Abstract
Purpose
Large inter-individual differences in warfarin maintenance dose are mostly due to the effect of genetic polymorphisms in multiple genes, including vitamin K epoxide reductase complex 1 (VKORC1), cytochromes P450 2C9 (CYP2C9), and cytochrome P450 4F2 (CYP4F2). Thus, several algorithms for predicting the warfarin dose based on pharmacogenomics data with clinical characteristics have been proposed. Although these algorithms consider these genetic polymorphisms, the formulas have different coefficient values that are critical in this context. In this study, we assessed the mutual validity among these algorithms by specifically considering racial differences.
Methods
Clinical data including actual warfarin dose (AWD) of 125 Japanese patients from our previous study (Eur J Clin Pharmacol 65(11):1097–1103, 2009) were used as registered data that provided patient characteristics, including age, sex, height, weight, and concomitant medications, as well as the genotypes of CYP2C9 and VKORC1. Genotyping for CYP4F2*3 was performed by the PCR method. Five algorithms that included these factors were selected from peer-reviewed articles. The selection covered four populations, Japanese, Chinese, Caucasian, and African-American, and the International Warfarin Pharmacogenetics Consortium (IWPC).
Results
For each algorithm, we calculated individual warfarin doses for 125 subjects and statistically evaluated its performance. The algorithm from the IWPC had the statistically highest correlation with the AWD. Importantly, the calculated warfarin dose (CWD) using the algorithm from African-Americans was less correlated with the AWD as compared to those using the other algorithms. The integration of CYP4F2 data into the algorithm did not improve the prediction accuracy.
Conclusion
The racial difference is a critical factor for warfarin dose predictions based on pharmacogenomics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Warfarin is one of the most widely used oral anticoagulants. It competitively inhibits vitamin K epoxide reductase complex subunit 1 (VKORC1) that catalyzes the recycling of vitamin K in the liver. The inhibition of vitamin K epoxide reductase (VKOR) impairs the activity of coagulation factors II, VII, IX, and X by depleting reduced vitamin K [1]. The pharmacological difficulties of warfarin treatment are associated with its narrow effective range and the large inter-individual differences in maintaining its effective dose. Due to these features, repeated monitoring of the coagulation ability defined as the prothrombin time–international normalized ratio (PT-INR) is required to determine individually the adequate dose at each time point [2], which lowers the risk for adverse events such as ischemic or hemorrhagic stroke [3].
Warfarin is a racemic mixture of S- and R-warfarin. S-warfarin is approximately three to five times more potent as an anticoagulant than R-warfarin. S-warfarin is predominantly metabolized by cytochrome P450 2C9 (CYP2C9) [4]. Cumulative evidence indicates that genetic polymorphisms of VKORC1 and CYP2C9 are closely associated with the optimal dose of warfarin [5]. The genetic polymorphisms of the CYP2C9, CYP2C9*2 (430C>T) and CYP2C9*3 (1075A>C) reduce the CYP2C9 activity to 12% and 5% of the wild-type activity, respectively. As a result, the maintenance warfarin dose in subjects with these genetic polymorphisms is strongly decreased [6]. In contrast, VKORC1 (-1639G>A), a genetic polymorphism in the promoter region of the VKORC, decreases the expression level of VKORC1, leading to a decreased maintenance warfarin dose [7]. Furthermore, it has been reported that the cytochrome P450 4F2 (CYP4F2) polymorphism affects warfarin dose [8]. CYP4F2 is involved in the accumulation of vitamin K in the liver by catalyzing the production of hydroxylated vitamin K [9]. The genetic polymorphism of the CYP4F2, CYP4F2*3 (1297C>T) decreases the enzymatic activity of CYP4F2, leading to increased hepatic vitamin K. Therefore, the genetic polymorphism of CYP4F2 increases the requirement for warfarin [10].
So far, several algorithms for predicting the warfarin dose have been proposed to determine the optimal dose prior to warfarin administration [11, 12]. Most of these algorithms typically use patient characteristics such as age, weight, height, concomitant drugs, and the genotypes of CYP2C9, VKORC1, and/or CYP4F2 [13]. These genotype-guided warfarin dosing algorithms were considered a rational approach to optimize warfarin dosing and, potentially, reduce adverse events. However, these algorithms are not widely used in the real world, because at least partially, the algorithm formulas propose coefficient values that differ strongly for each factor, and in addition, mutual validity among these algorithms has not been addressed [14].
The clinical significance of CYP2C9, VKORC1, and CYP4F2 genetic polymorphisms has been recognized in many populations with different races. The reduction in CYP2C9 activity caused by the genetic polymorphism of CYP2C9 (dominated by CYP2C9*2 and CYP2C9*3) varies significantly depending on racial differences; thus, warfarin responsiveness related with the frequency of these genetic polymorphisms for racial differences has attracted attention [15]. Among Caucasians, the allele frequencies of CYP2C9*2 and CYP2C9*3 are in the range of 8–12% and 6–10%, respectively, but they are lower in Southeast Asians. Variant allele frequencies are higher in Caucasians than Japanese, which may contribute, at least partially, to the racial differences. [16]. The frequency of the VKORC1 (-1639 G>A) minor allele is around 90% in Asian, around 40% in Caucasians, and around 9% in African-Americans [17]. The frequency of the CYP4F2*3 minor allele is approximately 30% in Asians and Caucasians and approximately 7% in African-Americans [18]. Because of the differences between the algorithms, it is unknown whether the significance of the genetic polymorphisms differs among populations. Previously, we demonstrated the importance of the CYP2C9 and VKORC1 genetic polymorphisms in a Japanese population and created a corresponding algorithm to predict the maintenance warfarin dosage [19]. In this study, we examined the mutual validity among the algorithms by using the previously collected clinical data of the Japanese population as a test set.
Methods
The Ethical Review Committee of Osaka University approved this study (approval number, 766).
Study subjects
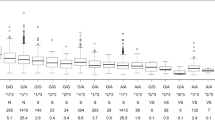
Clinical data of the subjects from our previous study [19] were used as registered data. Briefly, the study subjects were 125 Japanese patients with stable anticoagulant warfarin therapy, who provided written informed consent to allow their samples and clinical data to be used for secondary analyses. As shown in Table 1, the patients’ information on age, weight, height, PT-INR value, and genotypes of CYP2C9, VKORC1, and CYP4F2 were used for this study. Warfarin maintenance dose was defined as the dose that controlled the PT-INR range between 1.5 and 3.0 during the last three clinic visits and designated as actual warfarin dose (AWD) in this study. Patients were excluded if they had hepatic or renal dysfunction or if they had the concomitant medication by amiodarone, bucolome, fluconazole, miconazole, and sulfamethoxazole in addition to warfarin because these drugs affect the metabolism of warfarin.
Selection criteria for algorithms
As shown in Table 2, the algorithms, designated as Original and algorithms I (IWPC) to V, were selected for this study according to the following criteria: (1) The algorithms should include genetic polymorphisms of CYP2C9, VKORC1, and/or CYP4F2 as variables. (2) The articles with the algorithms should provide the patients’ information related to race and disease. (3) The algorithms should include both weight and height as their variables. (4) The algorithms should be designed to calculate the maintenance dose rather than the initial dose and should not require the initial dose as a variable. (5) The algorithms should be in articles with free access to the full text in the PubMed Central database. We excluded algorithms as candidates if they required the patients’ information on chronic kidney disease, smoking, and drinking of alcoholic beverages as variables.
The warfarin doses were calculated using the information on 125 patients and algorithms that fulfilled all selection criteria.
Statistical analyses
The performance of each algorithm was evaluated by the mean absolute error (MAE), relative error as root mean square error (RMSE), and root mean square percentage error (RMSPE) to the AWD calculated using the following formulas:
In addition, we confirmed the distribution of the difference between the AWD and the calculated warfarin dose (CWD) by Bland-Altman plots. Limits of agreement (LA) were defined as the symmetric range encompassing 95% of the data. Assuming a normal error distribution, the upper and lower limits of agreement (ULA and LLA, respectively) were calculated as the mean difference (MD) ± 1.96 times the standard deviation (S.D.) of the differences. We also calculated the percentage of patients whose CWD was within 20% of the AWD to evaluate the potential clinical value of each algorithm according to a previous report [20]. Finally, the one-way ANOVA and the Bonferroni correction method for multiple comparisons were performed, and the value of p < 0.05 was considered significantly different. IBM SPSS® Statistics Version 21 (IBM Corp.) was used for statistical analyses.
Results
As shown in Fig. 1, to analyze the correlations between the AWD and the CWD derived from the algorithms designated as Original (a), I (IWPC) (b), II (c), III (d), IV (e), and V (f), we created corresponding scatter plots between the AWD and the CWD with the line indicating the perfect prediction. In addition, we statistically analyzed the performance of each algorithm (Table 3). Because the Original algorithm was created based on the clinical data of the study subjects, we considered the errors (MAE, 0.63; RMSE, 0.84; RMSPE, 42) and the MD between the AWD and the CWD (MD, − 0.01; LLA, − 1.66; ULA, 1.63) from the Original algorithm as control values and evaluated the other algorithms. Interestingly, the algorithms I (IWPC) and II had MAE values (I, 0.66; II, 0.67) that were the closest to that of the Original algorithm (0.63), and their scatter plots were similar to each other (Fig. 1b and c). In addition, the percentages of patients whose AWD was within 20% of the CWDs derived from algorithms I and II (I, 49.6%; II, 50.4%) were higher than those from the other algorithms. In contrast, the algorithm V created with an African-American population had MAE (1.18) and RMSE values (1.34) that were higher than those of the Original algorithm. The algorithm V also had by far the highest values of RMSPE (99%) and the MD between the AWD and the CWD (MD, + 0.98; LLA, − 0.81; ULA, + 2.77) among all algorithms at a significant level and its scatter plot appeared to have the least correlation among them (Fig. 1f). We have noticed that this study cohort included the population that had PT-INR between 1.5 and 2, which is not the range that the algorithms I through V were developed for. Therefore, we created scatter plots between the AWD and the CWD for 51 subjects with PT-INR between 2.0 and 3.0, and we got the similar plots (Supplementary Figure 1).
Scatter plot of the actual warfarin dose (AWD) vs. the calculated warfarin dose (CWD). Scatter plots were applied to examine the correlation between the AWD and the CWD derived from each algorithm. The solid line indicates the line of equivalence, which shows that the AWD and the CWD are perfectly matched. The x-axis represents the AWD (mg/day) and the y-axis represents the CWD (mg/day). a Original algorithm. b Algorithm I (IWPC). c Algorithm II. d Algorithm III. e Algorithm IV. f Algorithm V
Importantly, the algorithm V showed significantly higher MD than the Original algorithm and the range of ± 1.96 S.D. shifted to the positive direction. To confirm visually the distribution of each difference between the AWD and the CWDs derived from the Original algorithm and algorithm V, the Bland-Altman plot was applied [21] (Supplementary Figure 2). In addition, the mean value of CWD derived from the algorithm V (mean ± S.D.; 3.65 ± 0.79) was significantly higher than those of the AWD (2.67 ± 1.25) and the CWD (2.66 ± 0.93) derived from the Original algorithm. As will be understood, the percentage of patients whose AWD was within 20% of the CWD derived from the algorithm V (24.0%) was the smallest among those with other algorithms.
Next, we focused on the algorithms III and IV, which use the CYP4F2 genotype information for the CWDs. Interestingly, as shown in Table 3, each algorithm had values of MAE (III, 0.72; IV, 0.75), RMSE (III, 0.91; IV, 0.94), RMSPE (III, 62%; IV, 53%), and the MD between the AWD and the CWD (III: MD, + 0.22; LLA, − 1.51; ULA, + 1.94; IV: MD, + 0.31; LLA, − 1.43; ULA, + 2.05) that were higher than those of the Original algorithm, although the algorithm III was created using the Japanese population data. These results indicated that the correlation between the AWD and the CWD derived from the algorithms III or IV may not be significantly improved by integrating the CYP4F2 genetic information into the algorithm.
Finally, to evaluate the mutual validity among these algorithms, as shown in Fig. 2, we created the scatter plots between the CWD derived from the Original algorithm (Original CWD) and the CWDs derived from the algorithms with the designation I (IWPC) to V. We also analyzed the MDs based on the Original CWD and the CWD from each test algorithm. As shown in Fig. 2, algorithm I (IWPC) appeared to have the best correlation among all scatter plots (Fig. 2a). It also showed the smallest value of MD (MD, − 0.20; LLA, − 0.68; ULA, + 0.27) as compared to those derived for the other algorithms (II, − 0.25; III, 0.23; IV, 0.32; V, 0.99) (Table 3). To exclude the possibility that the results were influenced by the data from the subjects with PT-INR 1.5–2.0, we created the scatter plots between the Original CWD and the CWDs from the other algorithms using the subjects with PT-INR between 2 and 3 (Supplementary Figure 2) and confirmed that the algorithm V has a tendency to overestimate the dose.
Scatter plot of the calculated warfarin dose (CWD) derived from each of the algorithms I (IWPC) to V vs. CWD derived from the Original algorithm. Scatter plots were applied to examine the correlation between the CWD derived from each algorithm I (IWPC) to V and the CWD derived from the Original algorithm (Original CWD). The solid line indicates the line of equivalence, which shows that the Original CWD and the CWD derived from each algorithm are perfectly matched. The x-axis designated as Original represents the Original CWD (mg/day). The y-axis represents the CWD derived from each algorithm (mg/day). a Algorithm I (IWPC). b Algorithm II. c Algorithm III. d Algorithm IV. e Algorithm V
Finally, as the algorithm I (IWPC) was applied for some retrospective studies and it also showed the close performance to the Original algorithm, in order to confirm if the algorithm I (IWPC) is the best applicable for Japanese population among other algorithms, we created corresponding scatter plots (Fig. 3) and calculated MD (shown as MD3 in Table 3) between the CWD derived from the algorithm I (IWPC) and the CWDs from the other algorithm II to V with our cohort, which was used to develop the Original algorithm. As a result, each scatter plot titled algorithms II (Fig. 3a), III (Fig. 3b), IV (Fig. 3c), and V (Fig. 3d) is similar to the plot with the corresponding figure title in Fig. 2 b to e. As MD3 showed in Table 3, the algorithms III, IV, and V were overestimated than the algorithm I (IWPC).
Scatter plot of the calculated warfarin dose (CWD) derived from each algorithm II to V vs. CWD derived from the algorithm I (IWPC). Scatter plots were applied to examine the correlation between the CWD derived from each algorithm II to V and the CWD derived from the algorithm I (CWD from algorithm I). The solid line indicates the line of equivalence, which shows that the CWD from algorithm I and the CWD from each algorithm are perfectly matched. The x-axis designated as algorithm I represents the CWD from algorithm I (IWPC) (mg/day). The y-axis represents the CWD derived from each algorithm (mg/day). a Algorithm II. b Algorithm III. c Algorithm IV. d Algorithm V
Discussion
The FDA-approved drug label for warfarin states that CYP2C9 and VKORC1 genotype information can contribute to the prediction of the warfarin dose, and multiple algorithms have been proposed to predict the warfarin maintenance dose. Since the warfarin maintenance dose is influenced by genetic variants; demographic parameters, including weight, height, and age; and environmental exposures, it is likely that algorithm performance is associated with the characteristics of the study cohorts; however, the mutual validity among algorithms remains to be addressed, though verification of the algorithms is a critical process in the precision medicine for warfarin therapy. Previously, we proposed the Original algorithm based on the clinical data of 125 Japanese patients, and in this study, we analyzed the mutual validity among the five select algorithms using the same clinical data as a test set. Moreover, we examined whether adding the CYP4F2 genotype information improves the prediction of the warfarin dose in the Japanese population, as compared to that using only the CYP2C9 and VKORC1 genotypes.
In the preliminary stage of this study, we found that both body height and weight are essential to get a good correlation between the AWD and the CWD. We probed the performance of the selected algorithms by creating scatter plots and by calculating several statistical parameters. As a result, we found that the algorithm proposed by IWPC (algorithm I) had the best correlation between the CWD and the AWD as compared to that derived from the others except for the Original algorithm. Because algorithm I was based on data from Caucasian, Asian, African-American, and mixed populations and accounts for racial differences as a variable, it could be applied for the Asian population, suggesting that racial differences might influence the warfarin dosage independently of the CYP2C9 and VKORC1 genotypes. Interestingly, two patients with VKORC1 GG genotype had remarkably higher CWDs derived from the Original algorithm than those derived from the other algorithms, and the points deviated substantially from the line indicating the perfect prediction of every scatter plot in Fig. 2 (Although they appear to be one point, two points overlapped). Because their AWDs were more than 7 mg/day, other factors besides the genotypes might have affected the warfarin dose in these two patients.
In contrast to algorithm I, the CWD using algorithm V, which exclusively derived from an African-American population, were less coincident with the AWD associated with a RMSPE of 99%, although this algorithm includes the genotypes of VKORC1 and CYP2C9, age, and body surface area (BSA). Bland-Altman analyses showed that the difference is significant. These findings are consistent with the previously reported observation that the genetic polymorphism of VKORC1 contributes to the variability of the warfarin dose at different degrees, depending on the race. It could be proposed that one or more additional genetic factors reduce the sensitivity to warfarin independently of VKORC1 in the African-American population. In addition to the VKORC1 genetic polymorphism, the influence of the CYP2C9*5, *6, *8, and *11 alleles might be as significant in African-Americans as the influence of the CYP2C9*2 and *3 alleles in Caucasians or Asians [20], and the algorithm V includes the genotypes of CYP2C9 *2, *3, *5, *6, *8, and *11 as variables. However, the coefficient of the variable is defined as 0 for the patients with wild-type and 1 for those with mutant genotype of CYP2C9. Considering the difference in the enzyme activity among the genotypes, the algorithm V might have the intrinsic limitation to be applied for this study beyond the racial difference.
Regarding the CYP4F2 genotype, its integration into the algorithms resulted in a nonsignificant improvement of the warfarin dose prediction among our registered patients. The importance of the CYP4F2 genotype for the warfarin dose prediction is controversial. CYP4F2 is involved in vitamin K metabolism but warfarin does not alter its activity directly. Additional vitamin K metabolism-related factors might affect the influence of CYP4F2 activity on the warfarin dose. We noticed that both algorithms III and IV with the CYP4F2 genotype had the tendency to generate a slightly overestimated CWD as compared to the AWD. Therefore, this algorithm might be improved by some specific adjustments, depending on the population. However, further studies are required to test whether an algorithm with the CYP4F2 genotypes as a variable, possibly in combination with additional vitamin K metabolism-related factors, would further improve genotype-guided warfarin dosing in the real world than those using only the CYP2C9 and VKORC1 genotypes.
Our study has other limitations in addition to the number of subjects. We did not evaluate the reported algorithms with other variables such as smoking status, alcohol consumption, the APOE genotypes encoding apolipoprotein E and other factors which can affect the warfarin dose because we did not have those clinical data for all the registered patients. However, we believe that the algorithms using fewer variables to predict dosages are more applicable in the real world.
In summary, we validated the algorithms for warfarin dosing using the VKORC1, CYP2C9, and CYP4F2 genotypes as variables by applying the clinical data of a Japanese population as a test set. Despite the apparent differences in the formulas between the algorithms, the CWD derived from algorithms based on Asians appeared to be significantly consistent with the AWD. The IWPC algorithm showed the high accuracy for the Japanese population because the algorithm accounts for racial difference as a variable whereas an algorithm exclusively derived from the African-American population is less useful for warfarin dose prediction.
In conclusion, racial differences are critical for the pharmacogenomics-based prediction of warfarin dosing. We expect prospective clinical studies to propose and/or confirm the flowchart for warfarin therapy regimens administered as precision medicine adjusted for each racial population.
References
Limdi NA, Veenstra DL (2008) Warfarin pharmacogenetics. Pharmacotherapy 28(9):1084–1097. https://doi.org/10.1592/phco.28.9.1084
Johnson JA (2008) Warfarin: an old drug but still interesting. Pharmacotherapy 28(9):1081–1083. https://doi.org/10.1592/phco.28.9.1081
Lesko LJ (2008) The critical path of warfarin dosing: finding an optimal dosing strategy using pharmacogenetics. Clin Pharmacol Ther 84(3):301–303. https://doi.org/10.1038/clpt.2008.133
Kaminsky LS, Zhang ZY (1997) Human P450 metabolism of warfarin. Pharmacol Ther 73(1):67–74. https://doi.org/10.1016/S0163-7258(96)00140-4
Carlquist JF, Horne BD, Muhlestein JB, Lappe DL, Whiting BM, Kolek MJ, Clarke JL, James BC, Anderson JL (2006) Genotypes of the cytochrome p450 isoform, CYP2C9, and the vitamin K epoxide reductase complex subunit 1 conjointly determine stable warfarin dose: a prospective study. J Thromb Thrombolysis 22(3):191–197. https://doi.org/10.1007/s11239-006-9030-7
Taube J, Halsall D, Baglin T (2000) Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood 96(5):1816–1819
Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE (2005) Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 352(22):2285–2293. https://doi.org/10.1056/NEJMoa044503
Shahabi P, Scheinfeldt LB, Lynch DE, Schmidlen TJ, Perreault S, Keller MA, Kasper R, Wawak L, Jarvis JP, Gerry NP, Gordon ES, Christman MF, Dube MP, Gharani N (2016) An expanded pharmacogenomics warfarin dosing table with utility in generalised dosing guidance. Thromb Heamost 116 (2):337–348. https://doi.org/10.1160/TH15-12-0955
McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE (2009) CYP4F2 is a vitamin K1 oxidase: an explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharmacol 75(6):1337–1346. https://doi.org/10.1124/mol.109.054833
Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, King CR, Brower A, Schmelzer JR, Glurich I, Vidaillet HJ, Yale SH, Qi Zhang K, Berg RL, Burmester JK (2008) CYP4F2 genetic variant alters required warfarin dose. Blood 111(8):4106–4112. https://doi.org/10.1182/blood-2007-11-122010
Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, Milligan PE, Grice G, Lenzini P, Rettie AE, Aquilante CL, Grosso L, Marsh S, Langaee T, Farnett LE, Voora D, Veenstra DL, Glynn RJ, Barrett A, McLeod HL (2008) Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther 84(3):326–331. https://doi.org/10.1038/clpt.2008.10
Kaye JB, Schultz LE, Steiner HE, Kittles RA, Cavallari LH, Karnes JH (2017) Warfarin pharmacogenomics in diverse populations. Pharmacotherapy 37(9):1150–1163. https://doi.org/10.1002/phar.1982
van Schie RM, Wadelius MI, Kamali F, Daly AK, Manolopoulos VG, de Boer A, Barallon R, Verhoef TI, Kirchheiner J, Haschke-Becher E, Briz M, Rosendaal FR, Redekop WK, Pirmohamed M, van der Zee AH M (2009) Genotype-guided dosing of coumarin derivatives: the European pharmacogenetics of anticoagulant therapy (EU-PACT) trial design. Pharmacogenomics 10(10):1687–1695. https://doi.org/10.2217/pgs.09.125
Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, Kesteven P, Christersson C, Wahlstrom B, Stafberg C, Zhang JE, Leathart JB, Kohnke H, Maitland-van der Zee AH, Williamson PR, Daly AK, Avery P, Kamali F, Wadelius M (2013) A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 369(24):2294–2303. https://doi.org/10.1056/NEJMoa1311386
Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R (2002) Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther 72(6):702–710. https://doi.org/10.1067/mcp.2002.129321
Takahashi H, Wilkinson GR, Caraco Y, Muszkat M, Kim RB, Kashima T, Kimura S, Echizen H (2003) Population differences in S-warfarin metabolism between CYP2C9 genotype-matched Caucasian and Japanese patients. Clin Pharmacol Ther 73(3):253–263. https://doi.org/10.1067/mcp.2003.26a
Takahashi H, Wilkinson GR, Nutescu EA, Morita T, Ritchie MD, Scordo MG, Pengo V, Barban M, Padrini R, Ieiri I, Otsubo K, Kashima T, Kimura S, Kijima S, Echizen H (2006) Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics 16(2):101–110. https://doi.org/10.1097/01.fpc.0000184955.08453.a8
Dean L (2012) Warfarin therapy and VKORC1 and CYP genotype. Medical Genetics Summaries [Internet]. https://www.ncbi.nlm.nih.gov/books/NBK84174 . Accessed 11 June 2018
Ohno M, Yamamoto A, Ono A, Miura G, Funamoto M, Takemoto Y, Otsu K, Kouno Y, Tanabe T, Masunaga Y, Nonen S, Fujio Y, Azuma J (2009) Influence of clinical and genetic factors on warfarin dose requirements among Japanese patients. Eur J Clin Pharmacol 65(11):1097–1103. https://doi.org/10.1007/s00228-009-0685-9
Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA (2009) Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 360(8):753–764. https://doi.org/10.1056/NEJMoa0809329
Carkeet A (2015) Exact parametric confidence intervals for Bland-Altman limits of agreement. Optom Vis Sci 92(3):e71–e80. https://doi.org/10.1097/OPX.0000000000000513
Huang SW, Chen HS, Wang XQ, Huang L, Xu DL, Hu XJ, Huang ZH, He Y, Chen KM, Xiang DK, Zou XM, Li Q, Ma LQ, Wang HF, Chen BL, Li L, Jia YK, Xu XM (2009) Validation of VKORC1 and CYP2C9 genotypes on interindividual warfarin maintenance dose: a prospective study in Chinese patients. Pharmacogenet Genomics 19(3):226–234. https://doi.org/10.1097/FPC.0b013e328326e0c7
Cha PC, Mushiroda T, Takahashi A, Kubo M, Minami S, Kamatani N, Nakamura Y (2010) Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum Mol Genet 19(23):4735–4744. https://doi.org/10.1093/hmg/ddq389
Zambon CF, Pengo V, Padrini R, Basso D, Schiavon S, Fogar P, Nisi A, Frigo AC, Moz S, Pelloso M, Plebani M (2011) VKORC1, CYP2C9 and CYP4F2 genetic-based algorithm for warfarin dosing: an Italian retrospective study. Pharmacogenomics 12(1):15–25. https://doi.org/10.2217/pgs.10.162
Bress A, Patel SR, Perera MA, Campbell RT, Kittles RA, Cavallari LH (2012) Effect of NQO1 and CYP4F2 genotypes on warfarin dose requirements in Hispanic-Americans and African-Americans. Pharmacogenomics 13(16):1925–1935. https://doi.org/10.2217/pgs.12.164
Acknowledgments
We are grateful to all doctors, nurses, and subjects who participated in this study.
Funding
This study was funded by the Management Expenses Grants from MEXT, Japan.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: MO, HN, YF, MM. Performed the experiments: MS, YF, SN, MM. Analyzed the data: MO, SN, SH, SM, YF, YM, JY, YS, YF, MM. Contributed new methods or models: YF, MM. Wrote the paper: YF, MM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Ethical Review Committee of Osaka University (approval number, 766). Written informed consent was obtained from each participant to allow their samples and clinical data to be used for secondary analyses. For this type of study formal consent is not required. The study has been performed in accordance with The Code of Ethics of the 1964 Declaration of Helsinki and its later amendments. This study does not contain any studies with animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
Scatter Plot of the actual warfarin dose (AWD) v.s. the calculated warfarin dose (CWD) only for 51 subjects with PT-INR between 2 and 3. Scatter plots were applied to examine the correlation between the AWD and the CWD derived from each algorithm. The solid line indicates the line of equivalence, which shows that the CWD and the AWD are perfectly matched. The X-axis represents the AWD (mg/day) and the Y-axis represents the CWD (mg/day). a Original algorithm. b Algorithm I (IWPC). c Algorithm II. d Algorithm III. e Algorithm IV. f Algorithm V. (PPTX 120 kb)
Figure S2
Bland and Altman plot for the actual warfarin dose (AWD) and the calculated warfarin dose (CWD) with the representation of the limits of agreement. Bland and Altman’s plots were performed to describe the agreement between the CWD and the AWD. The Y-axis represents the difference between the CWD and the AWD (CWD-AWD) (mg/day), and the solid line indicates the mean difference. The X-axis represents the mean of the CWD and the AWD ((CWD+AWD)/2) (mg/day). The dotted lines indicate the ±1.96 SD of the mean difference (CWD - AWD) are shown a parallel to the X-axis. a Original algorithm. b Algorithm I (IWPC). c Algorithm II. d Algorithm III. e Algorithm IV. f Algorithm V. (PPTX 131 kb)
Figure S3
Scatter Plot of the calculated warfarin dose (CWD) derived from each algorithm I (IWPC) to V v.s. CWD derived from the Original algorithm only for 51 subject with PT-INR between 2 and 3. Scatter plots were applied to examine the correlation between the CWD derived from each algorithm I (IWPC) to V and the CWD derived from the Original algorithm (Original CWD). The solid line indicates the line of equivalence, which shows that the CWD and the Original CWD are perfectly matched. The X-axis designated as Original represents the Original CWD (mg/day). The Y-axis represents the CWD derived from each algorithm (mg/day). a Algorithm I (IWPC). b Algorithm II. c Algorithm III. d Algorithm IV. e Algorithm V. (PPTX 107 kb)
Rights and permissions
About this article
Cite this article
Sasano, M., Ohno, M., Fukuda, Y. et al. Verification of pharmacogenomics-based algorithms to predict warfarin maintenance dose using registered data of Japanese patients. Eur J Clin Pharmacol 75, 901–911 (2019). https://doi.org/10.1007/s00228-019-02656-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-019-02656-7