Abstract
Cell surface receptors, often called transmembrane receptors, are key cellular components as they control and mediate cell communication and signalling, converting extracellular signals into intracellular signals. Elucidating the molecular details of ligand binding (cytokine, growth factors, hormones, pathogens,...) to cell surface receptors and how this binding triggers conformational changes that initiate intracellular signalling is needed to improve our understanding of cellular processes and for rational drug design. Unfortunately, the molecular complexity and high hydrophobicity of membrane proteins significantly hamper their structural and functional characterization in conditions mimicking their native environment. With its piconewton force sensitivity and (sub)nanometer spatial resolution, together with the capability of operating in liquid environment and at physiological temperature, atomic force microscopy (AFM) has proven to be one of the most powerful tools to image and quantify receptor-ligand bonds in situ under physiologically relevant conditions. In this article, a brief overview of the rapid evolution of AFM towards quantitative biological mapping will be given, followed by selected examples highlighting the main advances that AFM-based ligand-receptor studies have brought to the fields of cell biology, immunology, microbiology, and virology, along with future prospects and challenges.

Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since its invention in 1986 as technique to contour the topography of solid state surfaces with atomic resolution [1], atomic force microscopy (AFM) has quickly evolved into a multifunctional nanoscopic platform with growing applications in diverse fields, ranging from physics, chemistry, and material science to biology and medicine [2, 3]. In AFM imaging, the interaction forces between a very sharp tip and the surface of a sample are exploited to reconstruct topographical information. A classical AFM setup consists of a flexible cantilever ending with a sharp stylus, a laser diode focused on the back of the cantilever and reflected onto a position-sensitive photodiode, and a piezoelectric scanner, which is connected to the photodiode through a feedback loop. Typically, during an image acquisition, the stylus of the cantilever is raster scanned across a sample surface through the piezoelectric scanner. Variations in interaction forces between the tip and the sample, reflecting atomic-scale differences in surface topography, will be sensed by the tip and will cause a deflection of the cantilever, which will be detected as change of the laser position on the photodiode. The feedback loop will then re-adjust in real time the relative height position of the sample and the tip in order to keep their parameters of interaction constant (e.g. force of interaction, oscillation amplitude, or frequency shift, depending of the AFM imaging mode), thus enabling the reconstruction of the surface topography with sub-nanometer precision. Alternatively, an atomic force microscope is used as a force sensor, in what is commonly called force spectroscopy mode. In this mode, the stylus is cyclically approached and retracted from the surface while monitoring the variation of the force with respect to the tip-sample distance. This enables the reconstruction of force-distance curves with piconewton sensitivity, from which a plethora of mechanical and physico-chemical sample properties, such as elasticity, stiffness, and adhesion, can be extracted [3].
Already in the first decade after the AFM invention, new applications exploiting the potential of the technique to detect properties other than surface topography started to emerge (Fig. 1). In particular, taking advantage of its ability to detect interaction forces, it was rapidly demonstrated that the cantilever tip could be functionalized to allow the detection of specific forces for simultaneously mapping the topography of the sample and to extract its physico-chemical properties quantitatively. In 1994, Lieber’s group elegantly illustrated this principle by functionalizing AFM tips with self-assembled alkanethiol monolayers terminated with hydrophobic or hydrophilic groups (Fig. 1a). Those tips acting as chemical sensors were used to evidence hydrophilic and hydrophobic domains on patterned surfaces otherwise topographically smooth, setting the bases for chemical force spectroscopy (CFS) and mapping applications [4]. Shortly afterwards, the concept was transferred to biomolecular interactions, with the widely explored avidin-biotin [5,6,7] and antibody-antigen systems [8] opening the doors to the sensing of specific biomolecular interactions (Fig. 1b, c).
In the context of biomolecular interactions, the functionalization of the AFM stylus is of key importance. Unlike small chemicals, the direct grafting of biomolecules onto the AFM tip could lead to steric effects, which de facto result in a decrease of the lateral resolution. Furthermore, direct contact between biomolecules and tip surface would result in a reduction of the molecule mobility and orientation preventing the specific interaction. In addition, it could lead to partial biomolecule denaturation and as a consequence to a loss of its proper functional state, which makes the grafting strategy particularly critical for AFM cantilever functionalization. In 1996, Hinterdorfer et al. [8] developed a protocol for the controlled functionalization of AFM tips with single biomolecules (Fig. 1c), enabling the study of molecular recognition processes at the level of single interaction events by single-molecule force spectroscopy (SMFS). By covalently coupling antibodies to AFM tips via a long flexible polyethylene glycol (PEG) spacer at a sufficiently low concentration, single ligands were enabled to interact with their antigen pair on the sample surface. The flexible linker increases the binding probability thanks to the higher orientation freedom of the antibody. Finally, the stretching of the long and flexible PEG spacer during the unbinding process displays a well-defined nonlinear pattern before bond rupture happens, therefore providing a specific signature that allows a straightforward discrimination of the probed antibody-antigen interactions from the unspecific adhesion events.
Biomolecular mapping along with the possibility to operate AFM in liquid environment and at ambient temperature opened the avenue to the development of AFM-based live cell applications (Fig. 1d). In 2000, Gaub’s group exploited the high specificity of Helix pomatia lectin for N-acetyl-galactosamine-terminated glycolipids present on the cellular membrane of group A red blood cells to map and discriminate between mixed red blood cell populations [9]. By functionalizing the tip with the lectin through a PEG spacer, they were able to correlate both topography- and affinity-based maps of mixed group O and group A red blood cells adsorbed on a glass surface, showing that AFM could be used to probe affinity forces in situ on biologically relevant samples. In parallel, single-cell force spectroscopy applications (SCFS) started to emerge [10] (Fig. 1e). These applications are based on the immobilization of single living cells on a tipless AFM cantilever, via specific receptor-ligand interactions or adhesive coatings, which are subsequently used to record force-distance curves between the cell probe and a substrate or the surface of another cell. This development allowed novel biological parameters to be measured, such as cell-cell adhesion directly, and at the high spatial and force resolution characteristic for AFM [11].
In the last decade, AFM-based biological applications have witnessed a new revolution thanks to a plethora of sophisticated technological developments [12]. Key breakthroughs towards biomolecular and live cell applications include the development of environmental chambers to keep cells alive, including mammalian cells, in physiological conditions for extended periods of time; the development of faster and more sophisticated force-distance curve–based AFM methodologies (FD-based AFM); and the coupling of AFM to optical microscopy techniques (ranging from epifluorescence to confocal and super-resolution microscopes) (Fig. 1f–j). The evolution of FD-based AFM has brought mapping of biomolecules and live cells to a new exciting level, allowing the simultaneous fast acquisition of topographical and quantitative multiparametric information of biological samples at high resolution. In previously developed FD-based modes, such as force-volume AFM (FV-AFM), the cantilever follows a two-step rectangular trajectory in which for each pixel of the image the tip is approached and retracted from the sample in the z-direction and subsequently moved horizontally to the next pixel to scan in the x-y direction while recording force-distance curves. Although this approach allows the simultaneous recording of force-distance curves and the topography reconstruction of the surface on a pixel-by pixel basis, the slow acquisition time severely limits the number of pixel per image that can be practically acquired. Recently, this limitation has been partially overcome with the development of triangular or sinusoidal modes, which greatly increased the speed of image acquisition and hence the resolution of biomolecular mapping and force-sensing. As a result, new applications, and in particular the simultaneous topographical and quantitative analysis of receptors-ligand bonds, are now made possible on living cells (Fig. 1f–h) or purified model membranes (Fig. 1i, j), at molecular or even in some cases submolecular resolution [13,14,15]. Furthermore, the instrumental developments allowing the coupling of AFM and optical techniques, such as epifluorescence and confocal microscopy, have opened up new exciting perspectives for the study of biological processes in situ in biologically relevant contexts through the combination of AFM and cell and molecular biology approaches [3, 12].
In the following sections, an overview of the most recent applications concerning the AFM-based quantitative study of cell receptor-ligand interactions in physiologically relevant conditions will be addressed, providing examples in the fields of cellular biology, immunology, microbiology, and virology, which have especially benefited from the most recent developments. An outlook discussing present and possible future contributions of AFM to the molecular understanding of cell surface phenomena will follow, to highlight future directions and perspectives this exciting field is able to offer.
Probing receptor-ligand bonds in situ by AFM: state of the art
Cell biology and immunology applications
Imaging and quantifying cell receptor-ligand binding dynamics in biologically relevant conditions are critical steps to understand and modulate cell surface receptors with biomedical perspectives. In addition to inherent difficulties in structural characterization associated with receptor size and hydrophobicity, crystal structures, where available, are still not fully representative of the ensemble of conformations explored by the receptor within its cellular environment. It is well known that their dynamic assembly on the membrane modulates the functional state of many receptors and that often receptors are able to interact with multiple ligands with different levels of affinity and binding-related conformational changes.
Classical SMFS and FV-AFM have been extensively used to characterize the binding free energy landscape of receptor-ligand pairs both on supported lipid bilayers and on living cells [16, 17]. Typically, ligands of interest are covalently immobilized on the AFM tips that are subsequently used to probe and quantify forces and kinetic parameters of ligand-receptor binding. The main limitations of these approaches are the acquisition time and the absence of high-resolution topographical information, which hampers a molecular attribution of the quantified forces as well as the detection of dynamical changes in receptor conformation or spatial distribution upon ligand binding. This can be particularly problematic when studying receptor-ligand binding on living cells, due to the intrinsic complexity and heterogeneity of cell membranes. Simultaneous acquisition of topographical and qualitative ligand binding information has been achieved with the invention of the AFM-based topography and recognition imaging (TREC), which has been extensively used to map the organization and binding of several ligand-receptor pairs, such as the human gonadotropin–releasing hormone receptor and the distribution of Hsp70 on the surface of cancer cells [18]. Although TREC does not allow the quantitative extraction of binding parameters from the ligand-receptor recognition, it can be coupled to SMFS to retrieve the quantitative parameters of binding on selected receptors [19].
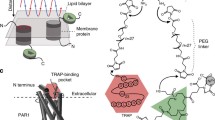
Recently, a major breakthrough in AFM-based ligand-receptor studies came from the advances in multiparametric FD-based AFM, which has proved to be a powerful approach to image cell surface receptors at the single-molecule level and to simultaneously quantifying the biophysical parameters describing the receptor binding process to a variety of ligands. Some of the most significant advances regard G protein–coupled receptors (GPCRs), which are the largest membrane receptor family and cover essential roles in cell homeostasis and responses to hormones and neurotransmitters [20]. In a recent study, FD-based AFM was used to reconstruct free energy landscapes of ligand binding of purified protease-activated receptor-1 (PAR1) dispersed in supported lipid bilayers (Fig. 2a–e). By functionalizing the AFM tip with its endogenous ligand, the thrombin receptor–activating peptide (TRAP), it was possible to image single human PAR1 molecules at nanometric resolution and simultaneously reconstruct the energy landscape of the ligand-receptor interaction under physiological conditions [21] (Fig. 2a). The FD-based AFM mode allows the reconstruction of the topographical information in a pixel-by-pixel manner, while simultaneously recording the interaction forces in force-distance curves between the ligand on the tip and the receptor on the surface over a wide range of loading rates (i.e. speed at which the force is applied) (Fig. 2b–d). Practically, the application of an external force on a ligand-receptor bond results in a loading rate–dependent deformation of the energy barrier separating the bound and unbound states, with a consequent reduction of the bond lifetime. This in turn leads to processes occurring at shorter time scales than in the absence of force, which enables the study of slow dissociation processes by SMFS, otherwise inaccessible by other techniques. Well-established biophysical models, such as the Bell-Evans model [22] and the Friddle-Noy-De Yoreo model [23], allow the extrapolation of the kinetic rate of bond dissociation (which is related to the bond lifetime) and of the free energy of unbinding at equilibrium (i.e. at zero applied force) respectively, leading to the reconstruction of the free energy landscape of the ligand-receptor bond interaction (Fig. 2e). The method was also applied in presence of a PAR1 inhibitor, and to a variety of ligands having slightly higher or lower affinity compared with TRAP, revealing subtle differences in the energy landscape of ligand-receptor binding and therefore proving itself as an effective approach to characterize accurate ligand binding profiles and potentially assist in drug testing and design. In a similar work, the same receptor was analyzed with bifunctionalized AFM tips, onto which the TRAP sequence and a tris-NTA group were used to detect either the extracellular binding site or the intracellular C-terminal end [24].
FD-based AFM for cell biology and immunology applications. (a–e) Multiparametric FD-based AFM is used to study TRAP binding to PAR1 receptor (panels adapted with permission from [21]). (a) AFM tip is functionalized with the TRAP peptide via a PEG linker and cyclically approached and retracted from PAR1 receptors reconstituted in a lipid bilayer. (b) Height AFM image (left) and corresponding adhesion image (right) of PAR1 receptors reconstituted in liposomes. (c) Overlay of height and adhesion image of PAR1, highlighting the specific interaction of TRAP with the receptors on the surface. (d) Representative force-distance curves. (e) Quantification of the TRAP-PAR1 binding free energy landscape from the fitting of the interaction forces vs loading rate plot. (f–j) FD-based AFM and its combination with optical and fluorescence microscopy is used to measure specific interactions between PRDX5 and TLR4 on living cells (panels adapted with permission from [25]). (f) PRDX5-functionalized tips are used to probe cocultures of living CHO cells overexpressing YFP-labelled TLR4 and control cells. (g) Height (left) and adhesion (right) images of CHO cells. The inset shows the corresponding overlay of phase and fluorescence images. Most of the adhesive events are observed on CHO cells overexpressing TLR4, indicating a specific interaction with PRDX5. (h) Macrophage-differentiated THP-1 cells are used as relevant model of innate immunity. (i) Variation of cell stiffness over time, indicating a PRDX5-induced mechano-response. (j) Extraction of biophysical parameters of PRDX5-TLR4 interaction from the force vs loading rate plot
Very recently, a similar FD-based AFM approach has been used to elucidate interactions between endogenous molecules and receptors of the innate immune system involved in inflammatory responses [25]. In response to infections and tissue damage, the innate immune system triggers inflammatory processes that aim to restore tissue homeostasis. Key receptors involved in this process are membrane-bound pattern recognition receptors (PRRs), such as Toll-like receptors, NOD-like receptors, and RIG-I-like receptors, which promote inflammation through their interaction with pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs). Knoops et al. [25] focused on the DAMP acting function of the antioxidant enzyme peroxiredoxin 5 (PRDX5) and on its specific recognition by Toll-like receptor 4 (TLR4) (Fig. 2f–j). After having proven the specificity of the interaction and reconstructed the kinetics of PRDX5 and TLR4 interaction by SMFS, the authors investigated this interaction on living CHO cells overexpressing TLR4 (Fig. 2f, g) and on macrophage-differentiated THP-1 cells, as a physiologically relevant model of immune system cells (Fig. 2h). Not only the role of PRDX5 to act as DAMP for TLR4 was confirmed on both cell models, but also, surprisingly, macrophage-differentiated THP1 cells also went through a significant increase of stiffness upon stimulation with PRDX5, suggesting that the interaction between PRDX5 and TLR4 induces a cellular mechano-response (Fig. 2i, j). FD-based AFM presents itself as very promising approach not only to map and characterize specific inflammation related interactions under physiological conditions but also to trigger and unveil in a temporally resolved manner-specific cellular responses connected with the innate immune system.
Microbiology applications
Since its invention and application to address biological question, a great deal of AFM research focused on the imaging and elucidation of pathogen-host interactions, both in terms of structural dynamics of cell walls and response to drugs and in terms of localization, adhesion, and mechanics of their individual components [26]. Pathogen infections are initiated by the strong microbial adhesion to host tissues and host protein–coated implanted devices and still represent a major global safety concern, which is increasing due to the growing emergence of multidrug-resistant strains. This process is largely controlled by microbial adhesins, which specifically interact with host proteins and tissues, through relatively poorly understood molecular mechanisms. AFM represents therefore an exceptional opportunity to gain insights into the mechanostability and molecular details of adhesin-mediated pathogen interactions, both at the single-molecule and the single-cell levels, and for the design of antiadhesion drugs that block cell adhesion and biofilm development. Over the years, several studies focused on the AFM-based characterization of the binding properties of fibronectin-binding proteins (FnBPs) from Staphylococcus aureus. By functionalizing the AFM tip with fibronectin, the molecular strength of fibronectin-S. aureus interaction was quantified on living bacteria [27, 28]. Interestingly, studies conducted on clinical isolates from patients with cardiovascular implants have shown a distinct force signature and a bond lifetime two times longer for bloodstream isolated bacteria from patients with infected devices, which were found to be associated with polymorphisms in FnBPA [29, 30]. Another adhesin that recently gained a lot of attention is SD-repeat protein G (SdrG) from Staphylococcus epidermidis. This adhesin binds the host target, a peptide of about 15 residues from human fibrinogen, through the “dock, lock, and latch” (DLL) mechanism, in which the peptide is first bound, then buried between two immunoglobulin-like domains N2 and N3, and finally “latched” with a strand connecting N3 to N2 [31]. In 2014, by using a combination of SMFS with fibrinogen-functionalized tips on living S. epidermis cells and SCFS on fibrinogen-coated surfaces, Dufrêne’s group identified in the single SdrG-fibrinogen interaction the strongest non-covalent bond known so far, in the order of 2 nN [32]. Later studies have shown similar forces for related adhesin-host protein interactions, such as clumping factors A and B [33, 34] (Fig. 3a–c). By combining SMFS and all-atom steered molecular dynamics simulations, Milles et al. elucidated the molecular mechanisms of this extreme mechanostability, which relies mainly on multiple hydrogen bonds with the peptide backbone and is further stabilized by calcium ions [35, 36]. Very recently, another extremely strong interaction has been identified to mediate the binding between the S. aureus surface protein A (SpA) and the plasma glycoprotein von Willebrand factor (vWF). During endovascular infections, S. aureus binds to endothelium under flow via vWF [37]. Using SMFS, the authors demonstrated that vWF binds to SpA with a ≈ 2 nN force and that these strong bonds are activated by mechanical stress. Interestingly, as opposed to previously studied adhesins, SpA is not interacting via the DLL mechanism. Instead the strong binding is seemingly ascribed to a force-induced conformational changes of the vWF, suggesting a new molecular mechanism of bacterial adhesion, which needs to be further elucidated in detail.
Overview of AFM-based receptor-ligand mapping for microbiology applications. (a–c) The combination of SCFS and SMFS is used to probe strong interactions between bacterial adhesins and host proteins (panels adapted with permission from [34]). (a) Schematics of SCFS and SMFS experimental setup to probe the DLL mechanism–guided interaction between clumping factor B (ClfB) from S. aureus and loricrin. (b) Adhesion force histogram from the interaction between S.aureus cells and loricrin measured by SCFS. The immobilization of single cells on the cantilever is followed by optical and fluorescence microscopy (inset). (c) Adhesion force histogram and map (inset) of loricrin-CLfB interaction recorded on living bacteria by SMFS. (d–h) Multiparametric FD-based AFM is used to localize phages on the surface of infected bacteria and to simultaneously map bacterial mechanical properties (panels adapted with permission from [13]). (d) Filamentous bacteriophages were engineered to expose a His6 tag on their pIX tail to facilitate their localization using Ni2+-NTA functionalized AFM tips. (e) Height image of E. coli cell infected with bacteriophages. The infection was followed by staining with Ni2+-NTA fluorescent conjugates (inset). (f) Simultaneously acquired adhesion image of the area in (e). The same area scanned in presence of EDTA is shown in the upper inset. (g) Representative force-distance curves. (h) Comparison between adhesion image (left) and elasticity map (right) of a zoomed area of an infected E. coli cell, showing the organization of the bacteriophages into soft domains surrounded by stiff material
Microbiology has also greatly benefited from the most recent advances of multiparametric FD-based AFM and its combination with optical microscopic techniques. While this approach has been extensively used to simultaneously image the topography and map physico-chemical, mechanical, and adhesive properties of bacterial proteins and cells, more recently the combination with functionalized tips allows to localize and force-probe specific surface biomolecules, providing more insights into ligand binding events at nanometric and sub-nanometric resolution [15, 38] and bacteriophage infection [13]. In one of the most significant examples on isolated proteins [15], the well-characterized bacteriorhodopsin (BR) from Halobacterium salinarum has been engineered to carry a C-terminal His5-tag, which is located at the cytoplasmic surface of BR trimers self-assembled in purple membrane. By functionalizing AFM tips with tris-nickel-nitriloacetate (Ni2+-trisNTA) groups as ligands, the authors managed to image and to simultaneously localize and quantify the Ni2+-trisNTA-His5-tag binding events with sub-nanometric precision, thus showing the applicability of FD-based AFM for epitope mapping and ligand-receptor binding studies at submolecular resolution. In a closely related work, the combination between confocal microscopy and AFM was exploited to sequentially localize extracellular and cytoplasmic surface of BR trimers by fluorescent labelling and to epitope map the His5-tag-carrying C-terminal by multiparametric AFM at submolecular resolution [14]. A similar strategy has been used to map the distribution of single bacteriophages on living Escherichia coli cells [13] (Fig. 3d–h). Filamentous bacteriophages were genetically engineered to express His6-tag on their pIX tail and subsequently used to infect the E. coli cells, while their escaping from the cells was blocked by deletion of pIII, causing the assembled phages to remain anchored to the bacterial cell wall (Fig. 3d). FD-based AFM with Ni2+-NTA functionalized tips enabled the multiparametric mapping of structure, adhesion, and elasticity of the cells (Fig. 3e–g), revealing that the sites of phage assembly form soft nanodomains at the bacterial septum (Fig. 3h), thus increasing our understanding of the infection process and its dynamics.
Virology applications
Viral infections are multistep processes initiated by the binding of viruses to host cell membrane moieties, comprising proteins, carbohydrates, and lipids, which ultimately triggers virus endocytosis, followed by the release of the viral genetic content into the cytoplasm, and progeny viral production and escape. Since viral replication cannot take place without a susceptible host cell, elucidating the first steps of virus-host cell binding is key to the design of effective antiviral measures. Although ensemble techniques, such as surface plasmon resonance and microscale electrophoresis, provide information of average thermodynamic and kinetic properties of the binding, single-molecule techniques are required to uncover the molecular properties of inhomogeneous systems and to obtain dynamic and statistical information, thus defining the detailed mechanism of virus-host cell binding steps under physiological conditions.
Single-molecule and single-virus AFM-based force spectroscopy approaches have been used to characterize the binding forces and kinetics of a variety of viruses, such as human immunodeficiency virus type 1 (HIV-1), rhinovirus (HRV), influenza virus (HA), and herpes simplex virus type (HSV) to some of their cognate ligands both on isolated molecules and on living cells [39,40,41,42,43], highlighting the dynamical nature, multivalency, and complexity of the first virus-host interactions. In two closely related studies, the interactions of the HIV-1 glycoprotein gp120 with the cellular receptor CD4 and the co-receptor CCR5 have been elucidated. First, gp120-functionalized AFM tips were used to study the binding to CD4 and CCR5 on both model surfaces and genetically engineered living cells, showing that a first unstable association of gp120 to CD4 is required for the subsequent and much more stable binding to CCR5 [39]. Then, by immobilizing HIV-1 viruses directly on the AFM tips, they showed this dynamic binding to be critically regulated by the interaction time with the cell, with the gp120-CD4 bonds undergoing rapid destabilization within 0.3 s, which is enhanced by CCR5, suggesting a co-receptor-induced conformational change in the gp120-CD4 bond [40]. Multivalency is one of the main features of virus binding to the host cell. Multiple weak interactions can add up to provide virus binding with nanomolar affinities. Using single-virus force spectroscopy, Rankl et al. [41] have probed the binding of HRV2 to very-low-density lipoprotein receptor, demonstrating a time-dependent transition of single to multiple parallel uncorrelated virus-receptor bonds. Multivalent virus attachment can also be very complex in both dynamics and molecular nature of the bonds established. Sieben et al. [42] elegantly combined optical tweezers, AFM-based single-virus force spectroscopy, and molecular dynamics simulations to study the binding of HA to cells presenting a different surface distribution of 2,3- and 2,6-α-linked sialic acid. The results uncovered a broad spectrum of multiple unbinding pathways and no clear preference for the different cell lines, indicating a complex energy landscape of the underlying interactions, which needs to be clarified in detail. Virus attachment to the cell surface needs to be tightly regulated, in relation to the contextual needs of the virus to form bonds that are strong enough to ensure the binding and entry, but at the same time the necessity of being able to migrate on the cell surface both during entry, to find its ligands, and during progeny escape. Using a combination of single-virus AFM–based force spectroscopy and single particle-tracking, Delguste et al. [43] identified in the mucin-like region of the HSV-1 glycoprotein gC an essential regulator of HSV binding to chondroitin sulfate and of virus diffusive behaviour.
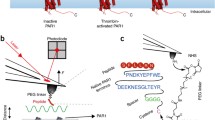
Although AFM alone has proved to be an effective tool to quantitatively characterize the forces involved in virus-cell interaction, it lacks the capacity of identifying the biomolecules and receptors taking part in the process. In this context, combination of optical and fluorescence microscopy techniques with AFM has recently brought significant advances in the elucidation of the first binding steps of viruses to human cells. Recently, Alsteens et al. developed an approach to measure virus-cell interactions by FD-based multiparametric AFM while benefitting from confocal live cell microscopy [44, 45]. To illustrate the potential of this approach, the authors studied the interaction between an engineered rabies virus carrying the envelope protein of avian sarcoma leucosis virus (EnvA-RABV) and living cells expressing the cognate avian tumour virus receptor A (TVA). By coculturing control cells and cells expressing fluorescently labelled TVA that were visualized by confocal microscopy during the AFM multiparametric acquisition, they were able to correlate the specific binding behaviour of the virus to TVA at high spatial resolution in situ on living cells. Furthermore, they developed a theoretical framework to extract mechanical, kinetic, and thermodynamic parameters controlling virus-receptor interactions, thus enabling the straightforward reconstruction of the free energy landscape of virus binding. The same approach has been very recently used to elucidate important aspects of the multivalent binding of human gammaherpesvirus to living cells during infection, unravelling a tight viral glycoprotein-controlled regulation of binding valency during attachment and release steps (Fig. 4a–f) [46]. As illustrated from the above-mentioned examples, although at its infancy multiparametric FD-based AFM in combination with fluorescence microscopy has already brought new important insights into the dynamics and regulation of the first steps of viral infections, with more outstanding applications foreseen in the near future.
Multiparametric FD-based AFM to probe the first binding steps of viruses to mammalian cells. (a) Single-virus functionalized tips are used to investigate specific interactions on model surfaces or living cells. (b) Comparison between forces of interaction of wild-type herpes virus and a mutant lacking the viral glycoprotein gp150 (gp150−) with heparin. (c–f) Funtional role of gp150 on herpes virus attachment to living cells. (c) AFM height image of a mCherry CHO cell (GAG+) and a CHO cell deficient of surface glycosaminoglycans (GAG−) and corresponding adhesion image acquired with the wild-type virus. The related fluorescence image is shown in the inset. (d) Zoom of the GAG+ cell adhesion map, highlighting the prevalence of interaction forces in the low range (100–200 pN). (e) AFM height image of a GAG+ and a GAG− cell and corresponding adhesion image acquired with the gp150− virus. The corresponding fluorescence image is shown in the inset. (f) Zoom of the GAG+ cell adhesion map, highlighting an increased abundance of interaction forces in the high range (200–300 pN) compared with the wild type, indicating a possible regulating role of gp150 for virus attachment and diffusion (panels reproduced with permission from [46])
Outlook
In recent years AFM has assumed a prominent role in life sciences and in particular has revealed itself as an invaluable tool to study the dynamics and biophysics underlying receptor-ligand bonds at the single-molecule level under conditions that closely resemble the in vivo scenario. Advanced FD-based AFM methodologies enable the high-resolution imaging of cell membrane proteins, while simultaneously mapping the mechanical, kinetics, and thermodynamic properties of binding sites and binding free energy landscape of ligand-receptor bonds. The combination of AFM with fluorescence microscopy techniques, nowadays routinely implemented in a variety of commercial instruments, has offered new insights regarding the direct visualization of the investigated cellular components, while simultaneously quantifying their binding properties, leading to an increased understanding of cell membrane regulated processes and dynamics. Few examples have been presented here, but they nevertheless testify the significant contribution of AFM-based approaches to a wide variety of fields, spanning from cell biology and immunological reactions to pathogen-host and virus-host interactions.
In this frame, important drawbacks that still limit the resolution capabilities of the technique are force sensitivity, temporal resolution, and thermal stability. The last in particular is responsible for the stability of the AFM measurement to vary over tip and is therefore especially critical to probe the very first steps of cellular receptor interactions in a dynamical manner. Thereby, recently developed ultrastable AFMs possessing sub-piconewton force resolution, high positional stability, and extremely low lateral thermal drift could be highly beneficial to ligand-receptor studies, particularly for time-resolved cellular processes [47].
Major breakthroughs in biomedical research will also arise from the systematic coupling of FD-based multiparametric AFM approaches with super-resolution microscopy techniques, such as STED and PALM/STORM [48]. Few innovative studies have already started to show the potential of this combined techniques for the field of mechanobiology, for instance to unveil the detailed correlation between membrane properties and cytoskeleton organization on living cells [49, 50]. The application of these combined approaches to the study of ligand-receptor interactions will enable to resolve the lateral organization of cellular receptors, such as clustering, and their impact on ligand binding at the level of single molecule. Furthermore, while mapping membrane biophysical properties, it will be possible to monitor cellular responses to controlled extracellular stimuli in a dynamical manner, getting precise insights into for example ligand binding-triggered intracellular cascades in real time.
In summary, AFM is now a well-established technique to monitor and study biological processes under physiological conditions with (sub)-nanometric resolution. Ultimately, ongoing technological developments and the further coupling of AFM with other biophysical and imaging platforms are expected to greatly enlarge the number of AFM biological applications, enabling to tackle unsolved biological questions from multiple angles simultaneously, thus greatly advancing our mechanistic understanding of biological phenomena.
References
Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56(9):930–3. https://doi.org/10.1103/PhysRevLett.56.930.
Pavliček N, Gross L. Generation, manipulation and characterization of molecules by atomic force microscopy. Nat Rev Chem. 2017;1:0005. https://doi.org/10.1038/s41570-016-0005.
Alsteens D, Gaub HE, Newton R, Pfreundschuh M, Gerber C, Müller DJ. Atomic force microscopy-based characterization and design of biointerfaces. Nat Rev Mat. 2017;2(5). doi:https://doi.org/10.1038/natrevmats.2017.8.
Frisbie CD, Rozsnyai LF, Noy A, Wrighton MS, Lieber CM. Functional group imaging by chemical force microscopy. Science (New York, NY). 1994;265(5181):2071–4. https://doi.org/10.1126/science.265.5181.2071.
Ludwig M, Dettmann W, Gaub HE. Atomic force microscope imaging contrast based on molecular recognition. Biophys J. 1997;72(1):445–8.
Lee GU, Kidwell DA, Colton RJ. Sensing discrete streptavidin-biotin interactions with atomic force microscopy. Langmuir. 1994;10(2):354–7. https://doi.org/10.1021/la00014a003.
Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science (New York, NY). 1994;264(5157):415–7. https://doi.org/10.1126/science.8153628.
Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc Natl Acad Sci U S A. 1996;93(8):3477–81. https://doi.org/10.1073/pnas.93.8.3477.
Grandbois M, Dettmann W, Benoit M, Gaub HE. Affinity imaging of red blood cells using an atomic force microscope. J Histochem Cytochem. 2000;48(5):719–24. https://doi.org/10.1177/002215540004800516.
Benoit M, Gabriel D, Gerisch G, Gaub HE. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat Cell Biol. 2000;2(6):313–7. https://doi.org/10.1038/35014000.
Alsteens D, Beaussart A, Derclaye S, El-Kirat-Chatel S, Park HR, Lipke PN, et al. Single-cell force spectroscopy of Als-mediated fungal adhesion. Anal Methods. 2013;5(15):3657–62. https://doi.org/10.1039/C3AY40473K.
Dufrêne YF, Ando T, Garcia R, Alsteens D, Martinez-Martin D, Engel A, et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat Nanotechnol. 2017;12(4):295–307. https://doi.org/10.1038/nnano.2017.45.
Alsteens D, Trabelsi H, Soumillion P, Dufrêne YF. Multiparametric atomic force microscopy imaging of single bacteriophages extruding from living bacteria. Nat Commun. 2013;4:2926. https://doi.org/10.1038/ncomms3926.
Laskowski PR, Pfreundschuh M, Stauffer M, Ucurum Z, Fotiadis D, Muller DJ. High-resolution imaging and multiparametric characterization of native membranes by combining confocal microscopy and an atomic force microscopy-based toolbox. ACS Nano. 2017;11(8):8292–301. https://doi.org/10.1021/acsnano.7b03456.
Pfreundschuh M, Harder D, Ucurum Z, Fotiadis D, Muller DJ. Detecting ligand-binding events and free energy landscape while imaging membrane receptors at subnanometer resolution. Nano Lett. 2017;17(5):3261–9. https://doi.org/10.1021/acs.nanolett.7b00941.
Fuhrmann A, Ros R. Single-molecule force spectroscopy: a method for quantitative analysis of ligand-receptor interactions. Nanomedicine (London, England). 2010;5(4):657–66. https://doi.org/10.2217/nnm.10.26.
Wildling L, Rankl C, Haselgrubler T, Gruber HJ, Holy M, Newman AH, et al. Probing binding pocket of serotonin transporter by single molecular force spectroscopy on living cells. J Biol Chem. 2012;287(1):105–13. https://doi.org/10.1074/jbc.M111.304873.
Chtcheglova LA, Hinterdorfer P. Simultaneous AFM topography and recognition imaging at the plasma membrane of mammalian cells. Semin Cell Dev Biol. 2018;73:45–56. https://doi.org/10.1016/j.semcdb.2017.08.025.
Koehler M, Macher G, Rupprecht A, Zhu R, Gruber HJ, Pohl EE, et al. Combined recognition imaging and force spectroscopy: a new mode for mapping and studying interaction sites at low lateral density. Sci Adv. 2017;9(1):128–34. https://doi.org/10.1166/sam.2017.3066.
Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356. https://doi.org/10.1038/nature08144.
Alsteens D, Pfreundschuh M, Zhang C, Spoerri PM, Coughlin SR, Kobilka BK, et al. Imaging G protein–coupled receptors while quantifying their ligand-binding free-energy landscape. Nat Methods. 2015;12:845. https://doi.org/10.1038/nmeth.3479.
Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor–ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397(6714):50–3. https://doi.org/10.1038/16219.
Friddle RW, Noy A, De Yoreo JJ. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc Natl Acad Sci U S A. 2012;109(34):13573–8. https://doi.org/10.1073/pnas.1202946109.
Pfreundschuh M, Alsteens D, Wieneke R, Zhang C, Coughlin SR, Tampé R, et al. Identifying and quantifying two ligand-binding sites while imaging native human membrane receptors by AFM. Nat Commun. 2015;6:8857. https://doi.org/10.1038/ncomms9857.
Knoops B, Becker S, Poncin MA, Glibert J, Derclaye S, Clippe A, Alsteens D. Specific interactions measured by AFM on living cells between peroxiredoxin-5 and TLR4: relevance for mechanisms of innate immunity. Cell Chem Biol 2018;25(5):550–559 e3. doi:https://doi.org/10.1016/j.chembiol.2018.02.006.
Dufrêne YF. Atomic force microscopy in microbiology: new structural and functional insights into the microbial cell surface. mBio. 2014;5(4):e01363–14. https://doi.org/10.1128/mBio.01363-14.
Mitchell G, Lamontagne CA, Brouillette E, Grondin G, Talbot BG, Grandbois M, et al. Staphylococcus aureus SigB activity promotes a strong fibronectin-bacterium interaction which may sustain host tissue colonization by small-colony variants isolated from cystic fibrosis patients. Mol Microbiol. 2008;70(6):1540–55. https://doi.org/10.1111/j.1365-2958.2008.06511.x.
Buck AW, Fowler VG, Yongsunthon R, Liu J, DiBartola AC, Que Y-A, et al. Bonds between fibronectin and fibronectin-binding proteins on Staphylococcus aureus and Lactococcus lactis. Langmuir. 2010;26(13):10764–70. https://doi.org/10.1021/la100549u.
Casillas-Ituarte NN, Lower BH, Lamlertthon S, Fowler VG Jr, Lower SK. Dissociation rate constants of human fibronectin binding to fibronectin-binding proteins on living Staphylococcus aureus isolated from clinical patients. J Biol Chem. 2012;287(9):6693–701. https://doi.org/10.1074/jbc.M111.285692.
Lower SK, Lamlertthon S, Casillas-Ituarte NN, Lins RD, Yongsunthon R, Taylor ES, et al. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc Natl Acad Sci U S A. 2011;108(45):18372–7. https://doi.org/10.1073/pnas.1109071108.
Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, et al. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115(2):217–28.
Herman P, El-Kirat-Chatel S, Beaussart A, Geoghegan JA, Foster TJ, Dufrêne YF. The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol Microbiol. 2014;93(2):356–68. https://doi.org/10.1111/mmi.12663.
Herman-Bausier P, Labate C, Towell AM, Derclaye S, Geoghegan JA, Dufrêne YF. Staphylococcus aureus clumping factor A is a force-sensitive molecular switch that activates bacterial adhesion. Proc Natl Acad Sci U S A. 2018;115(21):5564–9. https://doi.org/10.1073/pnas.1718104115.
Vitry P, Valotteau C, Feuillie C, Bernard S, Alsteens D, Geoghegan JA, Dufrêne YF. Force-induced strengthening of the interaction between Staphylococcus aureus clumping factor B and loricrin. MBio. 2017;8(6). https://doi.org/10.1128/mBio.01748-17.
Milles LF, Schulten K, Gaub HE, Bernardi RC. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science (New York, NY). 2018;359(6383):1527–33. https://doi.org/10.1126/science.aar2094.
Milles LF, Unterauer EM, Nicolaus T, Gaub HE. Calcium stabilizes the strongest protein fold. Nat Commun. 2018;9(1):4764. https://doi.org/10.1038/s41467-018-07145-6.
Viela F, Prystopiuk V, Leprince A, Mahillon J, Speziale P, Pietrocola G, Dufrêne YF. Binding of Staphylococcus aureus protein A to von Willebrand factor is regulated by mechanical force. MBio. 2019;10(2). https://doi.org/10.1128/mBio.00555-19.
Alsteens D, Dupres V, Yunus S, Latgé J-P, Heinisch JJ, Dufrêne YF. High-resolution imaging of chemical and biological sites on living cells using peak force tapping atomic force microscopy. Langmuir. 2012;28(49):16738–44. https://doi.org/10.1021/la303891j.
Chang MI, Panorchan P, Dobrowsky TM, Tseng Y, Wirtz D. Single-molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells. J Virol. 2005;79(23):14748–55. https://doi.org/10.1128/jvi.79.23.14748-14755.2005.
Dobrowsky TM, Zhou Y, Sun SX, Siliciano RF, Wirtz D. Monitoring early fusion dynamics of human immunodeficiency virus type 1 at single-molecule resolution. J Virol. 2008;82(14):7022–33. https://doi.org/10.1128/jvi.00053-08.
Rankl C, Kienberger F, Wildling L, Wruss J, Gruber HJ, Blaas D, et al. Multiple receptors involved in human rhinovirus attachment to live cells. Proc Natl Acad Sci U S A. 2008;105(46):17778–83. https://doi.org/10.1073/pnas.0806451105.
Sieben C, Kappel C, Zhu R, Wozniak A, Rankl C, Hinterdorfer P, et al. Influenza virus binds its host cell using multiple dynamic interactions. Proc Natl Acad Sci U S A. 2012;109(34):13626–31. https://doi.org/10.1073/pnas.1120265109.
Delguste M, Peerboom N, Le Brun G, Trybala E, Olofsson S, Bergström T, et al. Regulatory mechanisms of the mucin-like region on herpes simplex virus during cellular attachment. ACS Chem Biol. 2019;14(3):534–42. https://doi.org/10.1021/acschembio.9b00064.
Alsteens D, Newton R, Schubert R, Martinez-Martin D, Delguste M, Roska B, et al. Nanomechanical mapping of first binding steps of a virus to animal cells. Nat Nanotechnol. 2017;12(2):177–83. https://doi.org/10.1038/nnano.2016.228.
Newton R, Delguste M, Koehler M, Dumitru AC, Laskowski PR, Muller DJ, et al. Combining confocal and atomic force microscopy to quantify single-virus binding to mammalian cell surfaces. Nat Protoc. 2017;12(11):2275–92. https://doi.org/10.1038/nprot.2017.112.
Delguste M, Zeippen C, Machiels B, Mast J, Gillet L, Alsteens D. Multivalent binding of herpesvirus to living cells is tightly regulated during infection. Sci Adv. 2018;4(8):eaat1273. https://doi.org/10.1126/sciadv.aat1273.
Yu H, Siewny MGW, Edwards DT, Sanders AW, Perkins TT. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science (New York, NY). 2017;355(6328):945–50. https://doi.org/10.1126/science.aah7124.
Harke B, Chacko JV, Haschke H, Canale C, Diaspro A. A novel nanoscopic tool by combining AFM with STED microscopy. Opt Nanoscopy. 2012;1(1):3. https://doi.org/10.1186/2192-2853-1-3.
Chacko JV, Zanacchi FC, Diaspro A. Probing cytoskeletal structures by coupling optical superresolution and AFM techniques for a correlative approach. Cytoskeleton (Hoboken, NJ). 2013;70(11):729–40. https://doi.org/10.1002/cm.21139.
Curry N, Ghézali G, Kaminski Schierle GS, Rouach N, Kaminski CF. Correlative STED and Atomic Force Microscopy on Live Astrocytes Reveals Plasticity of Cytoskeletal Structure and Membrane Physical Properties during Polarized Migration. Front Cell Neurosci. 2017;11:104. https://doi.org/10.3389/fncel.2017.00104.
Funding
This work was supported by the Fonds National de la Recherche Scientifique (F.R.S.-FNRS grant number: PDR T.0090.15 to D.A.), the Research Department of the Communauté française de Belgique (Concerted Research Action), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the ‘MOVE-IN Louvain’ Incoming post-doc Fellowship programme, and the European Molecular Biology Organization (EMBO ALTF 542-2018 to C.L.). D.A. is Research Associate at the FNRS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lo Giudice, C., Dumitru, A.C. & Alsteens, D. Probing ligand-receptor bonds in physiologically relevant conditions using AFM. Anal Bioanal Chem 411, 6549–6559 (2019). https://doi.org/10.1007/s00216-019-02077-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-02077-6