Abstract
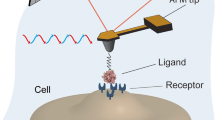
Ligand-receptor recognition on the cell membrane enables the communication of cells with the extracellular environment. Atomic force microscopy (AFM)-based single-molecule dynamic force spectroscopy represents one of the most powerful techniques available to directly investigate ligand-receptor recognition under physiological conditions without considerable disruption to cells. It provides important information for research on biological processes, disease pathogenesis, and mechanism of drugs. Here we describe an example of applying single-molecule dynamic force spectroscopy to study the binding of epidermal growth factor (EGF) to its receptor EGFR, as well as the effect of two clinical drugs, Pertuzumab and Trastuzumab, on the interaction of EGF and EGFR.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hinterdorfer P, Dufrene YF (2006) Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods 3:347–355
Wang C, Yadavalli VK (2014) Investigating biomolecular recognition at the cell surface using atomic force microscopy. Micron 60:5–17
Liu B, Chen W, Zhu C (2015) Molecular force spectroscopy on cells. Annu Rev Phys Chem 66:427–451
Shi X, Zhang X, Xia T, Fang X (2012) Living cell study at the single-molecule and single-cell levels by atomic force microscopy. Nanomedicine 10:1625–1637
Jiang Y, Zhu C, Ling L, Wan L, Fang X, Bai C (2003) Specific aptamer-protein interaction studied by atomic force microscopy. Anal Chem 75:2112–2116
Yang H, Yu J, Fu G, Shi X, Xiao L, Chen Y, Fang X, He C (2007) Interaction between single molecules of Mac-1 and ICAM-1 in living cells: an atomic force microscopy study. Exp Cell Res 313:3497–3504
O’Donoghue M, Shi X, Fang X, Tan W (2012) Single-molecule atomic force microscopy on live cells compares aptamer and antibody rupture forces. Anal Bioanal Chem 4 02:3205–3209
Bell G (1978) Models for specific adhesion of cells to cells. Science 200:618–627
Merkel R, Nassoy P, Leung A, Ritchie K, Evans E (1999) Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 397:50–53
Zhang X, Shi X, Xu L, Yuan J, Fang X (2013) Atomic force microscopy study of the effect of HER 2 antibody on EGF mediated ErbB ligand-receptor interaction. Nanomed Nanotechnol Biol Med 9:627–635
Liu J, Zhang X, Wang X, Xu L, Li J, Fang X (2016) Single-molecule force spectroscopy study of the effect of cigarette carcinogens on thrombomodulin-thrombin interaction. Sci Bull 61:1187–1194
Wei Y, Zhang X, Xu L, Yi S, Li Y, Fang X, Liu H (2012) The effect of cigarette smoke extract on thrombomodulin-thrombin binding: an atomic force microscopy study. Sci China Life Sci 55:891–897
Yu J, Wang Q, Shi X, Ma X, Yang H, Chen Y-G, Fang X (2007) Single-molecule force spectroscopy study of interaction between transforming growth factor beta 1 and its receptor in living cells. J Phys Chem B 111:13619–13625
Shi X, Xu L, Yu J, Fang X (2009) Study of inhibition effect of Herceptin on interaction between Heregulin and ErbB Receptors HER3/HER2 by single-molecule force spectroscopy. Exp Cell Res 315:2847–2855
Safenkova IV, Zherdev AV, Dzantiev BB (2012) Application of atomic force microscopy for characteristics of single intermolecular interactions. Biochemistry (Moscow) 77:1536–1552
Sader JE, Lu J, Mulvaney P (2014) Effect of cantilever geometry on the optical lever sensitivities and thermal noise method of the atomic force microscope. Rev Sci Instrum 85:113702
Noy A (2011) Force spectroscopy 101: how to design, perform, and analyze an AFM-based single molecule force spectroscopy experiment. Curr Opin Chem Biol 15:710–718
Evans EA, Calderwood DA (2007) Forces and bond dynamics in cell adhesion. Science 316:1148–1153
Ge L, Jin G, Fang X (2012) Investigation of the Interaction between a Bivalent Aptamer and Thrombin by AFM. Langmuir 28:707–713
Bieri O, Wirz J, Hellrung B, Schutkowski M, Drewello M, Kiefhaber T (1999) The speed limit for protein folding measured by triplet-triplet energy transfer. Proc Natl Acad Sci U S A 96:9597–9601
Odorico M, Teulon JM, Berthoumieu O, Chen SWW, Parot P, Pellequer JL (2007) An integrated methodology for data processing in dynamic force spectroscopy of ligand-receptor binding. Ultramicroscopy 107:887–894
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Liu, J., Li, W., Zhang, X., Feng, Y., Fang, X. (2019). Ligand-Receptor Binding on Cell Membrane: Dynamic Force Spectroscopy Applications. In: Santos, N., Carvalho, F. (eds) Atomic Force Microscopy. Methods in Molecular Biology, vol 1886. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-8894-5_8
Download citation
DOI: https://doi.org/10.1007/978-1-4939-8894-5_8
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-8893-8
Online ISBN: 978-1-4939-8894-5
eBook Packages: Springer Protocols