associated with cholesterol metabolism and coronary artery calcification in Japanese hemodialysis patients. J Ren Nutr 22:114–119
Kilic U, Gok O, Bacaksiz A, Izmirli M, Elibol-Can B, Uysal O (2014) SIRT1 gene polymorphisms affect the protein expression in cardiovascular diseases. PLoS One 9:e90428
Pizzinat N, Marchal-Victorion S, Maurel A, Ordener C, Bompart G, Parini A (2003) Substrate-dependent regulation of MAO-A in rat mesangial cells: involvement of dopamine D2-like receptors. Am J Physiol Renal Physiol 284:F167–F174
Manoli I, Le H, Alesci S, McFann KK, Su YA, Kino T, Chrousos GP, Blackman MR (2005) Monoamine oxidase-A is a major target gene for glucocorticoids in human skeletal muscle cells. FASEB J 19:1359–1361
Ammon HP, Müller AB (1985) Forskolin: from an ayurvedic remedy to a modern agent. Planta Med 51:473–477
Schlepper M, Thormann J, Mitrovic V (1989) Cardiovascular effects of forskolin and phosphodiesterase-III inhibitors. Basic Res Cardiol 84(Suppl 1):197–212
Dubey MP, Srimal RC, Nityanand S, Dhawan BN (1981) Pharmacological studies on coleonol, a hypotensive diterpene from Coleus forskohlii. J Ethnopharmacol 3:1–13
Wysham DG, Brotherton AF, Heistad DD (1986) Effects of forskolin on cerebral blood flow: implications for a role of adenylate cyclase. Stroke 17:1299–1303
Ou X-M, Chen K, Shih JC (2004) Dual functions of transcription factors, transforming growth factor-beta-inducible early gene (TIEG)2 and Sp3, are mediated by CACCC element and Sp1 sites of human monoamine oxidase (MAO) B gene. J Biol Chem 279:21021–21028
Chen K (2004) Organization of MAO A and MAO B promoters and regulation of gene expression. Neurotoxicology 25:31–36
Shih JC, Chen K (2004) Regulation of MAO-A and MAO-B gene expression. Curr Med Chem 11:1995–2005
Colbert MC, Hall DG, Kimball TR, Witt SA, Lorenz JN, Kirby ML, Hewett TE, Klevitsky R, Robbins J (1997) Cardiac compartment-specific overexpression of a modified retinoic acid receptor produces dilated cardiomyopathy and congestive heart failure in transgenic mice. J Clin Invest 100:1958–1968
Kotake D, Sato T, Hirasawa N (2014) Retinoid signaling in pathological remodeling related to cardiovascular disease. Eur J Pharmacol 729:144–147
Edelstein SB, Breakefield XO (1986) Monoamine oxidases A and B are differentially regulated by glucocorticoids and ‘aging’ in human skin fibroblasts. Cell Mol Neurobiol 6:121–150
Buttgereit F, Burmester G-R, Lipworth BJ (2009) Inflammation, glucocorticoids and risk of cardiovascular disease. Nat Clin Pract Rheumatol 5:18–19
Walker BR (2007) Glucocorticoids and cardiovascular disease. Eur J Endocrinol 157:545–559
Barceló F, Ortiz-Lombardía M, Martorell M, Oliver M, Méndez C, Salas JA, Portugal J (2010) DNA binding characteristics of mithramycin and chromomycin analogues obtained by combinatorial biosynthesis. Biochemistry 49:10543–10552
Choi ES, Nam JS, Jung JY, Cho NP, Cho SD (2014) Modulation of specificity protein 1 by mithramycin A as a novel therapeutic strategy for cervical cancer. Sci Rep 24:7162
Yao L, Dai X, Sun Y, Wang Y, Yang Q, Chen X, Liu Y, Zhang L, Xie W, Liu J (2018) Inhibition of transcription factor SP1 produces neuroprotective effects through decreasing MAO B activity in MPTP/MPP+ Parkinson’s disease models. J Neurosci Res 96:1663–1676
Yu Q, Huang Q, Du X, Xu S, Li M, Ma S (2018) Early activation of Egr-1 promotes neuroinflammation and dopaminergic neurodegeneration in an experimental model of Parkinson’s disease. Exp Neurol 302:145–154
Ahn SY, Cho CH, Park KG, Lee HJ, Park SK, Lee IK, Koh GY (2004) Tumor necrosis factor-alpha induces fractalkine expression preferentially in arterial endothelial cells and mithramycin A suppresses TNF-alpha-induced fractalkine expression. Am J Pathol 164:1663–1672
Orellana EA, Kasinski AL (2015) MicroRNAs in cancer: a historical perspective on the path from discovery to therapy. Cancers (Basel) 7:1388–1405
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906
Hartig SM, Hamilton MP, Bader DA, McGuire SE (2015) The miRNA interactome in metabolic homeostasis. Trends Endocrinol Metab 26:733–745
Barwari T, Joshi A, Mayr M (2016) MicroRNAs in cardiovascular disease. J Am Coll Cardiol 68:2577–2584
Titze-de-Almeida R, David C, Titze-de-Almeida SS (2017) The race of 10 synthetic RNAi-based drugs to the pharmaceutical market. Pharm Res 34:1339–1363
van Rooij E, Kauppinen S (2014) Development of microRNA therapeutics is coming of age. EMBO Mol Med 6:851–864
Chaudhuri AD, Yelamanchili SV, Fox HS (2013) MicroRNA-142 reduces monoamine oxidase A expression and activity in neuronal cells by downregulating SIRT1. PLoS One 8:e79579
Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K et al (2009) DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 37:W273–W276
Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36:D149–D153
Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43:D146–D152
Dweep H, Gretz N, Sticht C (2014) miRWalk database for miRNA-target interactions. Methods Mol Biol 1182:289–305
Krüger J, Rehmsmeier M (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34:W451–W454
Witkos TM, Koscianska E, Krzyzosiak WJ (2011) Practical aspects of microRNA target prediction. Curr Mol Med 11:93–109
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39:1278–1284
Miranda KC, Huynh T, Tay Y, Ang Y-S, Tam W-L, Thomson AM, Lim B, Rigoutsos I (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126:1203–1217
D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M et al (2010) Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J 31:2765–2773
D’Alessandra Y, Carena MC, Spazzafumo L, Martinelli F, Bassetti B, Devanna P, Rubino M, Marenzi G, Colombo GI, Achilli F et al (2013) Diagnostic potential of plasmatic MicroRNA signatures in stable and unstable angina. PLoS One 8:e80345
Roy S, Bantel H, Wandrer F, Schneider AT, Gautheron J, Vucur M, Tacke F, Trautwein C, Luedde T, Roderburg C (2017) miR-1224 inhibits cell proliferation in acute liver failure by targeting the antiapoptotic gene Nfib. J Hepatol 67:966–978
Niu Y, Mo D, Qin L, Wang C, Li A, Zhao X, Wang X, Xiao S, Wang Q, Xie Y et al (2011) Lipopolysaccharide-induced miR-1224 negatively regulates tumour necrosis factor-α gene expression by modulating Sp1. Immunology 133:8–20
Qian J, Li R, Wang Y-Y, Shi Y, Luan W-K, Tao T, Zhang J-X, Xu Y-C, You Y-P (2015) MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem 403:33–41
Nakano T, Nagatsu T, Higashida H (1985) Expression of A and B types of monoamine oxidase in differentiated neuroblastoma hybrid cells. J Neurochem 44:755–758
Rațiu C, Uțu D, Petruș A, Norbert P, Olariu S, Duicu O, Sturza A, Muntean DM (2018) Monoamine oxidase inhibition improves vascular function and reduces oxidative stress in rats with lipopolysaccharide-induced inflammation. Gen Physiol Biophys 37:687–694
Holschneider DP, Chen K, Seif I, Shih JC (2001) Biochemical, behavioral, physiologic, and neurodevelopmental changes in mice deficient in monoamine oxidase A or B. Brain Res Bull 56:453–462
Sims KB, de la Chapelle A, Norio R, Sankila EM, Hsu YP, Rinehart WB, Corey TJ, Ozelius L, Powell JF, Bruns G (1989) Monoamine oxidase deficiency in males with an X chromosome deletion. Neuron 2:1069–1076
Murphy DL, Sims KB, Karoum F, de la Chapelle A, Norio R, Sankila EM, Breakefield XO (1990) Marked amine and amine metabolite changes in Norrie disease patients with an X-chromosomal deletion affecting monoamine oxidase. J Neurochem 54:242–247
Holschneider DP, Scremin OU, Roos KP, Chialvo DR, Chen K, Shih JC (2002) Increased baroreceptor response in mice deficient in monoamine oxidase A and B. Am J Physiol Heart Circ Physiol 282:H964–H972
Harrap SB, Wong ZYH, Stebbing M, Lamantia A, Bahlo M (2002) Blood pressure QTLs identified by genome-wide linkage analysis and dependence on associated phenotypes. Physiol Genomics 8:99–105
Brummett BH, Boyle SH, Siegler IC, Zuchner S, Ashley-Koch A, Williams RB (2008) Lipid levels are associated with a regulatory polymorphism of the monoamine oxidase-A gene promoter (MAOA-uVNTR). Med Sci Monit 14:CR57–CR61
Camarena B, Santiago H, Aguilar A, Ruvinskis E, González-Barranco J, Nicolini H (2004) Family-based association study between the monoamine oxidase A gene and obesity: implications for psychopharmacogenetic studies. Neuropsychobiology 49:126–129
Need AC, Ahmadi KR, Spector TD, Goldstein DB (2006) Obesity is associated with genetic variants that alter dopamine availability. Ann Hum Genet 70:293–303
Fuemmeler BF, Agurs-Collins TD, McClernon FJ, Kollins SH, Kail ME, Bergen AW, Ashley-Koch AE (2008) Genes implicated in serotonergic and dopaminergic functioning predict BMI categories. Obesity (Silver Spring) 16:348–355
Deckert J, Catalano M, Syagailo YV, Bosi M, Okladnova O, Di Bella D, Nöthen MM, Maffei P, Franke P, Fritze J et al (1999) Excess of high activity monoamine oxidase A gene promoter alleles in female patients with panic disorder. Hum Mol Genet 8:621–624
Sabol SZ, Hu S, Hamer D (1998) A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet 103:273–279
Wu Y-H, Fischer DF, Swaab DF (2007) A promoter polymorphism in the monoamine oxidase A gene is associated with the pineal MAOA activity in Alzheimer’s disease patients. Brain Res 1167:13–19
Hotamisligil GS, Breakefield XO (1991) Human monoamine oxidase A gene determines levels of enzyme activity. Am J Hum Genet 49:383–392
Sauna ZE, Kimchi-Sarfaty C (2011) Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12:683–691
Feig DI, Kang D-H, Johnson RJ (2008) Uric acid and cardiovascular risk. N Engl J Med 359:1811–1821
Muiya NP, Wakil S, Al-Najai M, Tahir AI, Baz B, Andres E, Al-Boudari O, Al-Tassan N, Al-Shahid M, Meyer BF et al (2014) A study of the role of GATA2 gene polymorphism in coronary artery disease risk traits. Gene 544:152–158
Connelly JJ, Wang T, Cox JE, Haynes C, Wang L, Shah SH, Crosslin DR, Hale AB, Nelson S, Crossman DC et al (2006) GATA2 is associated with familial early-onset coronary artery disease. PLoS Genet 2:e139
Haigis MC, Sinclair DA (2010) Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5:253–295
Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L (2004) Mammalian SIRT1 represses forkhead transcription factors. Cell 116:551–563
Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW (2004) Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 23:2369–2380
Guarente L (2006) Sirtuins as potential targets for metabolic syndrome. Nature 444:868–874
Westphal CH, Dipp MA, Guarente L (2007) A therapeutic role for sirtuins in diseases of aging? Trends Biochem Sci 32:555–560
Jiang W (2008) Sirtuins: novel targets for metabolic disease in drug development. Biochem Biophys Res Commun 373:341–344
Griffith GC (1960) Amine oxidase inhibitors; their current place in the therapy of cardiovascular diseases. Circulation 22:1156–1165
Acknowledgments
The authors are thankful to the researchers who contributed to studies on the monoamine oxidases. This work was supported by a grant from the Department of Biotechnology, Government of India, to NRM (project number: BT/PR5017/MED/30/756/2012). VG and VA received research fellowships from the Ministry of Human Resource Development, Government of India, and Department of Science and Technology, Government of India, respectively.
Conflict of Interest Statement
The authors declare that there is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Gupta, V., Arige, V., Mahapatra, N.R. (2019). Role of Monoamine Oxidases in Heart Diseases. In: Chakraborti, S., Dhalla, N., Dikshit, M., Ganguly, N. (eds) Modulation of Oxidative Stress in Heart Disease. Springer, Singapore. https://doi.org/10.1007/978-981-13-8946-7_6
Download citation
DOI: https://doi.org/10.1007/978-981-13-8946-7_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8945-0
Online ISBN: 978-981-13-8946-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)