Abstract
Background
Warfarin is the most frequently prescribed oral anticoagulant worldwide. Due to its narrow therapeutic index and inter-patient variability in dose requirement, this drug has been considered an ideal target for personalised medicine. Several warfarin dosing algorithms have been proposed to tailor the warfarin dosage in the European, Asian and African-American populations. However, minimal interest was directed towards Middle East countries. The factors affecting warfarin dose requirement could be different in patients from different geographical and ethnic groups, limiting the value of published dosing algorithms.
Objective
The first objective of this study was to examine the contribution of genetic and nongenetic factors on the variability of warfarin dose requirements in the Egyptian population using an easy, cost-effective and rapid analysis of vitamin K epoxide reductase complex subunit 1 (VKORC1) and cytochrome P450 (CYP) 2C9 single nucleotide polymorphism (SNP) genotyping of patients. A second objective was to develop and validate an algorithm for warfarin dose prediction that is tailored to Egyptian patients.
Methods
Eighty-four patients, 41 males and 43 females, with a median (25th–75th percentiles) age of 39 (31–48) years were recruited in this study. Fifty patients whose international normalised ratio (INR) was in the range of 2–3 were allocated to a study cohort. SYBR Green-based multiplex allele-specific real-time PCR was used for genotyping of CYP2C9 (1075A>C) and VKORC1 (1173C>T) polymorphisms. Linear regression analysis, including the variables age, gender, CYP2C9 and VKORC1 SNP genotypes, was run to derive the best model for estimating the warfarin dose that achieves an INR of 2–3. The new warfarin dosing algorithm was examined in a second cohort of patients (n = 34) to check its validity. The predicted dose requirements for a subgroup of our patients were calculated according to Gage and International Warfarin Pharmacogenetics Consortium (IWPC) algorithms available at http://www.warfarindosing.org.
Results
In the study cohort, warfarin dose/week in VKORC1 TT subjects was statistically significantly lower than in VKORC1 CC/CT subjects (p = 0.032), while there was no statistically significant difference in warfarin dose/week between CYP2C9*1*1 and *1*3 (p = 0.925). A multivariate stepwise linear regression analysis revealed that age and VKORC1 had independent and significant contributions to the overall variability in warfarin dose with a p-value = 0.013 and 0.042, respectively. Maintenance dose (mg/week) = 65.226 − 0.422 × (age) − 9.474 × (VKORC1). The estimated regression equation was able to account for 20.5 % of the overall variability in warfarin maintenance dose. A significant positive correlation, with sufficient strength, was observed between the predicted warfarin dose and the actual prescribed dose (r = 0.453, p = 0.001). In the validation cohort, after application of the dosing algorithm, correlation between predicted and actual dose was statistically significant (p = 0.023). The equation was particularly successful among patients with a dose ≥35 mg/week. The correlation coefficient between the actual and predicted doses for IWPC and Gage were 0.304 and 0.276, respectively. When compared with our algorithm (r = 0.279), the difference was non-significant: p = 0.903 and 0.990, respectively.
Conclusion
VKORC1 (1173C>T) contributes to the warfarin dose variability. Patients’ age and genetic variants of VKORC1 account for nearly 20.5 % of the variability in warfarin dose required to achieve an INR of 2–3. The success of a prediction equation based on these variables was proved in a different cohort: the predicted dose correlated significantly with the maintenance dose and the equation was more successful among patients with a dose ≥35 mg/week. The results of the warfarin algorithm we developed were comparable with those of the IWPC and Gage algorithms with the advantage of using one SNP (VKORC1 1173C>T) only. This represents an economic advantage in our community. Replication of this study in a larger cohort of patients is necessary before translation of this knowledge into clinical guidelines for warfarin prescription.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
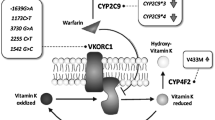
Warfarin is the most frequently prescribed oral anticoagulant worldwide [1]. Nevertheless, the US FDA rates warfarin among the top ten drugs with the greatest number of serious adverse reactions [2], mostly due to its narrow therapeutic window and high inter- and intra-individual variability in dose requirements. For these reasons, careful laboratory monitoring is necessary to maintain the international normalised ratio (INR) within the therapeutic range [3].
A number of factors have been identified to influence the anticoagulation effect of warfarin therapy, including age, sex, body weight, co-morbidity, concurrent medications, dietary intake, patient compliance level and genetic factors [4]. Warfarin is a racemic mixture of S- and R-enantiomers. The S-enantiomer is associated with 60–70 % of warfarin’s anticoagulant response, while the R-enantiomer accounts for 30–40 % of warfarin’s anticoagulant effect [5]. The S-enantiomer is metabolised at therapeutic concentration predominantly by cytochrome P450 (CYP) 2C9 [5]. The wild-type allele is CYP2C9*1 [5]. At least 33 clinically relevant variants of CYP2C9 (*2 through to *34) have been documented [6]. The most common variants CYP2C9*2 (430C>T, R144C, rs1799852) and CYP2C9*3 (1075A>C, I359L, rs1057910) generate enzymes with impaired hydroxylation of S-warfarin due to amino-acid changes, and several studies have shown that these variants have an effect on warfarin dose requirement [7–9].
Warfarin is a specific inhibitor of the vitamin K epoxide reductase (VKOR) encoded by the VKOR complex subunit 1 (VKORC1) gene [10]. Warfarin exerts its anticoagulant effects by preventing the ability of VKORC1 to regenerate reduced vitamin K from its epoxide form [11]. Reduced vitamin K is an essential cofactor for γ-glutamylcarboxylase (GGCX), the enzyme catalysing the post-translational γ-glutamyl carboxylation of the vitamin K-dependent clotting factors, II (prothrombin), VII, IX and X. Thus, warfarin prevents the functional maturation of vitamin K-dependent clotting factors, leading to reduced coagulation [12]. Genetic variations in VKORC1 are associated with altered sensitivity to warfarin [13]. Two single nucleotide polymorphisms (SNPs) of VKORC1 were found to estimate warfarin dosing phenotypes: −1639G>A (rs9923231) promoter polymorphism and 1173C>T (rs9934438) intronic polymorphism [9]. Studies of white and Asian patients have reported that CYP2C9 and VKORC1 polymorphisms influence the maintenance dose of warfarin [14, 15], but they appear to contribute to less than half of the inter-individual variability in the dose-response relationship to oral anticoagulants [14]. Several warfarin dosing algorithms have been proposed to tailor the warfarin dosing in the European, Asian and African-American populations. However, minimal interest was directed towards Middle East countries. The factors affecting warfarin dose requirement could be different in patients from different geographical and ethnic groups, limiting the value of published dosing algorithms.
The first aim of our study was to investigate the contribution of genetic and nongenetic factors on the variability of warfarin dose requirements in the Egyptian population using an easy, cost-effective and rapid analysis of VKORC1 and CYP2C9 genotyping of patients. The second objective was to develop and validate an algorithm for warfarin dose prediction that is tailored to Egyptian patients.
2 Subjects and Methods
2.1 Subjects
To create the study group, from October 2010 to November 2011, we investigated patients referred to Kasr Al-Ainy University Hospital (Cairo, Egypt) outpatient clinic for warfarin (Marevan™; GlaxoSmith Kline, Cairo, Egypt) therapy monitoring. The average number of patients attending the clinic is 60 patients/week. Dosing of warfarin is not computer assisted. The physicians prescribe warfarin following the empiric dosing method. Time in therapeutic range is 75 % on average. We selected 50 patients [23 males and 27 females, mean (standard deviation) age: 39.16 (12.00) years] who satisfied the following inclusion criteria: age >18 years, oral anticoagulation therapy (OAT) lasting for at least 3 months, and stable warfarin dose. A patient was considered to be on a stable warfarin dose if he or she had at least three consecutive INRs in the therapeutic (2–3) range for the same daily maintenance dose after at least 3 months of therapy. Exclusion criteria included cigarette smokers, subjects with abnormal liver function tests (alanine aminotransferase and aspartate aminotransferase ≥3 times the upper limit of normal) or thyroid function tests (definite hypothyroidism or hyperthyroidism), malnutrition, decompensated heart failure, bleeding diathesis, those receiving any drug known to have a major interaction with warfarin [e.g. amiodarone, statins (HMG-CoA reductase inhibitors), omeprazole, NSAIDs, anticonvulsants, sulfonamides, rifampin, azole antifungals and vitamin preparations containing vitamin K] and known noncompliance with OAT. Clinical data were obtained from the medical records compiled at the time of warfarin initiation after an interview by a physician. In the medical record, the presence of co-morbidities and the use of drugs apart from warfarin were also noted. Patients were excluded if clinical data were incomplete. Patients in the validation group (n = 34) were selected with the same inclusion/exclusion criteria used for the study group. The study was approved by the local Research Ethics Committee, and all patients provided informed consent.
2.2 Methods
For each selected patient, prothrombin time (PT) was measured by standard methods using the STA Compact® Hemostasis System (Diagnostica Stago, France). Genomic DNA was extracted from EDTA anticoagulated blood by means of QIAamp DNA blood Mini kit (Qiagen, Germany).
2.2.1 Genotyping Assay for Detection of Cytochrome P450 2C9 (1075A>C) (rs1057910) and Vitamin K Epoxide Reductase Complex Subunit 1 (1173C>T) (rs9934438) Polymorphisms by Using SYBR Green-Based Multiplex Allele-Specific PCR
CYP2C9 and VKORC1 genotyping were determined using SYBR Green-based multiplex allele-specific PCR and consequent melting curve analysis as previously described [16] by using StepOne™ Real-Time PCR Systems (Applied Biosystems, USA). Primers’ sequences are shown in Table 1 and they were designed by The Midland Certified Reagent Company (Texas, USA). For genotyping VKORC1 1173C>T locus, a 27-nt GC-tail was added to the 5′ end of the forward specific-primer that could only amplify the allele where the polymorphic site has a T residue. For the CYP2C9*3 polymorphism, a 28-nt GC-tail was added to the 5′ end of the forward specific-primer that could only amplify the *1 allele which has an A residue at position 1,075.
The 25 μL reaction mixture for both CYP2C9 and VKORC1 included 12.5 μL SYBR Green Mastermix (QuantiTect SYBR Green PCR Kits, Qiagen, USA) and 40 ng DNA. For CYP2C9, 0.6 μM C-primer forward, 0.15 μM A-primer forward (GC-tail) and 0.15 μM primer reverse were used. For VKORC1, 0.2 μM C-primer forward, 0.15 μM T-primer forward (GC-tail) and 0.15 μM primer reverse were used.
PCR amplification of CYP2C9 and VKORC1 was performed under the same conditions as follows: initial denaturation at 95 °C for 5 min, followed by 45 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 20 s and extension at 72 °C for 15 s, and a final extension at 72 °C for 2 min. After amplification, melting curve analysis was performed by heating the reaction mixture from 60 to 95 °C at a rate of 0.5 °C/min. The StepOne™ Real-Time PCR Systems automatically calculated the negative derivative of the change in fluorescence and generated a melting curve for each sample, as shown in Fig. 1. We attempted to develop a pharmacogenetic algorithm, choosing one CYP2C9 variant and one VKORC1 SNP, to reduce the costs to a minimum, and examine their impact on warfarin dose variation. It was decided to choose CYP2C9*3 and not CYP2C9*2 based on the fact that CYP2C9*3 has more effect (−21 to −49 %) compared with CYP2C9*2 (−14 to −20 %), as previously reported [17].
2.2.2 Application of International Warfarin Pharmacogenetics Consortium and Gage Algorithms to Compare Results of these Algorithms to the One Created in the Present Work
The predicted dose requirements for a subgroup of our patients for whom bodyweight and height were available (n = 35) were calculated according to Gage [18] and International Warfarin Pharmacogenetics Consortium (IWPC) algorithms [19] available at http://www.warfarindosing.org [20]. Parameters required for warfarin dose prediction by the two dosing algorithms were CYP2C9 and VKORC1 genotypes, age, height, bodyweight, gender, race, ethnicity, concomitant medications, target INR, smoking status and warfarin indication. Results of VKORC1 1173 C>T (rs9934438) were imputed to VKORC1 −1639G>A (rs9923231) as previously described [19].
2.2.3 Statistical Analysis
Numerical data were summarised as mean ± standard deviation when parametric and as median (25th–75th percentiles) when non-parametric. Differences between groups were detected using Student’s t-test for the former and Mann–Whitney test for the latter. Nominal data were summarised as number (percentage). Multivariate stepwise linear regression analysis was conducted using age, gender, VKORC1, CYP2C9 and combined VKORC1 and CYP2C9 as covariates in order to reach the minimum combination that can be used to predict the warfarin dose suitable to achieve an INR of 2–3. Correlation analysis was used to examine the strength of association between the predicted warfarin dose and the actual prescribed dose. The performance of the algorithm was assessed in two dose groups: participants requiring a dose <35 mg/week and those requiring a dose ≥35 mg/week for stable therapeutic anticoagulation. The threshold of 35 mg/week was chosen as the usual starting dose is 5 mg/day. Statistical analysis was run on SPSS for Windows, release 17.0 (SPSS Inc., Chicago, IL, USA). p-Values <0.05 were considered statistically significant.
3 Results
3.1 Demographic Data of the 84 Studied Subjects
Demographic data of the 84 studied subjects were subdivided into the study and validation cohorts. The median (25th–75th percentiles) age of the 84 patients was 39 (31–48) years. Forty-one were men and 43 were women. Most of the patients (75 %) received warfarin for valvular diseases. There was no significant difference in the demographic data of the two cohorts of patients as shown in Table 2.
3.2 Associations with Warfarin Dose
Our results show that the maintenance dose decreases, on average, by 0.4 mg/week for every 1 year increase in age. The warfarin weekly doses prescribed to patients were statistically significantly lower in VKORC1 TT when compared to VKORC1 CC/CT subjects in the study cohort (p = 0.032). There was no statistically significant difference in warfarin dose/week between CYP2C9*1*1 and *1*3 (p = 0.925) as shown in Table 3, and the Online Resource (Supplementary Figure IA and B).
a Melting curves for three different genotypes of vitamin K epoxide reductase complex subunit 1 (VKORC1) 1173C>T; profile with a single peak at 77.07 °C representing homozygote of CC, profile with a single peak at 81.39 °C representing homozygote of TT, and profile with a double peak at 77.07 and 81.39 °C representing heterozygote of CT. b Melting curves for three different genotypes of cytochrome P450 (CYP) 2C9 (1075A>C); profile with a single peak at 82.89 °C representing homozygote of AA (*1*1), profile with a single peak at 78.56 °C representing homozygote of CC (*3*3) and profile with a double peak at 82.89 and 78.56 °C representing heterozygote of AC (*1*3)
3.3 Warfarin Dosing Algorithm
To assess the effective factors for warfarin maintenance dose, a multivariate stepwise linear regression analysis was conducted with respect to age, gender, VKORC1, CYP2C9 and combined VKORC1 and CYP2C9 in the study group (n = 50). Analysis revealed that age and VKORC1 had an independent and significant contribution to the overall variability in warfarin dose (p = 0.013 and 0.042, respectively); other factors didn’t contribute to the prediction of warfarin dose (p > 0.05). The regression equation for warfarin is as follows:
where age is in years and VKORC1 is coded as 1 for TT and 0 for CT or CC. The estimated regression equation was able to account for 20.5 % (R 2 = 0.205) of the overall variability in warfarin maintenance dose. Age accounted for 13.2 % while VKORC1 polymorphism accounted for the extra 7.3 % of variation. A significant positive correlation, with sufficient strength, was observed between the predicted warfarin dose and the actual prescribed dose (r = 0.453, p = 0.001), as shown in Fig. 2.
This dosing algorithm was assessed in a second unrelated group of 34 patients on warfarin therapy and with stable control of anticoagulation (validation cohort); patient characteristics and demographics are shown in Table 2. Correlation between predicted and actual dose in the validation cohort was statistically significant (Spearman’s rho = 0.388, p = 0.023). The paired difference between the actual and predicted doses was statistically non-significant (Wilcoxon Signed Ranks Test, p = 0.317). The equation was more successful among patients with a dose ≥35 mg/week, where the predicted dose was less than 25 % different from the actual dose in 11 of 16 (68.8 %). When the predicted dose for a subgroup of our patients for whom bodyweight and height were available (n = 35) was calculated using IWPC and Gage algorithms, the correlation coefficient between the actual and predicted doses was 0.304 and 0.276, respectively. When compared with our algorithm (r = 0.279), the difference was non-significant: p = 0.903 and 0.990, respectively.
4 Discussion
Performing genotyping of CYP2C9 and VKORC1 is recommended among patients before initiation of warfarin [21]. In the present study, an algorithm for warfarin dosing was suggested in a cohort of patients who had achieved a therapeutic INR of 2–3. The algorithm was subsequently validated in a second cohort of patients.
The results of the present study show that the patients’ age and genetic variants of VKORC1 demonstrate significant correlations with warfarin dose and account for nearly 20.5 % of the variability in warfarin dose required to achieve an INR of 2–3. Age contributed 13.2 %, while VKORC1 genotypes contributed 7.3 % of the overall variability in warfarin maintenance dose. The results of our developed warfarin algorithm were comparable with those of the IWPC and Gage algorithms with the advantage of using one SNP (VKORC1 1173C>T) only. This represents an economic advantage in our community.
Recently, predictions of warfarin dose by multivariate analyses have been reported in several populations. In other reports, R 2 ranged from 54 to 63 % [15, 22]. Comparison of R 2 in the above-mentioned studies may not be possible because the criteria-considered covariates are different in each study. The IWPC [19] (n = 4,043 participants) reported that the contribution of each tested variable (age, VKORC1 and CYP2C9) in their proposed pharmacogenetic algorithm were 6.75, 13.3 and 7.2 %, respectively. Suriapranata et al. [23] concluded that non-genetic factors (age, bodyweight and height) contributed by 5.9 %, while VKORC1 −1639G/A and CYP2C9 rs17847036 accounted for 8.4 %, with a total contribution of 15.4 % to warfarin reactivity. Cho et al. [15] reported that covariates consisting of VKORC1 1173C>T SNP, age, BSA and CYP2C9 genetic variants accounted for 58.2 % of the overall variability. Herman et al. [24] reported that covariates consisting of bodyweight, age, concomitant drug influencing warfarin metabolism and CYP2C9 polymorphisms accounted for 37 % of the overall variability. Rusdiana et al. [25], found that the age and genetic variants of CYP2C9 and VKORC1 affected the variability of the warfarin response and was able to explain approximately 19.6 % of the PT-INR as a pharmacodynamic index and 16.3 % of the concentration of S-warfarin concentration as a pharmacokinetic index, in the setting of fixed low-dose warfarin therapy in the Indonesian population. As regards previous Egyptian studies, Shahin et al. [26] concluded that 31 % of variability in warfarin dose was explained by a combination of genetic and nongenetic factors; age contributed by 8.11 %, while CYP2C9 genetic variants accounted for 5.17 % of the overall variability. El Din et al. [27] reported that all variables together (age, sex, bodyweight, VKORC1 and CYP2C9*1/*2, *1/*3 and *2/*2 haplotypes) accounted for 61.3 % of the overall variability in warfarin dosages, while VKORC1 (1173C>T) accounted for 31.7 % and CYP2C9 accounted for 15.6 %. The variable contribution of the same SNP in the warfarin dosing algorithm in different ethnic populations may be attributed to different studied cohort sizes and ethnic differences, e.g. the VKORC1 1173TT genotype frequency (67.9 %) among our studied Egyptians was significantly more prevalent than that among Caucasians (12.5 %) [28].
In our study, the predicted dose achieved ideal estimation (less than 25 % difference from actual dose) in 47.1 % of patients in the validation cohort. Our findings were in agreement with the results reported by the IWPC [19] study, who reported that the predicted dose, calculated by a pharmacogenetic algorithm, showed an ideal estimation (20 % difference) in approximately 46 % of the validation cohort (n = 1,009). An interesting finding was the great increase in predictive accuracy among patients requiring a warfarin dose ≥35 mg/week (68.8 %). The good performance of the new algorithm in the upper dose range may not be useful to reduce the bleeding risk associated with warfarin therapy; however, it facilitates the detection of patients who need higher doses than the usual empirical starting dose. For these patients, identifying the dosage necessary to reach the therapeutic range may be cumbersome in current clinical practice, with the risk of inadequate anticoagulation and possible adverse thromboembolic events in the initial phase of the oral anticoagulant therapy, and with longer hospital stay.
The contribution of age and VKORC1 1173 SNP accounted for only 20.5 % of warfarin dose variability in our study cohort, while 79.5 % of the variability in dose requirement is left unaccounted for. The percentage of variation is relatively low compared with other models because besides VKORC1 1173C>T and CYP2C9*3 variants, there exist other SNPs, in both the VKORC1 and CYP2C9 genes as well as other genes, that control pharmacokinetics and pharmacodynamics of warfarin which may have an impact on warfarin dose variability and need to be examined in the Egyptian population. By increasing the number of SNPs studied, the contribution of each SNP, even if it were of small value, would have a cumulative effect that would increase the percentage of variation. Height, bodyweight or body surface area (BSA) were not included in the study as these data were not available for the entire group of 50 patients comprising the study cohort. However, height, bodyweight and BSA have shown variable contribution to the warfarin dosing algorithm, though these parameters did not prove to have significant contribution to warfarin dosing in previous studies that recruited an Egyptian population. Shahin et al. [26] concluded that BSA, height and bodyweight were not significant predictors of dose. They reported that it might be possible that this was because of the minimal variation in height, which varied in the population by less than 4 %. Furthermore, El Din et al. [27] reported that bodyweight showed no influence on dose variability. Moreover, the target INR in our study was 2–3. In other studies, the contribution of target INR, which was between 2 and 3.5 or 2 and 4, might have contributed to the percentage of variation. Also, this study had a relatively small number of participants, whereas studying a larger number may have contributed to a higher percentage of variation. Finally, variation could be attributed to co-morbidity, concurrent medication and low albumin levels, which were not included in the algorithm.
In Egypt, warfarin is the most widely prescribed anticoagulant for reducing thromboembolic events because of its proven efficacy and incomparable low cost compared with the new generation of anticoagulants such as thrombin inhibitor and activated factor X (FXa) inhibitors that have been developed and are available on the international market [29].
To our knowledge, there were no warfarin pharmacogenomic studies carried in Egypt, with the exception of only three studies [26, 27, 30] in the preceding 2 years. The novel attribute of our study was the development of a warfarin dosing algorithm based on the genetic and nongenetic factors of the study cohort, and the validation of the calculated warfarin dosing algorithm in a different cohort with a significant positive correlation observed between the predicted warfarin dose and the actual prescribed dose. In an earlier study, in which 195 Egyptian patients were enrolled, Shahin et al. [26] reported that genetic polymorphisms in VKORC1 3673G>A, CYP2C9 and APOE, along with nongenetic factors (age, smoking and pulmonary embolism), were determinants of warfarin dose requirements. El Din et al. [27] performed VKORC1 1173C>T and CYP2C9*1,*2,*3 genotyping in 46 Egyptian patients and their regression analysis explained about 61 % of the variation in stable dose. Their study design didn’t include validation of their regression model in a different cohort. Their regression model differed from ours in that it did not include the age variable and included the three variants of CYP2C9 (*1*2, *1*3 and *2*2). The recent study carried by Bazan et al. [30] compared the performance of two published clinical and pharmacogenetic algorithms-Gage [18] and IWPC [19]-as well as the warfarin dosing table comprising VKORC1 −1639G>A and CYP2C9 genotypes with empiric dosing in a dataset of 63 Egyptian patients.
Our results show that the maintenance dose decreases, on average, by 0.4 mg/week for every 1 year increase in age. Gage et al. [31] reported a decrease in dose requirements with age, owing to reduced clearance and/or increased responsiveness, by 8–10 % per decade of life. Increasing age of patients is associated with a higher sensitivity to warfarin, which may be caused by the fall in total hepatic content of VKOR because of the age-related decrease in hepatic mass [32]. Multiple linear regression models used to develop warfarin dosing algorithms have consistently found age to be a significant contributor to variability in dose requirements [7].
In accordance with the findings of this study, which show that gender did not contribute significantly to the observed variability in dose requirement, El Din et al. [27] and Teh et al. [33] concluded the same results. However, Cini et al. [34] and Dan et al. [35] found an association of male gender with higher warfarin requirements.
Previous studies [36, 37] concluded contradictory results regarding the influence of dietary factors such as alcohol consumption or vitamin K intake on warfarin dose requirements. Poor compliance with prescribed warfarin therapy may also cause either excessive bleeding or thrombosis response [37]. As all of the patients who were treated with warfarin in this study had received counselling, were put on stable diet plan and were monitored at least once monthly, it is unlikely that vitamin K had a significant impact on the warfarin dose requirements. To investigate the association between dietary factors and warfarin dosage, more controlled studies are needed to achieve reliable results. Among the different factors involved in dose variability are smoking, various illnesses and concurrent medications that were controlled in this study as part of the exclusion criteria.
As regards the frequency of VKORC1 (1173C>T) and CYP2C9 (1075A>C) gene polymorphisms, our results are similar to those of studies previously carried out on an Egyptian population [27, 30], while they were different from other ethnic groups as discussed in El Din et al. [27]. In the present study, the distribution of VKORC1 (1173C>T) show deviation from Hardy Weinberg Equilibrium, which may be explained by the high inbreeding, with an average inbreeding coefficient of 0.0145 in the Egyptian population, as previously reported [38].
In the current study, an association between the VKORC1 (1173C>T) SNP and the weekly warfarin maintenance dose was found. These results are consistent with other studies [27, 28]. The 1173C>T polymorphism in the VKORC1 is a nucleotide substitution in intron 1 of the gene. Presence of the variant T nucleotide is associated with a lower warfarin daily dose requirement, and the 1173 SNP might be in linkage disequilibrium (LD) with other variants that alter VKORC1 activity [28]. It is suggested that −1639A>G could be that variant. The −1639 promoter SNP is located in an E-Box in the 5′-untranslated region of the gene. The consensus sequence of this E-box is CANNTG. Changing the second base A to G, as observed in the −1639 site, would abolish the E-box consensus and would increase the promoter activity by 44 %. This suggested that E-box could function as a repressor binding site that represses transcription [39]. Moreover, a VKORC1 −1639 G>A SNP qualitatively changed the expression of the VKORC1 protein [40].
We did not observe significant differences in warfarin dose demands between CYP2C9*1*1 and CYP2C9*1*3. Our findings are in concordance with previous studies carried on a Japanese [14], Chinese [41] and Indonesian population [23]. This is in contrast to other reports [7–9]. This discordance might be due to the following causes: (1) intake of concomitant drugs or diets that might have overruled the reduced activity of CYP2C9 caused by the *3 allele; (2) the allele frequency of CYP2C9*3 affecting the enzyme activity was too low to allow detection of a significant difference in our study cohort; (3) a selection bias due to a comparatively small sample of a strictly defined group of warfarin-treated patients, who were selected based on their very stable INR values; and (4) the influence of additional genetic factors that we did not account for, including multidrug resistance 1 (MDR1) [42] genes encoding vitamin K-dependent clotting factors [43], GGCX encoding γ-glutamyl carboxylase in the vitamin K cycle [44], the γ-glutamyl carboxylase inhibitory protein calumenin [45], apolipoprotein E [26] and possible genes encoding additional components of the VKOR complex [7].
There is an international debate on the use of pharmacogenetic information in guiding warfarin treatment [46, 47] and the cost effectiveness of analyses on genotype-guided dosing has been reported [48, 49]. As technology has improved and costs have decreased, the increasing availability of genomic testing promises genotype-tailored medical care [50]. In the present study, a two-tube assay was used for the simultaneous genotyping of VKORC1 and CYP2C9 polymorphisms separately under identical thermocycling conditions based on T m-shift technology where the selected primers for both VKORC1 and CYP2C9 have a calculated T m of 55–60 °C (excluding GC tail) [16]. Compared to other current methods [51] for genotyping VKORC1 and CYP2C9 genes, T m-shift genotyping is simple and inexpensive. Primers are standard DNA oligonucleotides, to which generic SYBR Green fluorescent dye is added. Both alleles can be discriminated in a single closed-tube reaction simultaneously and results are obtained immediately after PCR. No post-PCR processing, such as enzymatic digestion, hybridisation or gel electrophoresis, is needed, which can eliminate one of the major causes of contamination in diagnostic PCR laboratories [16]. Although convenient allele discrimination by PCR using allele-specific TaqMan probes permits analysis of both alleles simultaneously, the cost of the probes should be taken into consideration [16]. Therefore, this technique is technically feasible for VKORC1 and CYP2C9 genotyping before beginning warfarin therapy. This allows a clinician to prescribe the first warfarin dose within the same day [16].
The study is, however, not without weaknesses. First, the relatively small sample size, which may not have detected other determinant factors of warfarin dose or may have produced a less precise estimate for the regression coefficients, with the consequence that the new elaborated dosing algorithm included only factors with a high prevalence or a strong effect on warfarin dose. Another possible consequence of the relatively small sample size was the absence of the CYP2C9*3*3 genotype in the study cohort and its presence in the validation group, though there was neither selection bias nor misclassification because patients were selected blindly of their genotype. In any case, the CYP2C9*3 variant didn’t contribute to the warfarin dose, so it was not included in our proposed dosing algorithm. Second, this study does not address whether a pharmacogenetically predicted dose of warfarin translates into better clinical endpoints, such as a reduction in the time to dose stabilization, fewer out-of-range INRs and/or a reduced incidence of bleeding episodes.
5 Conclusion
In Egyptian patients, VKORC1 (1173C>T) contributes to warfarin dose variability. Patients’ age and genetic variants of VKORC1 account for nearly 20.5 % of the variability in warfarin dose required to achieve an INR of 2–3. On validation of the suggested warfarin dosing algorithm in a different cohort, the predicted dose correlated significantly, with sufficient strength, with the maintenance dose and the equation was more successful among patients with a dose ≥35 mg/week; out of 16 patients with a dose ≥35 mg/week, 11 (68.8 %) showed dose agreement.
It appears to be appropriate to increase the number of candidate genes to include those such as CYP4F2, which has a moderately significant effect on warfarin dose as previously reported [52], and those involved in the metabolism of warfarin to set up a powerful tool that is easy for rapid use in all laboratories and clinical settings, to improve warfarin therapy management. In addition, differences in ethnic backgrounds should be taken into account in models producing quantitative dosing algorithms. Replication of this study in a larger cohort of patients is necessary before translating this knowledge into clinical guidelines for warfarin prescription. This would likely have a major impact on patient safety and the efficacy of warfarin during both the initiation and maintenance of therapy, reducing the burden of frequent INR measurements and improving safety by reducing the risk of over- or under-anticoagulation in Egyptian patients.
References
You J, Wong R, Waye M, et al. Warfarin dosing algorithm using clinical, demographic and pharmacogenetic data from Chinese patients. J Thromb Thrombolysis. 2011;31:113–8.
Sasaki T, Tabuchi H, Higuchi S, et al. Warfarin-dosing algorithm based on a population pharmacokinetic/pharmacodynamic model combined with Bayesian forecasting. Pharmacogenomics. 2009;10(8):1257–66.
Lazo-Langner A, Kovacs MJ. Predicting warfarin dose. Curr Opin Pulm Med. 2010;16(5):426–31.
Loebstein R, Yonath H, Peleg D, et al. Interindividual variability in sensitivity to warfarin—nature or nurture? Clin Pharmacol Ther. 2001;70:159–64.
Yin T, Miyata T. Warfarin dose and the pharmacogenomics of CYP2C9 and VKORC1 - rationale and perspectives. Thromb Res. 2007;120:1–10.
Crawford JM, Aspinall MG. The business value and cost–effectiveness of genomic medicine. Pers Med. 2012;9(3):265–86.
Shuen AY, Wong BY, Fu L, Selby R, et al. Evaluation of the warfarin-resistance polymorphism, VKORC1 Asp36Tyr, and its effect on dosage algorithms in a genetically heterogeneous anticoagulant clinic. Clin Biochem. 2012;45:397–401.
Skov J, Bladbjerg E-M, Leppin A, et al. The influence of VKORC1 and CYP2C9 gene sequence variants on the stability of maintenance phase warfarin treatment. Thromb Res. 2013;131:125–9.
Flockhart DA, O’Kane D, Williams MS, et al. On behalf of the ACMG Working Group on Pharmacogenetic Testing of CYP2C9, VKORC1 Alleles for Warfarin Use. Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med. 2008;10:139–50.
Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–41.
Suttie JW. The biochemical basis of warfarin therapy. Adv Exp Med Biol. 1987; 214:3–16.
Stenflo J, Fernlund P, Egan W, et al. Vitamin K dependent modifications of glutamic acid residues in prothrombin. Proc Natl Acad Sci U S A. 1974; 71:2730–3.
Rettie AE, Tai G. The pharmocogenomics of warfarin: closing in on personalized medicine. Mol Interv. 2006;6:223–7.
Yoshizawa M, Hayashi H, Tashiro Y, et al. Effect of VKORC1-1639 G>A polymorphism, body weight, age, and serum albumin alterations on warfarin response in Japanese patients. Thromb Res. 2009;124:161–6.
Cho HJ, Sohn KH, Park HM, et al. Factors affecting the interindividual variability of warfarin dose requirement in adult Korean patients. Pharmacogenomics. 2007;8:329–37.
Huang SW, Li Q, Zhu S-Y, Li L, et al. SYBR Green-based real-time PCR assay for detection of VKORC1 and CYP2C9 polymorphisms that modulate warfarin dose requirement. Clin Chem Lab Med. 2009;47(1):26–31.
Gage BF, Lesko LJ. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb Thrombolysis. 2008; 25:45–51. doi: 10.1007/s11239-007-0104-y.
Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–31.
The International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–64.
Gage BF. Warfarin dosing. Washington University, St Louis. http://www.warfarindosing.org/Source/Home.aspx. Accessed Jun 2013.
US Food and Drug Administration. Table of valid genomic biomarkers in the context of approved drug labels. http://www.fda.gov/cder/genomics/genomic_biomarkers_table.htm. Updated 10 Sep 2008.
Miao L, Yang J, Huang C, et al. Contribution of age, body weight, and CYP2C9 and VKORC1 genotype to the anticoagulant response to warfarin: proposal for a newdosing regimen in Chinese patients. Eur J Clin Pharmacol. 2007;63:1135–41.
Suriapranata IM, Tjong WY, Wang T, et al. Genetic factors associated with patient-specific warfarin dose in ethnic Indonesians. BMC Med Genet. 2011;12:80.
Herman D, Locatelli I, Grabnar I, Peternel P, et al. Influence of CYP2C9 polymorphisms, demographic factors and concomitant drug therapy on warfarin metabolism and maintenance dose. Pharmacogenomics J. 2005;5(3):193–202.
Rusdiana T, Araki T, Nakamura T, et al. Responsiveness to low-dose warfarin associated with genetic variants of VKORC1, CYP2C9, CYP2C19, and CYP4F2 in an Indonesian population. Eur J Clin Pharmacol. 2013;69:395–405.
Shahin MH, Khalifa SI, Gong Y, et al. Genetic and nongenetic factors associated with warfarin dose requirements in Egyptian patients. Pharmacogenet Genomics. 2011;21(3):130–5.
El Din MS, Amin DG, Ragab SB, et al. Frequency of VKORC1 (C1173T) and CYP2C9 genetic polymorphisms in Egyptians and their influence on warfarin maintenance dose: proposal for a new dosing regimen. Int J Lab Hematol. 2012;34:517–24.
D’Andrea G, D’Ambrosio R, Di Perna P, Chetta M, Santacroce R, Brancaccio V, Grandone E, Margaglione M. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose anticoagulant effect of warfarin. Blood. 2005;105:645–9.
Daly A, King B. Pharmacokinetics of oral anticoagulants. Pharmacogenetics. 2003;13:247–52.
Bazan NS, Sabry NA, Rizk A, et al. Validation of pharmacogenetic algorithms and warfarin dosing table in Egyptian patients. Int J Clin Pharm. 2012;34:837–44.
Gage BF, Eby C, Milligan PE, et al. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004;91:87–94.
D’Andrea G, D’Ambrosio R, Margaglione M. Oral anticoagulants: pharmacogenetics relationship between genetic and non-genetic factors. Blood Rev. 2008; 22(3):127–40.
Teh LK, Langmia IM, Fazleen Haslinda MH, et al. Clinical relevance of VKORC1 (G-1639A and C1173T) and CYP2C9*3 among patients on warfarin. J Clin Pharm Ther. 2012;37:232–6.
Cini M, Legnani C, Cosmi B, et al. A new warfarin dosing algorithm including VKORC1 3730 G >A polymorphism: comparison with results obtained by other published algorithms. Eur J Clin Pharmacol. 2012;68(8):1167–74.
Xu D, Liu Y, Zhong S-L, et al. Effect of demographic factors on warfarin dosing in patients after cardiac valve replacement. J Practical Med. 2010;5:750–753. http://www.123xu.com/gl/9009.htm. Accessed 20 May 2012.
Jonas DE, McLeod HL. Genetic and clinical factors relating to warfarin dosing. Trends Pharmacol Sci. 2009;30(7):375–86.
Namazi S, Azarpira N, Hendijani F, et al. The impact of genetic polymorphisms and patient characteristics on warfarin dose requirements: a cross-sectional study in Iran. Clin Ther. 2010;32(6):1050–60.
Habib Z. Haptoglobin polymorphism in Egyptians. Ann Hum Biol. 1983; 10(4):385–87.
Yuan H, Chen J, Lee M, Wung J, et al. A novel functional VKORC1 promoter polymorphism is associated with interindividual and interethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14:1745–51.
Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–93.
Gu Q, Kong Y, Schneede J, et al. VKORC1-1639G>A, CYP2C9, EPHX1691A>G genotype, body weight, and age are important predictors for warfarin maintenance doses in patients with mechanical heart valve prostheses in southwest China. Eur J Clin Pharmacol. 2010; 66:1217–27.
Wadelius M, Sörlin K, Wallerman O, et al. Warfarin sensitivity related toCYP2C9, CYP3A5, ABCB1 (MDR1) and other factors. Pharmacogenomics. 2004;4(1):40–8.
Wajih N, Sane DC, Hutson SM, et al. The inhibitory effect of calumenin on the vitamin K-dependent γ-carboxylation system. Characterization of the system in normal and warfarin resistant rats. J Biol Chem. 2004;279(24):25276–83.
Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5(4):262–70.
Voora D, Koboldt DC, King CR, et al. A polymorphism in the VKORC1-regulator calumenin predicts higher warfarin doses in African-Americans. Clin Pharmacol Ther. 2010;87(4):445–51.
Wadelius M. Use of pharmacogenetics in guiding treatment with warfarin. Clin Chem. 2009;55:709–11.
Rosove MH, Grody WW. Should we be applying warfarin pharmacogenetics to clinical practice? No, not now. Ann Intern Med. 2009;151:270–3.
Eckman MH, Rosand R, Greenberg SM, et al. Cost effectiveness of using pharmacogenetic information in warfarin dosing for patients with non-valvular atrial fibrillation. Ann Intern Med. 2009;150:73–83.
You JHS, Tsui KKN, Wong RSM, et al. Potential clinical and economic outcomes of CYP2C9 and VKORC1 genotype-guided dosing in patients initiating warfarin therapy. Clin Pharmacol Ther. 2009;86:540–7.
Vassy JL. Can genetic information change patient behavior to reduce type 2 diabetes risk? Pers Med. 2013;10(1):9–12.
Toriello M, Meccariello P, Mazzaccara C, et al. Comparison of the TaqMan and light cycler systems in pharmacogenetic testing: evaluation of CYP2C9*2/*3 polymorphisms. Clin Chem Lab Med. 2006;44:285–7.
Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–12.
Acknowledgments
No sources of external funding were used in the preparation of this article.
Disclosure statement
The authors declare that they have no conflicts of interest that are directly relevant to the content of this article.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ekladious, S.M.M., Issac, M.S.M., Sharaf, S.A.EA. et al. Validation of a Proposed Warfarin Dosing Algorithm Based on the Genetic Make-Up of Egyptian Patients. Mol Diagn Ther 17, 381–390 (2013). https://doi.org/10.1007/s40291-013-0046-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-013-0046-3