Abstract
In nature, microorganisms such as fungi or bacteria naturally degrade lignin, inspiring numerous attempts worldwide to investigate these reactions with bio-mimicking methods. The present chapter offers a critical overview of the latest concepts and achievements in lignin biological degradation, focussing on fungi, bacteria and enzymes as catalysts to produce chemicals and their use for novel applications. White-rot fungi Pleurotus ostreatus has been widely investigated due to its high delignification performances, however other strains such as Phanerochaete sp. and Phlebia sp. have proven to have similar potential. Due to their amenability to genetic modification, bacteria have opened the coveted path to chemical production from lignin. Vanillin, ferulic acid or muconate can already be obtained by genetically modified Pseudomonas putida or Rhodococcus jostii. Lignolytic enzymes, e.g. peroxidases or laccases, are yet to find viable applications. Mechanisms of lignin biodegradation and bonds cleavage are being better understood thanks to the study of these elemental reactions.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
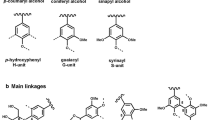
The word “Lignin ” comes from Latin, “lignum”, that can be literally translated as tree, timber or even firewood. Evolving from this, the word “lignin” was specifically used in 1813 by A.P. de Candolle, a Swiss botanist, to denote the product obtained after wood treatment with solvents and mild acid. Twenty-five years later, Anselme Payen reported on two products in wood : cellulose , and what would be later called “lignin” [1]. During the following decades, scientists investigated this peculiar product to identify its structure . The NMR studies [2] of Ludwig [3] and Nimz [4] have led to the commonly admitted representation of the lignin structure : a 3D polymer network, highly cross-linked, and resulting from the co-polymerisation of three different phenol derivative monomers (Fig. 6.1a, b). The reaction of these three monomers leads to the creation of a wide variety of linkages , among which six common bonds: the β-O-4, 5–5, β-5, 4-O-5, β-1 and β-β (Fig. 6.1b and Table 6.1). This large range of bonds and the high cross-linking density render lignin extremely recalcitrant to degradation [5]. It is this high degree of bonding and chemical heterogeneity that has prevented science from upgrading lignin into fine chemicals or bio-based polymers on industrial scale. As a result, 98 % of the lignin world production at present is currently simply burned for energy [6].
Numerous studies have described approaches to depolymerise lignin. Many approaches apply pretreatment and rely on chemical methods to depolymerise lignin. However, results are often difficult to compare as a consequence of the complicated 3D structure of lignin and the lack of standard analytical methods. In nature, organisms naturally degrade wood by different catabolic pathways involving enzymes.
The present chapter primarily focuses on how these organisms have been investigated and utilised to break down lignin into valuable chemicals and by-products . This review first analyses fungi as catalyst to synthesise fine chemicals from lignin. Second, the application of bacteria and the mechanism by which they operate is studied; last, the role of enzymes is analysed. It is the objective of this chapter to review the biodegradation approaches for selectively breaking down and converting lignin into fine chemicals, to provide a perspective on the promising strategies and to map the current scientific frontiers.
2 Fungal Degradation
When considering decaying wood , the first organisms to come to mind are fungi. Wood degrading fungi are mostly divided into three broad categories: white-rot, brown-rot and soft-rot fungi [10] (Table 6.2). Biomass degrading fungi described in the literature are mostly from two phyla (subdivision in biological classification): Basidiomycota and Ascomycota , both from the subkingdom of fungi Dikarya [11]. Although, other phyla are sometimes considered, these two are the most commonly reported for biomass degradation. In the literature, basidiomycetous white-rot fungi are the most studied, since they can selectively degrade lignin, leaving the cellulose and hemicellulose fraction rather intact. However, lignin degradation can also result from the action of other strains and even marine-derived fungi [12]. The term white-rot fungi results from the colour of the degraded wood ; as lignin is being digested by the organism, the only remains are the white cellulose and hemicellulose . This selectivity is particularly important for applications that upgrade cellulose , such as pulp or bioethanol production.
White-rot fungi represent a large group of species, each featuring a different mechanism to attack the lignin barrier directly with a cocktail of enzymes [13, 14]. Fungi produces various enzymes, some to degrade lignocellulose , and others with different roles but with essential role for the fungi to prosper on complex biomass . It has been shown that inhibition of the production of these enzymes can highly hinder or even prevent any growth of the fungi on ligneous and cellulosic mediums [15–17].
For the last 50 years, lignin degrading fungi have been studied from a microbiological perspective, focussing on the mechanisms of lignin degradation and identifying the enzymes involved. Three different types of fungi application have emerged [18]: (a) conversion of lignocellulosic biomass into animal feed and food [19–21] (b) pretreatment agent for delignification (c) biodegradation agent for some phenolic contaminants or various wastes. Transformation of woody biomass into edible mushrooms falls beyond the scope of this study. This review focuses on the two latter applications.
It is worthwhile to clarify some of the nomenclature that can be confusing. There is a clear distinction between delignification , which is the actual removal of lignin from biomass , and lignin degradation/depolymerisation , which represents the cleavage of C-C bonds or ether bonds within the lignin structure , yielding lignin oligomers [22]. Delignification , characterised, for example, by a smaller kappa number ,Footnote 1 is often due to increased solubility of lignin, either by modification or by radical grafting [24, 25]. Consequently, it is possible that lignin degradation also contributes to increased solubility through decreased molecular weight ; however delignification does not imply lignin depolymerisation . Both delignification and lignin bio-degradation are reviewed in this chapter. These two phenomena are critical for unlocking economically feasible bio-refineries capable of producing a full range of marketable chemicals and materials .
2.1 Delignification
The delignification process has two main domains of application: pulp and paper industry , and biofuel production. The pulp and paper industry converts wood into fibres for paper. There are two main pulping processes: mechanical and chemical pulping . In mechanical pulping , lignocellulosic fibres are separated from the wood structure by applying stress. An example is Thermo-Mechanical Pulping (TMP) which relies on steam to plasticise and heat wood above the glass transition temperature (T g) of lignin while applying defibrillating shear, thus reducing fibre damage and energy consumption. Chemical pulping relies on chemical agents and heat to dissolve the ligning-rich fraction of the lumen lamella binding fibres. There are two main chemical processes: Kraft pulping (alkaline) and sulphite pulping (acidic). Kraft pulping is the most important pulping process producing 70 % of all pulp. Pulping aims at developing good quality fibres; full delignification is not always required as it decreases pulp yield . Full fibre delignification or whitening requires bleaching, increasing cost and environmental impact [27–31]. White-rot fungi, with their mild reaction condition, have been investigated as low energy and environment-friendly treatment to reduce lignin content or to replace an existing step of the chemical/thermomechanical pretreatment ; this process is referred to as biopulping . In biofuel production processes, sugars from diverse lignocellulosic resources are fermented by microorganisms to yield targeted products . Hence, the first step of biomass processing is its conversion into simple sugars (glucose, xylose), usually by an enzymatic pathway. Although this process is easy for starch (glucose units linked by α-1,4 and α-1,6 glycosidic bonds) compounds, it is much more complicated for cellulose due to its regular structure (glucose unit linked by β-1,4 glycosidic bonds) which makes it highly crystalline and compact leading to a high resistance to biological degradation [26]. Moreover, the intertwining between cellulose , hemicellulose and lignin results in recalcitrance which hinders the action of the microorganisms [5]. Fungi represent a promising pretreatment agent to reduce this intertwining considering its mild reaction conditions [27, 28]. White-rot strains are of special consideration because of their low uptake of cellulose sugars.
It is estimated that enzymes account (sing.) for 4.5 % of the cost for biofuel production from corn starch and up to 20 % when the whole plant is involved [29–31]. Reviews note that mechanical and chemical pretreatment of biomass for biofuel production can become costly [5, 32–37]. Therefore pretreatment is a critical step for the economics of a process. Different approaches with fungi have been considered to promote delignification by pretreatment combinations.
Single Fungus Treatment
Physical and chemical pretreatment of biomass are notorious for the harsh reaction conditions required. On the contrary, biological pretreatment involves much milder conditions, representing a perfect substitute or complementary degrading agent for delignification [38]. Ge and co-workers have reported on the use of Ceriporiopsis subvermispora, a white-rot fungus, as treatment for Albizia moluccana (Albizia), a widespread invasive tree species in tropical and subtropical regions [39]. The white-rot fungus efficiently decreased the lignin content by 24 % while degrading cellulose and hemicellulose only by half this value. As a consequence, pretreatment with Ceriporiopsis subvermispora allowed a jump in cumulative methane yields from 33.9 L/kg of volatile solid for raw albizia to 123.9 L/kg for pretreated wood chips.
For biofuel applications, fungi could serve not only as delignification agents, but also as production vectors for various products . Indeed, some fungi can degrade lignin along with cellulose which can be useful for production of sugars (Table 6.2) White-rot fungus Phlebia sp. were shown by Kamei et al. to exhibit such combined behaviour for the direct production of bioethanol from cellulosic materials [40]. The same group later reported on the direct conversion of lignocellulosic biomass to bioethanol using this same fungus [41], then proving Phlebia sp. MG-60 to have the combined abilities of lignin degradation, cellulose saccharification, and ethanol fermentation .
Sugar cane bagasse was treated to improve bioethanol yield . Biomass and process variables such as moisture content, additives and presence of metallic cations were shown to greatly affect delignification efficacy [42]. Growth medium addition improved both delignification and ethanol production. Addition of Fe2+, Mn2+ or Cu2+ slightly decreased delignification but bioethanol production was improved by reducing bagasse carbohydrate degradation.
Following the same strategy, Xie et al. reported on the use of an oleaginous fungus strain as lignin degrading microorganism [43]. Cunninghamella echinulate FR3 was determined to be able to degrade cell wall lignin as efficiently as most Basidomycetes fungi. Two strains of sorghum , the wild-type and reduced-lignin-content type (genetically modified to feature improved saccharification efficiency ), were submitted to enzymatic hydrolysis followed by fungal biodegradation. Lignin loss reached 31 % for the wild-type sorghum , while toping up to 46 % for the mutant strain . However, up to 35 % wt of cellulose was degraded during the process. This oleaginous fungus was not targeted as pretreatment to increase sugar production, but was used for lipid bioaccumulation, potentially leading to a new way to provide feedstock for biodiesel refineries [43].
Fungus as Co-treatment
Even though the current chemical and thermomechanical pulping pretreatments are efficient, an additional bio-treatment stage could further increase delignification yield , decrease energy consumption, increase selectivity or fibre quality while decreasing environmental impact. With those benefits in mind, Baker et al. reported on the synergistic effect combining pressure refining, a common process for the pulp and paper industry , with three different white-rot fungi [44]. Ceriporiopsis subvermispora, Phlebiopsis gigantea and Phlebia radiata were fed with pressure refined Miscanthus wood chips. In this strategy, pressure refining concentrates lignin in pellets on the surface of the cellulose fibres [45]. This configuration was tested to increase delignification efficiency by improving the lignin accessibility to the degrading enzymes. After 28 days, the three strains reached their white-rot fungi expectation by decreasing the relative amount of lignin in Miscanthus medium by 10–20 %. However, it is the Ceriporiopsis subvermispora strain that exhibited the best lignin degrading properties by decreasing lignin by 70–75 % of the original content. Un-pressure refined Miscanthus could only reach 10 % of lignin content reduction, highlighting the cooperative action of physical and biological pretreatments.
Kamei et al. reported on the decrease of lignin content and increase in ethanol production when Phlebia sp. MG-60 was fed with alkaline-pretreated sugarcane bagasse compared to untreated sugarcane bagasse [46]. The final results depended on the initial alkaline concentration. After 10 days, the total production of ethanol was 210 mg/g of treated bagasse , providing an ethanol yield (production of ethanol compared to theoretical maximum) of 66 % for the bagasse pretreated with 0.8 % NaOH. Untreated bagasse only reached ca. 2 % of ethanol yield these demonstrating the synergistic effect of the two pretreatments.
Fungi Co-culture
Fungi co-cultures have been investigated to improve lignocellulosic ethanol production efficiency . The cumulative effects of pretreating biomass with two fungi can either improve delignification in the case of two lignolytic fungi [47], or significantly improve the release of reducing sugars by incubating white-rot and brown-rot fungi together [48]. Co-culture of white-rot Ceriporiopsis subvermispora and brown-rot Postia placenta fungi on Liriodendron tulipifera wood chips was reported by Parrow et al. [49]. Although each species increased reducing sugar production during the post saccharification process compared to sterile conditions, rising from 75 to 250 mg/g, the co-culture showed no benefits. Evaluation of interspecific growth interactions showed that an “inhibition” zone was created at the intersection of each taxon growth domain. The two fungi can coexist in the same medium, but did not feature interspecific stimulatory (or inhibitory) interactions . Better understanding of how different species can cooperate in the same biomass is required to improve delignification and saccharification efficiency .
Ma and Ruan reported that Coprinus comatus (producing lignolytic enzymes ) and Trichoderma reesei (producing hemi and cellulolytic enzymes) have synergistic interaction on corn stover [50]. No inhibition was observed at the intersection of the two growth domains, even with intertwining. The co-culture improved delignification by 10 % compared with the lignolytic fungus alone, giving a maximum of 66.5 % delignification after 72 h at optimum temperature conditions. Weight loss, glucan and xylan degradation were also synergistically improved by the co-cultivation of the two strains . A total reducing sugar yield of 82 % was achieved.
Table 6.3 summarises the current critical studies on fungal delignification. Comparison of results remains a challenge because of the different methods used to measure delignification; methods used range from lignin content measurement to IR spectroscopy [51] and even include wettability testing [52].
2.2 Waste Treatment
Lignin degrading fungi have been investigated for their waste degrading properties. Indeed, many fungi have phenol degrading properties suitable to treat a wide range of phenolic pollutants [69, 70]. This field is investigated not only for the treatment of oil -or paper-mill waste water for lignin removal, but also for the textile industry waste water which has a high content of toxic aromatic dyes [71, 72]. Martin and Manzanares reported on the delignification of straw alkaline-pulping liquors by Trametes versicolor [73]. The white-rot fungus was able to remove 75 % of the lignin in the effluent, bringing it from 2 g/L to 0.5 g/L. An important simultaneous decolourisation of the effluent resulted from the delignification .
2.3 Chemical Production
Some studies have considered fungi for lignin chemical modification to change its properties and also to produce chemicals from lignin derivatives . Falconnier et al. reported on the production of vanillin from ferulic acid by white-rot fungus Pycnoporus cinnabarinus I-937 [74]. Vanillin concentration up to 64 mg/L was achieved for a molar yield of 27.5 % w/w. However, recovering vanillin from this mixture proved to be challenging as the fungus produces laccase that repolymerises ferulic acid into a lignin-like polymer.
Work from Zou and co-workers detail lignin demethylation by two fungal strains , Cylindrocladium sp. and Aspergillus sp. [75]. Demethylation of lignin is of high interest for functionalising lignin by liberating reactive groups. The new hydroxyl groups can serve for grafting new functionalities , dangling chains, or even for resins [76, 77]. After 3 weeks in culture, 40 % of the methoxy groups were removed from lignin without causing significant degradation (10 % decrease in content). This work represents a promising start for using fungi as biological modifiers in industrial applications.
2.4 Perspectives
Even though engineering fungi is in its infancy, the current trend to engineer fungi as reactant or catalyst to convert lignin into valued products is very promising. Fungus can be genetically modified as has been proven in the past [15–17, 78], but there are presently only a small number of applications compared with those for bacteria.
White-rot fungi in general, and the Pleurotus ostreatus strain in particular, are of interest for their high delignification rate. However, most of the previous studies have been limited either to basic pretreatment process for lignocellulosic biomass or to waste biodegradation. Using fungi as individual species, co-culture or even combined with chemical reactions has tremendous potential, including the synthesis of fine chemicals and functionalisation of lignin into value added polymers. A clear understanding of the fungi reaction mechanisms is needed, along with their kinetics and the adoption of standard lignin analytical (for yield and content) methods for enabling unbiased comparisons among processes and studies.
3 Bacterial Degradation
In contrast to fungal lignin degradation, enzymology of bacterial lignin breakdown is currently not well understood [79]; extracellular peroxidase and laccase enzymes also appear to be involved. These bacteria can be found in soils, where decaying wood is present, but also in the digestive systems of herbivores, like cows rumens, or in xylophage insects guts, like termites [80]. These bacteria belong to three phyla , actinomycetes , α-proteobacteria and γ-proteobacteria [79]. One review reports on some lignin-degrading prokaryotes in the following phyla : firmicutes, β-proteobacteria , δ-proteobacteria , bacteroidetes and archaea [81].
As a lignin degrading organism, bacteria have been studied for delignification or bioremediation, just as fungi, but to a smaller extent, which is probably because bacterial enzymes have shown to have a lower redox potential [82, 83]. However bacteria have some interesting properties over those of fungi: they are stable over a wide range of pH [84], they can feed on lignin as the sole source of carbon and energy [85], and they are easy to genetically modify [86].
3.1 Delignification
Although considered less effective than fungi, lignin degrading bacteria are also able to utilise lignocellulosic biomass . They are used in delignification processes, either for biopulping or for the biofuel production to increase accessibility of cellulose , but also for bioremediation and waste management. Hacq et al. reported that Serratia liquefaciens could detoxify pulp and paper mill effluent by removing contaminants and lignin by up to 58 % [87]. Agricultural residues can also create environmental pollution, and as a lignin degrading organism, bacteria have received wide interest for treatment [88, 89].
Bacteria delignification research has focused onto Kraft pulp applications because of its high annual production [90]. Shi and co-workers showed that different bacteria strains could successfully degrade Kraft lignin with good yields [91–93]. Cupriavidus basilensis B-8 and Pandaroea sp. B6, two protobacteria, expressed high lignolytic enzymes (manganese peroxidase and laccase ) activity when reacted with Kraft lignin . After 7 days in optimum concentration and pH , a total lignin removal of ca. 45 % was achieved. By degrading Kraft lignin with no other source of carbon required, these two bacteria strains showed good potential for industrial delignification.
Priyadarshinee et al. reported improvement of eucalyptus Kraft pulping by bacterial treatment, resulting in a decrease in kappa number [23]. The raw eucalyptus Kraft pulp was inoculated with two different bacteria strains , Pseudomonas fluorescens NITDPY and Planococcus sp.TRC1. The two bacteria decreased the kappa number by 32 and 37 % in 7 days, respectively. The total amount of phenolic compounds was also reduced. The two different strains react differently to their new carbon source , with P. fluorescens NITDPY degrading lignin faster in the early stage of the experiment through phenolic compound release. Both strains released reducing sugar, but only in a small amount: 0.32 mg/g and 0.15 mg/g for Planococcus sp.TRC1 and P. fluorescens NITDPY, respectively. This indicates that cellulose is not much affected much by the bacteria which is important for pulp and paper applications. The Pseudomonas fluorescens NITDPY and Planococcus sp.TRC1 bacteria show good industrial potential, more than some fungi and they provide non-negligible cellulose degradation. Example of lignin degrading bacteria studied for delignification can be found in Table 6.4.
3.2 Chemical Production
Bacteria are considered to be merely as effective as fungi for delignification; however, they are much easier to genetically modify than fungi [86]. The ability of bacteria to accumulate some of the lignin degradation products as carbon source or energy is an interesting property for chemical production [98]. In the last few years, applications for lignin degrading bacteria have changed. Initially, microorganisms were mostly investigated to degrade and remove lignin from biomass to facilitate recovery of other compounds. Now some research groups are reporting their use for lignin valorisation. While procedures can be complex, the principles are simple. By genetically modifying some bacteria strains , disruption in the metabolic pathway is created, leading to the accumulation of a compound of interest in the medium.
Sainsbury and co-workers reported on the conversion of lignocellulose to vanillin by deleting one enzyme coding gene in Rhodococcus jostii RHA1 [99]. R. jostii degrades lignin derivatives using a biological funnelling process, creating a few intermediates from multiple substrates. The genetic material coding the mechanism responsible for the degradation of vanillin and vanillic acid intermediates were removed from the bacterium DNA . The resulting suppression of vanillin dehydrogenase production causes vanillin to accumulate along with by-products in the reaction medium (Fig. 6.2). After 144 h in a medium containing 2.5 % wheat straw lignocellulose and 0.05 % glucose, the genetically modified bacterium accumulated vanillin up to 96 mg/L. An important amount of ferulic acid could also be observed after 168 h of reaction. When the substrate was changed to Kraft lignin , which is a major industrial by-product of the pulp and paper , an amount of 13 mg/L of vanillin could still be obtained, indicating, however, that lignocellulose remains a better substrate than Kraft lignin . With a similar objective, Graf et al. reported the identification of the gene coding vanillin dehydrogenase in Bacillus subtilis 3NA [100], proving the versatility of this technique.
Catabolic pathways of R. jostii RHA1 used for lignin depolymerisation . Italicised names are gene coding the elementary reaction involved. Vanillin accumulation is due to the aldehyde dehydrogenase gene vdh deletion (Adapted with permission from Sainsbury et al. [99]. Copyright 2013 American Chemical Society. NAD: Nicotinamide adenine dinucleotide)
Linger et al. reported on the biological funnelling behaviour of a bacteria, Pseudomonas putida KT2440 . This organism converts a heterogeneous substrate, such as lignin derivatives , into a sole product , in this case a medium chain length polyhydroxyacid (mcl-PHA). The process was tested on alkaline pretreated liquor (APL), a highly concentrated depolymerised lignin mixture: 32 % lignin made mostly of monomers , dimers and trimers . The result was an accumulation of mcl-PHA into P. putida at high concentration (0.252 g/L at 32 % cell dry weight). Mcl-PHA has a wide range of possible use, from depolymerisation to alkenoic acids, via the thermal pathway, to alkane productions; these reactions were reported in the article [101].
Vardon et al. highlighted an innovative pathway to produce fine chemicals from lignin [102]. Through genetic modification , the natural degradation pathway of bacterium Pseudomonas putida KT2440 was reshaped to yield the production of muconate . P. putida was engineered to transform lignin derived aromatics into catechol and to prevent muconate degradation (Fig. 6.3). Basically, the pcaHG gene that encodes the degradation of protocatechuate is replaced by aroY, a gene from another bacterium , Enterobacter cloacae, which allows decarboxylation instead. The genomic portion that promotes degradation of muconate is then deleted and the transformation of phenol to catechol is allowed through the addition of genomic material dmpKLMNOP from Pseudomonas sp. CF600. The overall production of muconate was reported to reach 0.70 g/L after 24 h [102].
Protocatechuate and catechol branch of the β-ketoadipate pathway in P. putida KT2440 disrupted by deletion of the genes encoding PcaHG and CatBC (crossed arrow). Italicised names are genes coding the elementary reaction involved. Insertion of genes encoding AroY and DmpKLMNOP (double line arrow) yielded muconate accumulation (Adapted from Vardon et al. 2015 [102] with permission of The Royal Society of Chemistry)
Similarly, Johnson and Beckham reported on the genetic modification of P. putida to create pyruvate from lignin derivatives [103]. This was achieved by removing the undesired endogenous reactions in P. putida, the ortho (intradiol) degradation pathway of catechol and protocatechuate and replacing it with the meta (extradiol) cleavage , from another bacterium , Sphingobium sp., to increase the production of pyruvate which is ultimately converted into L-lactate. To avoid the pyruvate from undergoing side reactions, the gene encoding pyruvate dehydrogenase was deleted, preventing any reaction into acetyl-CoA. Moreover, the addition of an external genetic material coding bovine lactate dehydrogenase into Pseudomonas putida was part of a strategy to provide greater competition for pyruvate that might otherwise react within the TCA cycle. However, the efficiency of this method was only tested with lignin model compounds such as benzoate or p–coumarate. No experiments have been performed with lignin.
3.3 Perspectives
Delignification by bacteria is still very poorly understood and less studied than fungal degradation. An attractive strategy currently being explored is to rely on microbial consortia consisting of mixtures of bacteria and fungi to synergistically further lignin biodegradation [104, 105]. Another promising avenue to explore is the bacteria’s bio-funnelling behaviour for chemical production. Through advances in genetic engineering, the synthesis of valuable products from lignin is now achievable; reactions rates, control of competitive reaction and selectivity will determine the economics.
4 Enzymatic Degradation
Both fungi and bacteria involve complex mechanisms of enzymes and intermediates to degrade lignin [13]. However, there is a major drawback for their deployment toward bio-refineries. The control over the inner process is poor and often non-existent, making it difficult to recover the intermediate species of interest. Using enzymes directly and individually in a controlled way is an attractive alternative to deconstructing the degradation process into elementary reactions, allowing a better understanding of the depolymerisation mechanisms and control, which can improve the isolation of the selected chemicals [106].
Enzymes are macro-proteins which can be described as biological catalyst s of high selectivity . Each enzyme accepts a very defined range of substrates. Hence, to degrade the different lignin bonds, organisms require multiple types of enzymes classified into two families: peroxidases and laccases . Peroxidases represent a large group of enzymes involving hydrogen peroxide as the electron acceptor for the specific oxidative reaction they catalyse, such as lignin peroxidases (LiP, EC 1.11.1.14), manganese peroxidase (MnP , EC 1.11.1.13), versatile peroxidase (VP, EC 1.11.1.16), and horseradish peroxidase (HRP, EC 1.11.1.7). Peroxidases belong to the super family of heme-dependent peroxidases . Laccases represents the other major enzyme family for lignin degradation. (Lac, EC 1.10.3.2) Laccase forms its own group by itself belonging to the family of multidomain cupredoxin, and the superfamily of cupredoxins. Laccases use oxygen as electron acceptor. That process avoids any deactivation by hydrogen peroxide that sometimes happens with peroxide enzymes.
Lignolytic enzymes have complex mechanisms . Their origin, structure of substrate (type of lignin) and exterior condition play a major role in their actions. Laccase native from plants, for example, tend to polymerise lignin while fungal or bacterial laccase rather catalyse lignin degradation [107]. Laccases by themselves tend to polymerise low phenolic lignin [108–111] while depolymerisation is favoured with the presence of mediator molecules or with high phenolic ratio lignin [110–114]. Versatile peroxidase from Pleurotus eryngii have been demonstrated to polymerise low molecular weight compounds by inducing cross-linking [115].
Lignolytic enzymes can be produced by bacteria or fungi in a stimulating medium, then extracted, purified and concentrated. The optimisation of this costly production influences industrial viability of enzyme processes. It has been well investigated along with improving enzymes performance (temperature stability , pH sensitivity , etc.) [116–121].
Enzymes can be considered for similar application as those of fungi or bacteria, such as delignification or waste treatment [122–127], but they also have been widely used to better understand the mechanism of lignin degradation by organisms. A series of model lignin molecules, each representing a specific bond in lignin (Fig. 6.1) have been investigated [128, 129].
4.1 Laccases
Laccase represents one of the most reported lignolytic enzymes in the literature [22, 130, 131]. For a long time, laccase was ignored for lignin degradation for two reasons: (i) Phanerochaete chrysosporium , considered a model lignin degrading organism was thought to be unable to produce laccase (ii) its low redox potential (0.5–0.8 V versus normal hydrogen electrode [6]) only allows it to oxidise a small portion of lignin components (the phenolic parts). The role of laccase in lignin degradation is now well demonstrated [17, 22, 128, 132, 133]. The laccase reaction happens around four different copper ions explaining its blue colour [6]. The indirect mechanism of degradation was highlighted in 1990 by Bourbonnais and Paice [132] which who showed the importance of small intermediate molecules, called mediators . Laccase uses molecular oxygen to oxidise the mediator , which then acts as chemical oxidant for lignin. This process allows enzymes to overcome their steric limitation that otherwise eliminates bulky molecules, such as lignin, as potential substrates. These intermediates allow laccases to overcome their phenolic-substrate restriction, thus expanding the range of potential oxidation of the enzyme . Laccase mediators are usually small phenolic compounds such as lignin phenolic components, vanillin , syringaldehyde , veratryl alcohol , or even synthetic mediators , 1-hydroxybenzotriazole (1-HBT), 2,2′-azinobis(3-ethylbenzthiazoline-6-sulphonate) (ABTS ), 2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPO ) , violuric acid (VLA) [134]. The choice of mediator can influence the oxidative potential of laccase and also induce stereo-preference in the substrate. Bohlin et al. showed laccase to exhibit different behaviours with a mixture of diastereoisomers of a β-O-4 model bond molecule [135]. The oxidation is maximum with HBT and shows no preferential isomer, while it is slightly lower with ABTS which exhibits the threo diastereoisomer to be less reactive than the erythro isomer.
The source of laccase , the mediator and the substrate significantly influence the products and reaction rate. Heap et al. reported on the reaction between laccase from Trametes versicolor (white-rot fungus) and various model compounds of β-O-4 bonds with and without mediator [136]. For phenolics, dimerisation can happen with or without the mediator 1-HBT, while Cα oxidation (oxidation of the hydroxyl group on the α position to ketone, see Fig. 6.1b) proceeded with the two non-phenolic compounds (Table 6.5). Bond degradation can happen through ring cleavage for the most electron-rich aromatic ring dimer due to the addition of a methoxy group; for both these non-phenolic molecules, the suppression of mediator prevents any reaction and yields unreacted dimers. The same couple laccase -HBT improved the saccharification of wheat straw. Combining 150 U/g of laccase with 5 % w/w 1-HBT as mediator increased the dilute acid pretreated wheat straw saccharification process by increasing the glucose concentration by 35 % (6.6–8.9 g/L) [136].
The degradation of the β-O-4 bonds were extensively studied as it is the most prevalent lignin bond. Kawai et al. reviewed the mechanism of a non-phenolic β-O-4 model bond , 1,3-dihydroxy-2-(2,6-dimethoxyphenoxy)-1-(4-ethoxy-3-methoxyphenyl)propane, to identify the resulting products [129, 137, 138]. After characterisation of the products , multiple reactions were identified. From 1000 nmol of initial β-O-4 model bond molecule, 120 nmol of products from Cα-Cβ cleavage (cleavage of the bond between carbon α and carbon β, see Fig. 6.1) were collected, along with 99 nmol from β-ether cleavage , 64 nmol from aromatic ring degradation and 43 nmol from Cα oxidation highlighting the complexity of enzymatic lignin degradation.
4.2 Peroxidases
Peroxidases represent the second main category of lignolytic enzymes . Lignin peroxidase (LiP), manganese peroxidase (MnP ) , versatile peroxidase (VP) and horseradish peroxidase (HRP) belong to this group, with a respective redox potential of 1.2 V, 0.8 V, 1.4 V and 0.95 V versus standard hydrogen electrode [6]. Their reaction mechanism are extensively described in two reviews [6, 139].
The high redox potential of lignin peroxidase allows it to oxidise both the phenolic and non-phenolic moieties of lignin. Its heme ion is a ferric one contained inside a ferric protoporphyrin [6]. In the past, lignin peroxidase was shown able to cleave the C-C bond in synthetic ligninFootnote 2 [140]. Lim and co-workers presented the C-C cleavage in different β-1 model bond s [141]. The different model bond s had various substituent groups on the two aromatic rings , and showed different reactivity towards lignin peroxidase , with a conversion ranging from 46 % for the less substituted dimer to 14 % for the dimer with two methoxy groups on each ring. A slight stereo-preference was also highlighted with erythro dimers preferred for degradation. Product analysis suggests a single electron transfer on one of the aromatic rings , followed by a Cα-Cβ cleavage .
Lignin peroxidase can degrade ether model bond s [142]. Lim et al. studied the action of lignin peroxidase of β-O-4 dimers and tetramers with different amounts of methoxy substituent on aromatic rings [143]. Similar to the β-1 bond models , the less substituted model underwent faster bond cleavage . The mechanism combines single electron transfer from dimer or trimer followed by bond cleavage . These two studies highlight different lignin degradation for the two different structures of lignin.
Manganese peroxidase is one of the most common lignolytic enzymes in organisms. It uses hydrogen peroxide just as LiP. Although it can function without [144], MnP requires manganese ions Mn2+ in solution to be fully efficient. In acidic medium, Mn(II) is oxidised by hydrogen peroxide , creating free Mn3+ ions which diffuse and oxidise the lignin phenolic moieties (Eq. 6.1). MnP structure is very similar to LiP, both having ferric ions in their heme group [6].
After reporting biobleaching abilities of Bjerkandera sp. strain BOS55 with a high concentration of MnP , Moreira et al. isolated this enzyme and tested it for delignification properties [145]. After 6 h, and under optimal conditions of pH , H2O2 and Mn2+, a maximum reduction of kappa number of 13 % was obtained for eucalyptus unbleached Kraft pulp. This work highlights the potential application of manganese peroxidase as a bio-bleaching additive.
Versatile peroxidase (VP) is another lignolytic enzyme that can be found in some lignin degrading fungi such as Pleurotus eryngii or Bjerkandera spp. [146]. Its high redox potential (E0 > +1.4 V versus standard hydrogen electrode) allows it to accept a wide range of potential substrate compared to other lignolytic enzymes [6]. VP can degrade β-O-4 model bond s and synthetic lignin [147, 148]. Fernández-Fueyo and co-workers successfully isolated versatile peroxidase from Pleurotus ostreatus [147]. It was shown that this fungus lacked any lignin peroxidase activity; VP assumed this role instead. A phenolic β-O-4 model bond was exposed to VP and both Cα-Cβ cleavage and Cα oxidation occurred. A few products from Cβ-O-C4 were detected. A significant depolymerisation was measured by gel permeation chromatography when the same enzyme was fed to synthetic lignin (dehydrogenation polymer).
Horseradish peroxidase (HRP) is considered to be part of the phenoloxidase responsible for lignin degradation [13]. However, it is also an important lignin and aromatic compounds polymerisation promoter [149, 150]. For that reason, HRP is usually considered as a polymerisation biocatalyst or as a lignin modifier agent [151, 152]. Xia et al. reported horseradish peroxidase to depolymerise two synthetic highly phenolic lignin-based polymers (lignophenols) [153]. Upon continuous addition of hydrogen peroxide , HRP degraded products at a yield of 17 % for lignocatechol and 33 % for lignocresol . Average molecular weight was significantly decreased by about 4–8 fold. For comparison laccase was able to degrade the two lignophenols but at lower conversions, probably due to a low activity of laccase on the lignin moieties in lignophenols.
4.3 Cocktails
From the research perspective, using only one type of enzyme with one well-defined substrate is ideal to elucidate the fundamental mechanism behind delignification and lignin degradation. However, it is a costly operation since it requires production and isolation of the enzyme . Millions of years of natural evolution have lead microorganisms to use not a single enzyme but a cocktail of enzymes to degrade lignin. Mixture of lignolytic enzymes might represent a cheaper and more effective alternative to achieve lignin degradation.
Schroyen and co-workers investigated peroxide enzyme VP from Bjerkandera adusta and laccase from Trametes versicolor to improve production of phenolic compounds and biomethane potential from various lignocellulosic substrates [154]. The two enzymes created a significant increase in phenolic compounds and showed potential as enzymatic pretreatment .
Afrida et al. studied extracellular enzymes from two fungi, Irpex lacteus KB-1.1 and Lentinus tigrinus LP-7 for biobleaching [155]. No separation, isolation or purification of those enzymes mixture was performed, allowing the full enzyme broth to participate to biobleaching. After 4 days, the kappa number was reduced by 4.4 % and 6.7 %, respectively, while the combination of the two enzymes reduced the kappa number by up to 7.4 %. This small improvement demonstrates a synergistic interaction between the extracellular enzymes and has applicability on the industrial scale to significantly decrease chlorine dioxide for bleaching. A similar study showed the use of xylanase and laccase to save 15 % and 25 % of ClO2 respectively, even reaching 35 % when used one after the other (xylanase then laccase ) [127]. However, the co-operation of the two enzymes has not been investigated.
4.4 Bioinspired Enzyme-Like Synthetic Compounds
Enzyme production is often complex and costly, thus limiting industrial applications. After identification of lignin degrading enzymes in early 1980s in Phanerochaete chrysosporium , numerous research groups tried to biomimic these “ligninases ”, by synthesising competitive enzyme -like complexes. Zucca et al. brought these compounds back to the front scene with an extensive review [156]. Natural metalloporphines, such as hemin or heme group in hemoglobin, are well known to exhibit peroxidase or catalase-like activity and inspired research on new oxidation catalyst . The ferriheme in lignolytic peroxidases was shown to degrade high-potential lignin structure [156]. After several generations of synthetic heme groups, the stability became higher than enzymes with higher catalytic activity [156–159]. Immobilisation on clay or other substrate can significantly increase its stability [160, 161]. Metalloporphines quickly proved similar efficiency than enzymes at degrading common lignin model bond molecules [162–164], and even more. Compared to enzymes, the absence of protein scaffold reduced hindrance, allowing a wider range of substrate. Some lignin model bonds usually not considered for enzymatic degradation (e.g. 5–5, β-5, or diphenyl methane DPM bond) can be cleaved, oxidised or undergo ring cleavage reaction with metalloporphines [160, 165, 166].
Farell and Skerker analysed four metalloporphines/metalloporphyrin (Fig. 6.4) and their catalytic degradation of different model bonds [166]. Similar to natural enzymes, these biomimic catalyst s were deactivated by high H2O2 concentration, and their activity was pH dependent. Veratryl alcohol , a typical substrate for lignolytic enzymes , could be readily oxidised by the two catalysts. Metalloporphine compound with iron metallic ion can oxidise β-1 and β-O-4 model bonds , but also induce C-C cleavage in β-5 model bond and aromatic ring cleavage in 5–5 model bonds , which has never been observed with enzyme of fungal systems. For metalloporphyrin with manganese for active centre, clear delignification was identified, with a reduction of 40 % in the kappa number in just 15 min.
Structure of a metalloporphine, M being the metallic ion, with the eight β positions and four meso one. IUPAC nomenclature [168] defines porphyrins as porphine derivatives where organic side chains are substituted for all the eight hydrogen atoms in the porphine pyrrole rings (the β positions). Although most of the synthetic heme catalyst s are porphines by definition, they are nevertheless they are usually misleadingly referred as porhyrins [156]
Crestini and co-workers highlighted the catalytic activity between manganese and iron porphyrin [165]. As Kraft lignin contains a significant amount of DPM and 5–5 substructure, two model molecules representing these bonds were studied for degradation. The manganese porphines exhibited higher conversions than the iron one, with a particular high activity for manganese meso-tetra(N-methylpyridinio)porphine pentaacetate. Higher activity of Mn porphines was attributed to higher stability of these complexes. When residual Kraft lignin was submitted to the different catalyst s, results indicated that manganese and iron porphines oxidised the lignin, but the latter induced a high amount of coupling reactions probably yielding higher molecular weight lignin. In general, manganese porphines proved superior to iron porphines for delignification and model bond cleavage .
A delignification of wood sawdust by metalled phthalocyanine or porphyrin was reported by Barbat and co-workers [167]. This pretreatment is considered for replacement of chemical process, sodium chlorite solution followed by alkaline extraction , for holocellulose recovery. Although this is a promising technique, with 1 % w/w phenolic compounds release from biomass , quantitative delignification amount has not been measured.
4.5 Perspectives
An emerging pathway for enzymes (natural or synthetic) is the catabolic treatment of biomass for delignification . To this end, enzymes represent a more manageable alternative to fungi or bacteria as their degradation mechanism is much simpler. Despite significant progress, there is still no commercial application of lignolytic enzymes for lignin degradation. The lack of efficient production systems and the poor understanding of the degradation pathways have prevented the development of efficient systems implementable on the industry industrial scale. A good comprehension of the interaction mechanism between enzymes and the role of the different mediators (natural and synthetic) is needed for critical breakthrough in lignin bio-treatment. This knowledge can enable engineering microorganisms with high efficient lignin degradation properties tailored for specific industrial application, including fine chemical production.
5 Conclusion and Future Outlook
Lignin is the second most abundant polymer on earth, just behind cellulose . It represents a widely available, low cost and sustainable feedstock offering tremendous opportunities for the production of phenolic bio-based fine chemicals and monomers . Despite numerous studies devoted to its degradation, lignin still remains a most recalcitrant polymer to break down into oligomers and reproducible monomers . This is because of the multitude of chemical bonds involved and the variability of lignin chemical composition which is a function of the lignocellulosic source and the extraction process. This chapter has highlighted many of the shortcomings in fundamental knowledge restricting development; a methodical and comprehensive study on the principles and mechanism of biodegradation is required to unleash lignin as a controlled source for conversion into fine chemicals. Alternatives mimicking nature are of special interest. Many fungi and bacteria can degrade lignin, inspiring a plethora of schemes and processes for delignification and production of fine chemicals. Of those currently studied, white-rot fungi appear as the most promising delignification microorganism. The choice to genetic engineer – or not – has to be considered as it offers new routes for increased yields and selectivities needed for fine chemical production. Enzymatic degradation of lignin still faces high costs, low reaction rates, and poorly known bond selectivity . The inhibition mechanisms typical to enzymatic degradation are not well understood for “ligninases ”, as well as the effect of temperature resistant enzymes and reactions in a solvent . A promising avenue is to rely on enzyme cocktails or sequential enzymatic reaction schemes, because purification is typically an important cost of the process, and a wide array of products can be expected from lignin biodegradation into monomers/short oligomer due to the multitude of bonds. As lignin represents the best and most renewal natural source of phenol , it is well worth investing into the fundamental biodegradation studies that will lead to breakthroughs for process commercialization .
Notes
- 1.
Kappa number is a titration process representative of the pulp colour and approximatively proportional to lignin content at low concentration [23]; it can be used to assess delignification efficiency.
- 2.
Synthetic polymer is a polymer with structure similar to lignin. It can vary from simple polyphenol to more elaborate structure .
Abbreviations
- ABTS:
-
2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)
- Acetyl COA:
-
Acetyl coenzyme A
- Aff.:
-
“affinis”, Latin form of “related”
- CTMP:
-
Chemithermomechanical Pulping
- DPM:
-
Diphenyl Methane bond
- EC:
-
Enzyme Commission number
- GFC:
-
Gel Filtration Chromatography
- 1-HBT:
-
1-Hydroxybenzotriazole
- Lac:
-
Laccase
- LC-MS:
-
Liquid Chromatograph-Mass Spectrometry
- LiP:
-
Lignin Peroxidase
- mcl-PHA:
-
Medium Chain Length Polyhydroxyalkanoate
- MnP:
-
Manganese Peroxidase
- NMR:
-
Nuclear Magnetic Resonance
- Sp.:
-
Species (sing.)
- Spp.:
-
Species (plur.)
- TCA cycle:
-
Tricarboxylic Acid cycle
- TEMPO :
-
2,2,6,6-Tetramethyl-1-piperidinyloxy
- VLA:
-
Violuric acid
- VP:
-
Versatile peroxidase
References
Zakzeski J, Bruijnincx PC, Jongerius AL, Weckhuysen BM. The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev. 2010;110(6):3552–99.
Laurichesse S, Avérous L. Chemical modification of lignins: towards biobased polymers. Prog Polym Sci. 2014;39(7):1266–90.
Ludwig CH, Nist BJ, McCarthy JL. Lignin. XII. 1 the high resolution nuclear magnetic resonance spectroscopy of protons in compounds related to lignin. J Am Chem Soc. 1964;86(6):1186–96.
Nimz H. Beech lignin—proposal of a constitutional scheme. Angew Chem Int Ed Engl. 1974;13(5):313–21.
Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–7.
Pollegioni L, Tonin F, Rosini E. Lignin‐degrading enzymes. FEBS J. 2015;282(7):1190–213.
Adler E. Lignin chemistry—past, present and future. Wood Sci Technol. 1977;11(3):169–218.
Hon DN-S, Shiraishi N. Wood and cellulosic chemistry, revised, and expanded. CRC Press; 2000.
Capanema EA, Balakshin MY, Kadla JF. Quantitative characterization of a hardwood milled wood lignin by nuclear magnetic resonance spectroscopy. J Agric Food Chem. 2005;53(25):9639–49.
Arora DK. Fungal biotechnology in agricultural, food, and environmental applications. New York: CRC Press; 2003.
Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous OW, Dai Y-C, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson K-H, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo J-M, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüßler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao Y-J, Zhang N. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111(5):509–47.
Atalla MM, Zeinab HK, Eman RH, Amani AY, Abeer A. Screening of some marine-derived fungal isolates for lignin degrading enzymes (LDEs) production. Agric Biol J N Am. 2010;1(4):591–9.
Leonowicz A, Matuszewska A, Luterek J, Ziegenhagen D, Wojtaś-Wasilewska M, Cho N-S, Hofrichter M, Rogalski J. Biodegradation of lignin by white-rot fungi. Fungal Genet Biol. 1999;27(2–3):175–85.
Salame TM, Knop D, Levinson D, Mabjeesh SJ, Yarden O, Hadar Y. Inactivation of a Pleurotus ostreatus versatile peroxidase‐encoding gene (mnp2) results in reduced lignin degradation. Environ Microbiol. 2014;16(1):265–77.
Bourdais A, Bidard F, Zickler D, Berteaux-Lecellier V, Silar P, Espagne E. Wood utilization is dependent on catalase activities in the filamentous fungus Podospora anserina. PLoS One. 2012;7(4):e29820.
Xie N, Ruprich‐Robert G, Silar P, Chapeland‐Leclerc F. Bilirubin oxidase‐like proteins from Podospora anserina: promising thermostable enzymes for application in transformation of plant biomass. Environ Microbiol. 2015;17(3):866–75.
Xie N, Chapeland‐Leclerc F, Silar P, Ruprich‐Robert G. Systematic gene deletions evidences that laccases are involved in several stages of wood degradation in the filamentous fungus Podospora anserina. Environ Microbiol. 2014;16(1):141–61.
Kirk TK, Chang H-M. Potential applications of bio-ligninolytic systems. Enzym Microb Technol. 1981;3(3):189–96.
van Kuijk SJA, Sonnenberg ASM, Baars JJP, Hendriks WH, Cone JW. Fungal treated lignocellulosic biomass as ruminant feed ingredient: a review. Biotechnol Adv. 2015;33(1):191–202.
Khan NA, Hussain S, Ahmad N, Alam S, Bezabhi M, Hendriks WH, Yu P, Cone JW. Improving the feeding value of straws with Pleurotus ostreatus. Anim Prod Sci. 2015;55(2):241–5.
Colavolpe MB, Albertó E. Cultivation requirements and substrate degradation of the edible mushroom Gymnopilus pampeanus-A novel species for mushroom cultivation. Sci Hortic. 2014;180:161–6.
Munk L, Sitarz AK, Kalyani DC, Mikkelsen JD, Meyer AS. Can laccases catalyze bond cleavage in lignin? Biotechnol Adv. 2015;33(1):13–24.
Priyadarshinee R, Kumar A, Mandal T, Dasguptamandal D. Improving the perspective of raw eucalyptus Kraft pulp for industrial applications through autochthonous bacterial mediated delignification. Ind Crop Prod. 2015;74:293–303.
Lund M, Ragauskas AJ. Enzymatic modification of Kraft lignin through oxidative coupling with water-soluble phenols. Appl Microbiol Biotechnol. 2001;55(6):699–703.
Moldes D, Vidal T. New possibilities of Kraft pulp biobleaching with laccase and sulfonated mediators. Process Biochem. 2011;46(3):656–60.
Gray KA, Zhao L, Emptage M. Bioethanol. Curr Opin Chem Biol. 2006;10(2):141–6.
Palli L, Gullotto A, Tilli S, Gori R, Lubello C, Scozzafava A. Effect of carbon source on the degradation of 2-naphthalenesulfonic acid polymers mixture by Pleurotus ostreatus in petrochemical wastewater. Process Biochem. 2014;49(12):2272–8.
Shrestha P, Szaro TM, Bruns TD, Taylor JW. Systematic search for cultivatable fungi that best deconstruct cell walls of Miscanthus and sugarcane in the field. Appl Environ Microbiol. 2011;77(15):5490–504.
Kazi FK, Fortman J, Anex R, Kothandaraman G, Hsu D, Aden A, Dutta A. Techno-economic analysis of biochemical scenarios for production of cellulosic ethanol. NREL/TP-6A2-46588, National Renewable Energy Laboratory; June 2010; 2010
Klein‐Marcuschamer D, Oleskowicz‐Popiel P, Simmons BA, Blanch HW. The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol Bioeng. 2012;109(4):1083–7.
Shrestha P, Ibáñez AB, Bauer S, Glassman SI, Szaro TM, Bruns TD, Taylor JW. Fungi isolated from Miscanthus and sugarcane: biomass conversion, fungal enzymes, and hydrolysis of plant cell wall polymers. Biotechnol Biofuels. 2015;8(1):38.
Harmsen P, Huijgen W, Bermudez L, Bakker R. Literature review of physical and chemical pretreatment processes for lignocellulosic. Biomass. 2010; 1–49.
Donohoe BS, Decker SR, Tucker MP, Himmel ME, Vinzant TB. Visualizing lignin coalescence and migration through maize cell walls following thermochemical pretreatment. Biotechnol Bioeng. 2008;101(5):913–25.
Kumar P, Barrett DM, Delwiche MJ, Stroeve P. Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res. 2009;48(8):3713–29.
McMillan JD. Pretreatment of lignocellulosic biomass. In: ACS symposium series (USA), 1994.
Agbor VB, Cicek N, Sparling R, Berlin A, Levin DB. Biomass pretreatment: fundamentals toward application. Biotechnol Adv. 2011;29(6):675–85.
Alvira P, Tomás-Pejó E, Ballesteros M, Negro MJ. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: a review. Bioresour Technol. 2010;101(13):4851–61.
Castoldi R, Bracht A, de Morais GR, Baesso ML, Correa RCG, Peralta RA, Moreira RDFPM, Polizeli MDLTDM, de Souza CGM, Peralta RM. Biological pretreatment of Eucalyptus grandis sawdust with white-rot fungi: study of degradation patterns and saccharification kinetics. Chem Eng J. 2014;258:240–6.
Ge X, Matsumoto T, Keith L, Li Y. Fungal pretreatment of albizia chips for enhanced biogas production by solid-state anaerobic digestion. Energy Fuels. 2015;29(1):200–4.
Kamei I, Hirota Y, Mori T, Hirai H, Meguro S, Kondo R. Direct ethanol production from cellulosic materials by the hypersaline-tolerant white-rot fungus Phlebia sp. MG-60. Bioresour Technol. 2012;112:137–42.
Kamei I, Hirota Y, Meguro S. Integrated delignification and simultaneous saccharification and fermentation of hard wood by a white-rot fungus, Phlebia sp. MG-60. Bioresour Technol. 2012;126:137–41.
Khuong LD, Kondo R, De Leon R, Anh TK, Meguro S, Shimizu K, Kamei I. Effect of chemical factors on integrated fungal fermentation of sugarcane bagasse for ethanol production by a white-rot fungus, Phlebia sp. MG-60. Bioresour Technol. 2014;167:33–40.
Xie S, Qin X, Cheng Y, Laskar D, Qiao W, Sun S, Reyes LH, Wang X, Dai SY, Sattler SE, Kao K, Yang B, Zhang X, Yuan J. Simultaneous conversion of all cell wall components by an oleaginous fungus without chemi-physical pretreatment. Green Chem. 2015;17(3):1657–67.
Baker PW, Charlton A, Hale MD. Increased delignification by white-rot fungi after pressure refining Miscanthus. Bioresour Technol. 2015;189:81–6.
Gustafsson J, Lehto JH, Tienvieri T, Ciovica L, Peltonen J. Surface characteristics of thermomechanical pulps; the influence of defibration temperature and refining. Colloids Surf A Physicochem Eng Asp. 2003;225(1–3):95–104.
Khuong LD, Kondo R, De Leon R, Anh TK, Shimizu K, Kamei I. Bioethanol production from alkaline-pretreated sugarcane bagasse by consolidated bioprocessing using Phlebia sp. MG-60. Int Biodeterior Biodegrad. 2014;88:62–8.
Cook C, Francocci F, Cervone F, Bellincampi D, Bolwell PG, Ferrari S, Devoto A. Combination of pretreatment with white-rot fungi and modification of primary and secondary cell walls improves saccharification. Bioenergy Res. 2015;8(1):175–86.
Giles RL, Galloway ER, Zackeru JC, Naithani V, Parrow MW. Two stage fungal biopulping solubilizes lignocellulosic carbohydrates without supplemental enzymatic hydrolysis. Int Biodeterior Biodegrad. 2014;86:265–71.
Giles RL, Zackeru JC, Galloway ER, Elliott GD, Parrow MW. Single versus simultaneous species treatment of wood with Ceriporiopsis subvermispora and Postia placenta for ethanol applications, with observations on interspecific growth inhibition. Int Biodeterior Biodegrad. 2015;99:66–72.
Ma K, Ruan Z. Production of a lignocellulolytic enzyme system for simultaneous bio-delignification and saccharification of corn stover employing co-culture of fungi. Bioresour Technol. 2015;175:586–93.
Gai YP, Zhang WT, Mu ZM, Ji XL. Involvement of ligninolytic enzymes in degradation of wheat straw by Trametes trogii. J Appl Microbiol. 2014;117(1):85–95.
Liu L, Qian C, Jiang L, Yu H-Q. Direct three-dimensional characterization and multiscale visualization of wheat straw deconstruction by white-rot fungus. Environ Sci Technol. 2014;48(16):9819–25.
TAPPI. Acid-insoluble lignin in wood and pulp. T222 om-88. Atlanta: TAPPI Press; 1988.
Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D. Determination of structural carbohydrates and lignin in biomass. Version 2010. National Renewable Energy Laboratory, USA; 2008.
Kirk TK, Obst JR. Lignin determination. In: Methods in enzymology-biomass, part b, lignin, pectin, and chitin. vol 161. Academic Press. Inc.;1988. p. 87–101.
Deswal D, Gupta R, Nandal P, Kuhad RC. Fungal pretreatment improves amenability of lignocellulosic material for its saccharification to sugars. Carbohydr Polym. 2014;99:264–9.
Knežević A, Milovanović I, Stajić M, Lončar N, Brčeski I, Vukojević J, Ćilerdžić J. Lignin degradation by selected fungal species. Bioresour Technol. 2013;138:117–23.
Karp SG, Faraco V, Amore A, Letti LAJ, Soccol VT, Soccol CR. Statistical optimization of laccase production and delignification of sugarcane bagasse by Pleurotus ostreatus in solid-state fermentation. BioMed Res Int. 2015;12(16):19.
Knežević A, Milovanović I, Stajić M, Vukojević J. Potential of Trametes species to degrade lignin. Int Biodeterior Biodegrad. 2013;85:52–6.
Van Soest PJ, Robertson JB, Lewis BA. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74(10):3583–97.
López MJ, Suárez-Estrella F, Vargas-García MC, López-González JA, Verstichel S, Debeer L, Wierinck I, Moreno J. Biodelignification of agricultural and forest wastes: effect on anaerobic digestion. Biomass Bioenergy. 2013;58:343–9.
Liu S, Wu S, Pang C, Li W, Dong R. Microbial pretreatment of corn stovers by solid-state cultivation of Phanerochaete chrysosporium for biogas production. Appl Biochem Biotechnol. 2014;172(3):1365–76.
TAPPI. Kappa number of pulp. T236 cm-85. Atlanta: TAPPI Press; 1985.
Singh P, Sulaiman O, Hashim R, Peng LC, Singh RP. Evaluating biopulping as an alternative application on oil palm trunk using the white-rot fungus Trametes versicolor. Int Biodeterior Biodegrad. 2013;82:96–103.
Fukushima RS, Hatfield RD. Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. J Agric Food Chem. 2001;49(7):3133–9.
Ghorbani F, Karimi M, Biria D, Kariminia H, Jeihanipour A. Enhancement of fungal delignification of rice straw by Trichoderma viride sp. to improve its saccharification. Biochem Eng J. 2015;101:77–84.
Cianchetta S, Di Maggio B, Burzi PL, Galletti S. Evaluation of selected white-rot fungal isolates for improving the sugar yield from wheat straw. Appl Biochem Biotechnol. 2014;173(2):609–23.
Mohanram S, Rajan K, Carrier DJ, Nain L, Arora A. Insights into biological delignification of rice straw by Trametes hirsuta and Myrothecium roridum and comparison of saccharification yields with dilute acid pretreatment. Biomass Bioenergy. 2015;76:54–60.
Haroune L, Saibi S, Bellenger J-P, Cabana H. Evaluation of the efficiency of Trametes hirsuta for the removal of multiple pharmaceutical compounds under low concentrations relevant to the environment. Bioresour Technol. 2014;171:199–202.
García-Delgado C, Alfaro-Barta I, Eymar E. Combination of biochar amendment and mycoremediation for polycyclic aromatic hydrocarbons immobilization and biodegradation in creosote-contaminated soil. J Hazard Mater. 2015;285:259–66.
Namhyun C, Lee I-S, Song H-S, Bang W-G. Mechanisms used by white-rot fungus to degrade lignin and toxic chemicals. J Microbiol Biotechnol. 2000;10(6):737–52.
Han Y, Shi L, Meng J, Yu H, Zhang X. Azo dye biodecolorization enhanced by Echinodontium taxodii cultured with lignin. PLoS ONE. 2014;9(10):7185–8.
Martin C, Manzanares P. A study of the decolourization of straw soda-pulping effluents by Trametes versicolor. Bioresour Technol. 1994;47(3):209–14.
Falconnier B, Lapierre C, Lesage-Meessen L, Yonnet G, Brunerie P, Colonna-Ceccaldi B, Corrieu G, Asther M. Vanillin as a product of ferulic acid biotransformation by the white-rot fungus Pycnoporus cinnabarinus I-937: identification of metabolic pathways. J Biotechnol. 1994;37(2):123–32.
Zou L, Ross BM, Hutchison LJ, Christopher LP, Dekker RF, Malek L. Fungal demethylation of Kraft lignin. Enzym Microb Technol. 2015;73:44–50.
Qin J, Woloctt M, Zhang J. Use of polycarboxylic acid derived from partially depolymerized lignin as a curing agent for epoxy application. ACS Sustain Chem Eng. 2014;2(2):188–93.
Hilburg SL, Elder AN, Chung H, Ferebee RL, Bockstaller MR, Washburn NR. A universal route towards thermoplastic lignin composites with improved mechanical properties. Polymer (United Kingdom). 2014;55(4):995–1003.
Brun S, Malagnac F, Bidard F, Lalucque H, Silar P. Functions and regulation of the Nox family in the filamentous fungus Podospora anserina: a new role in cellulose degradation. Mol Microbiol. 2009;74(2):480–96.
Bugg TDH, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol. 2011;22(3):394–400.
Singh A, Singh DP, Tiwari R, Kumar K, Singh RV, Singh S, Prasanna R, Saxena AK, Nain L. Taxonomic and functional annotation of gut bacterial communities of Eisenia foetida and Perionyx excavatus. Microbiol Res. 2015;175:48–56.
Tian J-H, Pourcher A-M, Bouchez T, Gelhaye E, Peu P. Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Appl Microbiol Biotechnol. 2014;98(23):9527–44.
Brown ME, Chang MCY. Exploring bacterial lignin degradation. Curr Opin Chem Biol. 2014;19(1):1–7.
Ahmad M, Taylor CR, Pink D, Burton K, Eastwood D, Bending GD, Bugg TDH. Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol Biosys. 2010;6(5):815–21.
Mathews SL, Pawlak J, Grunden AM. Bacterial biodegradation and bioconversion of industrial lignocellulosic streams. Appl Microbiol Biotechnol. 2015;99(7):2939–54.
L-y C, Y-h C, C-j T, Yang Z-h, Zheng Y, Shi Y. Depolymerization and decolorization of Kraft lignin by bacterium Comamonas sp. B-9. Appl Microbiol Biotechnol. 2014;98(4):1907–12.
Chang Y-C, Choi D, Takamizawa K, Kikuchi S. Isolation of Bacillus sp. strains capable of decomposing alkali lignin and their application in combination with lactic acid bacteria for enhancing cellulase performance. Bioresour Technol. 2014;152:429–36.
Haq I, Kumar S, Kumari V, Singh SK, Raj A. Evaluation of bioremediation potentiality of ligninolytic Serratia liquefaciens for detoxification of pulp and paper mill effluent. J Hazard Mater. 2016;305:190–9.
Buraimoh OM, Ilori MO, Amund OO, Michel FC, Grewal SK. Assessment of bacterial degradation of lignocellulosic residues (sawdust) in a tropical estuarine microcosm using improvised floating raft equipment. Int Biodeterior Biodegrad. 2015;104:186–93.
Liang J, Peng X, Yin D, Li B, Wang D, Lin Y. Screening of a microbial consortium for highly simultaneous degradation of lignocellulose and chlorophenols. Bioresour Technol. 2015;190:381–7.
Gellerstedt G. Softwood Kraft lignin: raw material for the future. Ind Crop Prod. 2015;77:845–54.
Shi Y, Chai L, Tang C, Yang Z, Zheng Y, Chen Y, Jing Q. Biochemical investigation of Kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst Eng. 2013;36(12):1957–65.
Shi Y, Chai L, Tang C, Yang Z, Zhang H, Chen R, Chen Y, Zheng Y. Characterization and genomic analysis of Kraft lignin biodegradation by the beta-proteobacterium Cupriavidus basilensis B-8. Biotechnol Biofuels. 2013;6(1):1.
Chen Y, Chai L, Tang C, Yang Z, Zheng Y, Shi Y, Zhang H. Kraft lignin biodegradation by Novosphingobium sp. B-7 and analysis of the degradation process. Bioresour Technol. 2012;123:682–5.
Kumar M, Singhal A, Thakur IS. Comparison of submerged and solid state pretreatment of sugarcane bagasse by Pandoraea sp. ISTKB: enzymatic and structural analysis. Bioresour Technol. 2016;203:18–25.
Huang XF, Santhanam N, Badri DV, Hunter WJ, Manter DK, Decker SR, Vivanco JM, Reardon KF. Isolation and characterization of lignin‐degrading bacteria from rainforest soils. Biotechnol Bioeng. 2013;110(6):1616–26.
Chen J, Xu L, Wu Y, Tong J, Chen Y. Production, characterization of acetyl esterase from a rumen bacteria strain RB3, and application potential of the strain in biodegradation of crop residues. Renew Energy. 2014;68:134–9.
Singhal A, Jaiswal PK, Thakur IS. Biopulping of bagasse by Cryptococcus albidus under partially sterilized conditions. Int Biodeterior Biodegrad. 2015;97:143–50.
Salvachúa D, Karp EM, Nimlos CT, Vardon DR, Beckham GT. Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem. 2015;17(11):4951–67.
Sainsbury PD, Hardiman EM, Ahmad M, Otani H, Seghezzi N, Eltis LD, Bugg TDH. Breaking down lignin to high-value chemicals: the conversion of lignocellulose to vanillin in a gene deletion mutant of Rhodococcus jostii RHA1. ACS Chem Biol. 2013;8(10):2151–6.
Graf N, Wenzel M, Altenbuchner J. Identification and characterization of the vanillin dehydrogenase YfmT in Bacillus subtilis 3NA. Appl Microbiol Biotechnol. 2016;100(8):3511–21.
Linger JG, Vardon DR, Guarnieri MT, Karp EM, Hunsinger GB, Franden MA, Johnson CW, Chupka G, Strathmann TJ, Pienkos PT. Lignin valorization through integrated biological funneling and chemical catalysis. Proc Natl Acad Sci U S A. 2014;111(33):12013–8.
Vardon DR, Franden MA, Johnson CW, Karp EM, Guarnieri MT, Linger JG, Salm MJ, Strathmann TJ, Beckham GT. Adipic acid production from lignin. Energy Environ Sci. 2015;8(2):617–28.
Johnson CW, Beckham GT. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab Eng. 2015;28:240–7.
Jiménez DJ, Dini-Andreote F, Van Elsas JD. Metataxonomic profiling and prediction of functional behaviour of wheat straw degrading microbial consortia. Biotechnol Biofuels. 2014;7(1):1–18.
Brzonova I, Kozliak E, Kubátová A, Chebeir M, Qin W, Christopher L, Ji Y. Kenaf biomass biodecomposition by basidiomycetes and actinobacteria in submerged fermentation for production of carbohydrates and phenolic compounds. Bioresour Technol. 2014;173:352–60.
Bugg TDH, Rahmanpour R. Enzymatic conversion of lignin into renewable chemicals. Curr Opin Chem Biol. 2015;29:10–7.
Awasthi M, Jaiswal N, Singh S, Pandey VP, Dwivedi UN. Molecular docking and dynamics simulation analyses unraveling the differential enzymatic catalysis by plant and fungal laccases with respect to lignin biosynthesis and degradation. J Biomol Struct Dyn. 2015;33(9):1835–49.
Grönqvist S, Viikari L, Niku-Paavola M-L, Orlandi M, Canevali C, Buchert J. Oxidation of milled wood lignin with laccase, tyrosinase and horseradish peroxidase. Appl Microbiol Biotechnol. 2005;67(4):489–94.
Areskogh D, Li J, Gr G, Henriksson G. Investigation of the molecular weight increase of commercial lignosulfonates by laccase catalysis. Biomacromolecules. 2010;11(4):904–10.
Shleev S, Persson P, Shumakovich G, Mazhugo Y, Yaropolov A, Ruzgas T, Gorton L. Interaction of fungal laccases and laccase-mediator systems with lignin. Enzym Microb Technol. 2006;39(4):841–7.
Kondo R, Iimori T, Imamura H, Nishida T. Polymerization of DHP and depolymerization of DHP-glucoside by lignin oxidizing enzymes. J Biotechnol. 1990;13(2–3):181–8.
Xia Z, Yoshida T, Funaoka M. Enzymatic synthesis of polyphenols from highly phenolic lignin-based polymers (lignophenols). Biotechnol Lett. 2003;25(1):9–12.
Fernaud JRH, Carnicero A, Perestelo F, Cutuli MH, Arias E, Falcón MA. Upgrading of an industrial lignin by using laccase produced by Fusarium proliferatum and different laccase-mediator systems. Enzym Microb Technol. 2006;38(1–2):40–8.
Eggert C, Temp U, Dean JFD, Eriksson K-EL. A fungal metabolite mediates degradation of non-phenolic lignin structures and synthetic lignin by laccase. Febs Lett. 1996;391(1–2):144–8.
Salvachúa D, Prieto A, Mattinen M-L, Tamminen T, Liitiä T, Lille M, Willför S, Martínez AT, Martínez MJ, Faulds CB. Versatile peroxidase as a valuable tool for generating new biomolecules by homogeneous and heterogeneous cross-linking. Enzym Microb Technol. 2013;52(6–7):303–11.
Yamasaki Y, Yamaguchi M, Yamagishi K, Hirai H, Kondo R, Kamei I, Meguro S. Expression of a manganese peroxidase isozyme 2 transgene in the ethanologenic white-rot fungus Phlebia sp. strain MG-60. SpringerPlus. 2014;3(1):699.
Vaithanomsat P, Sangnam A, Boonpratuang T, Choeyklin R, Promkiam-on P, Chuntranuluck S, Kreetachat T. Wood degradation and optimized laccase production by Resupinate white-rot fungi in Northern Thailand. BioResources. 2013;8(4):6342–60.
Feng H, Zhang D, Sun Y, Zhi Y, Mao L, Luo Y, Xu L, Wang L, Zhou P. Expression and characterization of a recombinant laccase with alkalistable and thermostable properties from Streptomyces griseorubens JSD-1. Appl Biochem Biotechnol. 2015;176(2):547–62.
Kannaiyan R, Mahinpey N, Kostenko V, Martinuzzi RJ. Nutrient media optimization for simultaneous enhancement of the laccase and peroxidases production by coculture of Dichomitus squalens and Ceriporiopsis subvermispora. Biotechnol Appl Biochem. 2015;62(2):173–85.
Lim S-J, Jeon S-J. Optimal conditions for laccase production from the white-rot fungus Marasmius scorodonius. Korean J Microbiol Biotechnol. 2014;42(3):225–31.
Gorska EB, Jankiewicz U, Dobrzynski J, Galazka A, Sitarek M, Gozdowski D, Russel S, Kowalczyk P. Production of ligninolytic enzymes by cultures of white-rot fungi. Pol J Microbiol. 2014;63(4):461–5.
Yadav M, Yadav HS. Applications of ligninolytic enzymes to pollutants, wastewater, dyes, soil, coal, paper and polymers. Environ Chem Lett. 2015;13(3):309–18.
Chandrasekaran G, Choi S-K, Lee Y-C, Kim G-J, Shin H-J. Oxidative biodegradation of single-walled carbon nanotubes by partially purified lignin peroxidase from Sparassis latifolia mushroom. J Ind Eng Chem. 2014;20(5):3367–74.
Moreira S, Milagres AMF, Mussatto SI. Reactive dyes and textile effluent decolorization by a mediator system of salt-tolerant laccase from Peniophora cinerea. Sep Purif Technol. 2014;135:183–9.
Kuhar F, Castiglia V, Levin L. Enhancement of laccase production and malachite green decolorization by co-culturing Ganoderma lucidum and Trametes versicolor in solid-state fermentation. Int Biodeterior Biodegrad. 2015;104:238–43.
Inoue S, Igarashi Y, Yoneda Y, Kawai S, Okamura H, Nishida T. Elimination and detoxification of fungicide miconazole and antidepressant sertraline by manganese peroxidase-dependent lipid peroxidation system. Int Biodeterior Biodegrad. 2015;100:79–84.
Sharma A, Thakur VV, Shrivastava A, Jain RK, Mathur RM, Gupta R, Kuhad RC. Xylanase and laccase based enzymatic Kraft pulp bleaching reduces adsorbable organic halogen (AOX) in bleach effluents: a pilot scale study. Bioresour Technol. 2014;169:96–102.
Kawai S, Umezawa T, Higuchi T. Degradation mechanisms of phenolic β-1 lignin substructure model compounds by laccase of Coriolus versicolor. Arch Biochem Biophys. 1988;262(1):99–110.
Kawai S, Nakagawa M, Ohashi H. Degradation mechanisms of a nonphenolic β-O-4 lignin model dimer by Trametes versicolor laccase in the presence of 1-hydroxybenzotriazole. Enzym Microb Technol. 2002;30(4):482–9.
Youn H-D, Hah YC, Kang S-O. Role of laccase in lignin degradation by white-rot fungi. FEMS Microbiol Lett. 1995;132(3):183–8.
Mayer AM, Staples RC. Laccase: new functions for an old enzyme. Phytochemistry. 2002;60(6):551–65.
Bourbonnais R, Paice MG. Oxidation of non-phenolic substrates: an expanded role for laccase in lignin biodegradation. FEBS Lett. 1990;267(1):99–102.
Ander P, Eriksson K-E. The importance of phenol oxidase activity in lignin degradation by the white-rot fungus Sporotrichum pulverulentum. Arch Microbiol. 1976;109(1–2):1–8.
Moldes D, Díaz M, Tzanov T, Vidal T. Comparative study of the efficiency of synthetic and natural mediators in laccase-assisted bleaching of eucalyptus Kraft pulp. Bioresour Technol. 2008;99(17):7959–65.
Bohlin C, Andersson P-O, Lundquist K, Jönsson LJ. Differences in stereo-preference in the oxidative degradation of diastereomers of the lignin model compound 1-(3, 4-dimethoxyphenyl)-2-(2-methoxyphenoxy)-1, 3-propanediol with enzymic and non-enzymic oxidants. J Mol Catal B Enzym. 2007;45(1–2):21–6.
Heap L, Green A, Brown D, Van Dongen B, Turner N. Role of laccase as an enzymatic pretreatment method to improve lignocellulosic saccharification. Catal Sci Technol. 2014;4(8):2251–9.
Kawai S, Asukai M, Ohya N, Okita K, Ito T, Ohashi H. Degradation of a non-phenolic β-O-4 substructure and of polymeric lignin model compounds by laccase of Coriolus versicolor in the presence of 1-hydroxybenzotriazole. FEMS Microbiol Lett. 1999;170(1):51–7.
Kawai S, Nakagawa M, Ohashi H. Aromatic ring cleavage of a non-phenolic β-O-4 lignin model dimer by laccase of Trametes versicolor in the presence of 1-hydroxybenzotriazole. FEBS Lett. 1999;446(2–3):355–8.
Wong DWS. Structure and action mechanism of ligninolytic enzymes. Appl Biochem Biotechnol. 2009;157(2):174–209.
Hammel KE, Jensen K, Mozuch MD, Landucci LL, Tien M, Pease EA. Ligninolysis by a purified lignin peroxidase. J Biol Chem. 1993;268(17):12274–81.
Lim SH, Lee WS, Kim Y-I, Sohn Y, Cho DW, Kim C, Kim E, Latham JA, Dunaway-Mariano D, Mariano PS. Photochemical and enzymatic SET promoted C–C bond cleavage reactions of lignin β-1 model compounds containing varying number of methoxy substituents on their arene rings. Tetrahedron. 2015;71(24):4236–47.
Baciocchi E, Fabbri C, Lanzalunga O. Lignin peroxidase-catalyzed oxidation of nonphenolic trimeric lignin model compounds: fragmentation reactions in the intermediate radical cations. J Org Chem. 2003;68(23):9061–9.
Lim SH, Nahm K, Ra CS, Cho DW, Yoon UC, Latham JA, Dunaway-Mariano D, Mariano PS. Effects of alkoxy groups on arene rings of lignin β-O-4 model compounds on the efficiencies of single electron transfer-promoted photochemical and enzymatic C–C bond cleavage reactions. J Org Chem. 2013;78(18):9431–43.
Moreira MT, Feijoo G, Mester T, Mayorga P, Sierra-Alvarez R, Field JA. Role of organic acids in the manganese-independent biobleaching system of Bjerkandera sp. strain BOS55. Appl Environ Microbiol. 1998;64(7):2409–17.
Moreira MT, Sierra-Alvarez R, Lema JM, Feijoo G, Field J. Oxidation of lignin in eucalyptus Kraft pulp by manganese peroxidase from Bjerkandera sp. strain BOS55. Bioresour Technol. 2001;78(1):71–9.
Hofrichter M, Ullrich R, Pecyna MJ, Liers C, Lundell T. New and classic families of secreted fungal heme peroxidases. Appl Microbiol Biotechnol. 2010;87(3):871–97.
Fernández-Fueyo E, Ruiz-Dueñas FJ, Martínez MJ, Romero A, Hammel KE, Medrano FJ, Martínez AT. Ligninolytic peroxidase genes in the oyster mushroom genome: heterologous expression, molecular structure, catalytic and stability properties, and lignin-degrading ability. Biotechnol Biofuels. 2014; 7(3).
Caramelo L, Martínez MJ, Martínez ÁT. A search for ligninolytic peroxidases in the fungus Pleurotus eryngii involving α-Keto-γ-thiomethylbutyric acid and lignin model dimers. Appl Environ Microbiol. 1999;65(3):916–22.
Guerra A, Ferraz A, Cotrim AR, Da Silva FT. Polymerization of lignin fragments contained in a model effluent by polyphenoloxidases and horseradish peroxidase/hydrogen peroxide system. Enzym Microb Technol. 2000;26(5–6):315–23.
SCtälä H, Pajunen A, Rummakko P, Sipilä J, Brunow G. A novel type of spiro compound formed by oxidative cross coupling of methyl sinapate with a syringyl lignin model compound. A model system for the β-1 pathway in lignin biosynthesis. J Chem Soc Perkin Trans. 1999;1(4):461–4.
Liu R, Dong A, Fan X, Wang Q, Yu Y, Cavaco-Paulo A. HRP-mediated polyacrylamide graft modification of raw jute fabric. J Mol Catal B Enzym. 2015;116:29–38.
Zhou H, Chang Y, Wu X, Yang D, Qiu X. Horseradish peroxidase modification of sulfomethylated wheat straw alkali lignin to improve its dispersion performance. ACS Sustain Chem Eng. 2015;3(3):518–23.
Xia Z, Yoshida T, Funaoka M. Enzymatic degradation of highly phenolic lignin-based polymers (lignophenols). Eur Polym J. 2003;39(5):909–14.
Schroyen M, Vervaeren H, Vandepitte H, Van Hulle SWH, Raes K. Effect of enzymatic pretreatment of various lignocellulosic substrates on production of phenolic compounds and biomethane potential. Bioresour Technol. 2015;192:696–702.
Afrida S, Tamai Y, Watanabe T, Osaki M. Biobleaching of Acacia Kraft pulp with extracellular enzymes secreted by Irpex lacteus KB-1.1 and Lentinus tigrinus LP-7 using low-cost media. World J Microbiol Biotechnol. 2014;30(8):2263–71.
Zucca P, Rescigno A, Rinaldi AC, Sanjust E. Biomimetic metalloporphines and metalloporphyrins as potential tools for delignification: molecular mechanisms and application perspectives. J Mol Catal A Chem. 2014;388–389:2–34.
Meunier B. Metalloporphyrins as versatile catalysts for oxidation reactions and oxidative DNA cleavage. Chem Rev. 1992;92(6):1411–56.
Zakavi S, Mojarrad AG, Rayati S. Substituent effects on the catalytic activity of a series of manganese meso-tetra (aryl) porphyrins:(2-, 3-, 4)-Pyridyl, 4-sulfonatophenyl and 3-sulfonato-4-methoxyphenyl groups compared to phenyl and 4-methoxyphenyl ones. J Mol Catal A Chem. 2012;363–364:153–8.
Tsuchiya S, Seno M. Novel synthetic method of phenol from benzene catalysed by perfluorinated hemin. Chem Lett. 1989;2:263–6.
Crestini C, Pastorini A, Tagliatesta P. Metalloporphyrins immobilized on motmorillonite as biomimetic catalysts in the oxidation of lignin model compounds. J Mol Catal A Chem. 2004;208(1–2):195–202.
Crestini C, Pastorini A, Tagliatesta P. The immobilized porphyrin‐mediator system Mn (TMePyP)/clay/HBT (clay‐PMS): a lignin peroxidase biomimetic catalyst in the of lignin and lignin model compounds. Eur J Inorg Chem. 2004;2004(22):4477–83.
Cui F, Dolphin D. Metallophthalocyanines as possible lignin peroxidase models. Bioorg Med Chem. 1995;3(5):471–7.
Artaud I, Ben-Aziza K, Mansuy D. Iron porphyrin-catalyzed oxidation of 1, 2-dimethoxyarenes: a discussion of the different reactions involved and the competition between the formation of methoxyquinones or muconic dimethyl esters. J Org Chem. 1993;58(12):3373–80.
Labat G, Meunier B. Factors controlling the reactivity of a ligninase model based on the association of potassium monopersulfate to manganese and iron porphyrin complexes. J Org Chem. 1989;54(21):5008–11.
Crestini C, Saladino R, Tagliatesta P, Boschi T. Biomimetic degradation of lignin and lignin model compounds by synthetic anionic and cationic water soluble manganese and iron porphyrins. Bioorg Med Chem. 1999;7(9):1897–905.
Cui F, Wijesekera T, Dolphin D, Farrell R, Skerker P. Biomimetic degradation of lignin. J Biotechnol. 1993;30(1):15–26.
Barbat A, Gloaguen V, Sol V, Krausz P. Aqueous extraction of glucuronoxylans from chestnut wood: new strategy for lignin oxidation using phthalocyanine or porphyrin/H 2 O 2 system. Bioresour Technol. 2010;101(16):6538–44.
The Merck index. 12th ed. Whitehouse Station: Merck and Co. Inc; 1996.
Rochefort D, Bourbonnais R, Leech D, Paice MG. Oxidation of lignin model compounds by organic and transition metal-based electron transfer mediators. Chem Commun. 2002;11:1182–3.
Nousiainen P, Kontro J, Manner H, Hatakka A, Sipilä J. Phenolic mediators enhance the manganese peroxidase catalyzed oxidation of recalcitrant lignin model compounds and synthetic lignin. Fungal Genet Biol. 2014;72:137–49.
Acknowledgements
The financial support of the ARC Industrial Transformation Research Hub – Bioprocessing Advanced Manufacturing Initiative (BAMI) and Monash University is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media Singapore
About this chapter
Cite this chapter
Longe, L., Garnier, G., Saito, K. (2016). Lignin Biodegradation with Fungi, Bacteria and Enzymes for Producing Chemicals and Increasing Process Efficiency. In: Fang, Z., Smith, Jr., R. (eds) Production of Biofuels and Chemicals from Lignin. Biofuels and Biorefineries. Springer, Singapore. https://doi.org/10.1007/978-981-10-1965-4_6
Download citation
DOI: https://doi.org/10.1007/978-981-10-1965-4_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-1964-7
Online ISBN: 978-981-10-1965-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)