Abstract
Background Warfarin remains a difficult drug to use due to the large variability in dose response. Clear understanding of the accuracy of warfarin pharmacogenetic dosing methods might lead to appropriate control of anticoagulation. Objective This study aims to evaluate the accuracy of warfarin dosing table and two pharmacogenetic algorithms, namely the algorithms of Gage et al. (Clin Pharmacol Ther 84:326–331, 2008), and the International Warfarin Pharmacogenetics Consortium algorithm (IWPC) in a real Egyptian clinical setting. Additionally, three non-pharmacogenetic dosing methods (the Gage, IWPC clinical algorithms and the empiric 5 mg/day dosing) were evaluated. Setting Sixty-three Egyptian patients on a stable therapeutic warfarin dose were included. Patients were recruited from the outpatient clinic of the critical care medicine department. Methods CYP2C9 and VKORC1 polymorphisms were genotyped by real time PCR system. Predicted doses by all dosing methods were calculated and compared with the actual therapeutic warfarin doses. Results The Gage algorithm (adjusted R2 = 0.421, and mean absolute error (MAE) = 3.3), and IWPC algorithm (adjusted R2 = 0.419, MAE = 3.2) produced better accuracy than did the warfarin dosing table (adjusted R2 = 0.246, MAE = 3.5), the two clinical algorithms (R2 = 0.24, MAE = 3.7) and the fixed dose approach (MAE = 3.9). However, all dosing models produced comparable clinical accuracy with respect to proportion of patients within 1 mg/day of actual dose (ideal dose). Non-pharmacogenetic methods severely over-predicted dose (defined as ≥2 mg/day more than actual dose) compared to the three pharmacogenetic models. In comparison to non-pharmacogenetic methods, the three pharmacogenetic models performed better regarding the low dose group in terms of percentage of patients within ideal dose. In the high dose group, none of the dosing models predicted warfarin doses within ideal dose. Conclusion Our study showed that genotype-based dosing improved prediction of warfarin therapeutic dose beyond that available with the fixed-dose approach or the clinical algorithms, especially in the low-dose group. However, the two pharmacogenetic algorithms were the most accurate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Impact of findings on practice
-
The Gage and IWPC Pharmacogenetic algorithms are less accurate in Egyptian patients than in white Caucasian and Asian patients.
-
The use of pharmacogenetic algorithms for dosing warfarin are an improvement over empiric dosing and the clinical algorithms.
-
Other clinical factors like certain diseases and vitamin K consumption and genes other than VKORC1 and CYP2C9 probably have a significant impact on warfarin dosing in Egyptian patients with high therapeutic dose requirements.
Introduction
Warfarin is the most commonly prescribed anticoagulant drug for the prophylaxis and treatment of thromboembolic disorders [1]. Over the past decade, there has been substantial progress in our understanding of genetic contributions to warfarin response, particularly with regard to warfarin dose requirements [2].
Since the Food and Drug Administration (FDA) changed the labeling of warfarin in 2007 to reflect the impact of polymorphisms of the CYP2C9 and VKORC1 genes on warfarin metabolism and required dose [3], warfarin pharmacogenetics has drawn substantial attention towards “personalized” patient care. In January 2010, the FDA added specific instructions on how to use genotype to predict individualized doses: the new label provides a concise table of dosing recommendations, stratified by genotype [4].
The warfarin dosing methods can be divided into pharmacogenetic and non-pharmacogenetic methods. Non-pharmacogenetic dosing methods include empiric dosing strategies (e.g., fixed dose of 5 mg/day) and clinical dosing algorithms. Pharmacogenetic methods include the dosing table in the warfarin labeling and pharmacogenetic dosing algorithms. Several genotype-based dosing algorithms have been published [5–10]. However, the dosing estimation performed by Gage et al. [8] and the International Warfarin Pharmacogenetics Consortium (IWPC) dosing algorithms have been consistently identified as the most accurate of the algorithms [11–14]. On the other hand, providing an estimated dose in a table, such as the one in the new warfarin label, renders a genotype specific dose readily accessible and easier to implement in clinical practice.
In order to establish genetic strategies of warfarin therapy in common practice, it is essential to first demonstrate the validity of warfarin dosing algorithms [15–17] and to quantify the accuracy of warfarin dosing table in real clinical practice [18]. We chose to compare the performance of the two most accurate algorithms, namely the Gage et al. [8] and the IWPC pharmacogenetic dosing algorithms and the warfarin dosing table in a dataset comprising only Egyptian patients.
Aim of the study
The aim was to find out if the degree of accuracy provided by the pharmacogenetic methods would allow for personalized warfarin treatment in Egyptian patients.
Methods
Study population
Sixty-three Egyptian patients were recruited from the outpatient clinic of the critical care medicine department, Cairo University Teaching Hospitals. The recruited patients had a stable warfarin dose requirement for at least three consecutive times with dose titration to an international normalized ratio (INR) target range of 2–3.5.
The study was performed in accordance with the principles of the Declaration of Helsinki and its appendices [19]. Approval was obtained from the local Institutional Review Board and Ethics Committee, and written informed consent was obtained from all cases.
A baseline blood sample (10 ml) was withdrawn from each patient, 3 ml on EDTA tube for genotyping of the CYP2C9*2, CYP2C9*3 and VKORC1-1639 G > A alleles, 3 ml blood clotted sample for routine laboratory investigation, 2 ml on EDTA tube for complete blood count (CBC), and 2 ml citrated blood for anticoagulation tests.
Patient’s screening included patient’s demographics, indications for warfarin therapy, smoking status, additional medical problems, and concurrent medications.
Genotyping
DNA was extracted using Qiacube supplied by (Qiagen, USA) for automatic DNA extraction according to the manufacturer’s guide. The allelic discrimination was carried out using TaqMan® Probe-based fluorogenic 5′nuclease chemistry. Genotypes for CYP2C9 and VKORC1 were determined using TaqMan® Drug metabolism genotyping assays following the standard assay protocol on the Applied Biosystems® StepOne™ v 2.1 real-time PCR system. For VKORC1-1639 > A genotyping, the assay ID for Taqman® SNP genotyping assay was C_30403261_20 and the dbSNP ID: rs9923231. For CYP2C9*2 Assay ID: C_25625805_10 and dbSNP ID: rs1799853. For CYP2C9*3 Assay ID: C_27104892_10 and dbSNP ID: rs1057910.
Dose prediction methods
Empiric dose
This approach was chosen because a 5 mg empiric starting dose regimen is the most common method of warfarin induction at our institution.
Clinical algorithms
The predicted dose requirements for each patient were calculated according to Gage et al. [8] and IWPC [10] algorithms. The calculations were done using the website http://www.warfarindosing.org [11], without incorporating genetic information. Parameters required for warfarin dose prediction were age, height, weight, gender, race or ethnicity, concomitant medications, target INR, smoking status, and warfarin indication.
Pharmacogenetic algorithms
The predicted dose requirements for each patient were calculated according to Gage et al. [8] and IWPC algorithms [10]. The Gage et al. [8] algorithm is similar to the algorithm available at www.warafrindosing.org [11]; however, the online version has been expanded since the time we started this study to also accommodate newer single-nucleotide polymorphisms that have minor effects on dose. Parameters required for warfarin dose prediction by the two dosing algorithms were CYP2C9 and VKORC1 genotypes, age, height, weight, gender, race or ethnicity, concomitant medications, target INR, smoking status, and warfarin indication.
Warfarin label
The predicted dose requirements for each patient were calculated by taking the midpoint of the daily dose range provided in the table in the newly revised warfarin label (Table 1) [4, 18]. The exact methods used by the FDA to derive this table are not publicly available.
Statistical analysis of data
We compared dose prediction of the pharmacogenetic and non-pharmacogenetic dosing methods with the observed warfarin therapeutic dose in the real clinical setting. The two metrics used to evaluate the predictive accuracy of the dosing models were: the mean absolute error (MAE); and the coefficient of determination (R2). To evaluate the clinical accuracy of the dosing methods predictions, we calculated the percentage of patients whose predicted dose of warfarin was within ±1 mg/day of the actual stable therapeutic dose (ideal dose). Also, we calculated the percentage of patients for whom the predicted dose according to each model was at least 1 mg/day higher than the actual dose (overestimation) or at least 1 mg/day lower than the actual dose (underestimation). Here we adopted the 1 mg/day as a difference that clinicians would likely to define as clinically relevant [1, 10, 20]. We also determined whether each of the dosing methods severely over-predicted dose, defined as predicted dose ≥2 mg/day more than actual dose [14], or severely under-predicted dose defined as ≥2 mg/day less than actual dose.
To further compare the performance of the dosing methods, based on the IWPC model [10], we tested different cut-off values to create low (≤21 mg/week), intermediate (21–49 mg/week), and high dose groups (≥49 mg/week). We analyzed the predicted dose and the observed therapeutic dose and then determined the proportion of patients for whom the dose was underestimated, ideal or overestimated. All the data were analysed using PASW version 18 and graphics utilizing MS Excel.
Results
A total of 63 Egyptian patients agreed to participate in the study. All were turbinate living in Cairo or other surrounding cities. The patients’ demographics and clinical characteristics are shown in Table 2. Co-morbidities were present in 60 patients; the most frequent co-morbidity was coronary heart disease, followed by dyslipidemia and hypertention. In total, 32 patients had drugs with a potential of increasing warfarin’s effect. All 63 patients were genotyped. The minor allele frequencies of CYP2C9*2 and CYP2C9*3 were 0.071 and 0.095, respectively, whereas of VKORC1-1639G > A was 0.51. The observed frequencies for VKORC1 fitted Hardy–Weinberg Equilibrium (HWE), whereas for CYP2C9, (HWE) could not be tested due to the small sample size.
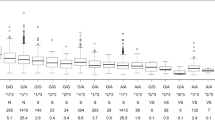
Maintenance dose versus number of variants
The study showed that the number of variant alleles carried by a patient, not considering the type of mutation, can have a linear association with the therapeutic warfarin dose. The relationship between the observed stable warfarin dose of each patient with respect to the number of variants in CYP2C9 and VKORC1 is shown in Fig. 1. The average observed daily therapeutic dose for the wild type group was 11.91 ± 4.44 mg, while the average observed daily therapeutic dose for patients carrying 4 variant alleles was 2.25 ± 0.35.
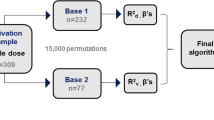
Validation of the pharmacogenetic algorithms and the warfarin dosing table
The predicted dose for each patient was calculated with the two pharmacogenetic algorithms, the warfarin dosing table and the two clinical algorithms. Comparison of the validity of all dosing models in the present cohort is presented in Fig. 2. The stable therapeutic dose was predicted with very close accuracy for both the algorithm of Gage et al. [8] with MAE 3.3 mg/day (SE = 0.46) and adjusted R2 of 42.1 %, and the IWPC algorithm with MAE 3.2 mg/day (SE = 0.46) and adjusted R2 of 41.9 %. However, compared to the two pharmacogenetic algorithms, warfarin dosing table showed a higher MAE of 3.5 mg/day (SE = 0.49) with lower adjusted R2 of 24.6 %.
The two clinical algorithms were less accurate than the two pharmacogenetic algorithms and the warfarin dosing table with respect to adjusted R2 and MAE, however they were more accurate than fixed dose approach regarding MAE.
Clinical accuracy
Table 3 shows that, the three pharmacogenetic models were statistically comparable with each other and with the non-pharmacogenetic methods regarding the proportion of patients whose predicted dose was ideal. Moreover, the three pharmacogenetic models showed significantly lower percentage of patients with severely over-predicted dose compared to the non-pharmacogenetic methods. On the other hand, all models performed similarly regarding severely under-predicted dose.
Also, as shown in Table 4, the three pharmacogenetic methods created less dose overestimation (above 1 mg/day of the actual dose) than would any of the non-pharmacogenetic methods. However, Gage algorithm produced the least overestimated dose predictions (15.9 %), compared with IWPC algorithm (17.5 %) and warfarin dosing table (17.5 %).
Comparison of algorithms and the warfarin dosing table in patient subgroups
Results of the validation analysis performed in subgroups of patients with low, medium, and high therapeutic dose requirements of warfarin are summarized in Table 4. Overall, all models tended to overestimate the dose for patients who required low amounts of warfarin and underestimated the dose for individuals requiring high doses of warfarin at therapeutic INR. All models performed best in the intermediate dose group with respect to percentage of patients within ideal dose. Compared to the non-pharmacogenetic models, the three pharmacogenetic models performed better in the low dose group. In low dose group, Gage’s algorithm provided the highest proportion of patients with an ideal dose (47.1 %). On the other hand, the non-pharmacogenetic methods performed better in the intermediate dose group. In the high dose group (≥7 mg/day), none of the dosing models predicted warfarin doses within ideal dose.
Discussion
Independent external validation of representative, if not all, warfarin dosing models is indispensable for choosing the best algorithm/formula for warfarin dosing prediction [12]. In the present study; based on MAE and R2, it was concluded that, the two pharmacogenetic dosing algorithms showed similar performance, and both performed better than the warfarin dosing table and the two clinical algorithms. However, all models out-performed the 5 mg fixed dose in terms of MAE. This is in accordance with Finkelman et al. [18] study, which showed that dosing based on genetic tables, was somewhat more accurate than that based on non-genetic methods; however, formal pharmacogenetic algorithms were the most accurate. Moreover, our study results are similar to other studies, which showed that, both pharmacogenetic algorithms were similar with respect to MAE and R2, and both outperformed the fixed dose approach although those studies showed lower MAE for both Gage et al. [8], and IWPC algorithms than ours demonstrating better accuracy in terms of MAE [14, 15, 20, 21].
In the present study, all dosing models produced comparable performance with respect to percentage of patients within ideal dose. However, the warfarin dosing table predicted the largest proportion of patients within ideal dose (28.6 %), followed by Gage algorithm (27 %), IWPC algorithm (25.4 %), Gage clinical algorithm (23.8 %) and both IWPC clinical algorithm and fixed dose approach (22.2 %) were the least predicting. The latter percentages of predicted doses that fell within ideal dose by the pharmacogenetic methods were much less than those reported by Finkelman et al. [18] study comprising patients mostly from the IWPC cohort and those reported by Shin and Kayser [22] study. The latter two studies showed that, a pharmacogenetic dosing algorithm predicted more doses within ideal dose (52 %) than a clinical dosing algorithm (39 %), and the dosing table in warfarin label (43 %), and an empiric dosing method (37 %). Also, Marin-Leblanc et al. [15] study on French white patients reported that, predicted doses using Gage or IWPC algorithms were likely to fall within ideal doses approximately 40 % of the time.
In the present study, all dosing models under-predicted dose in more than 50 % of the patients. The latter percentage is much more than other studies in which pharmacogenetic algorithms under-predicted dose in only <30 % of the patients [12, 20]. On the other hand, similar to our study, Shaw et al. [14] study found that fixed dose approach severely over-predicted dose in higher percentage of patients compared to Gage et al. [8] and IWPC algorithms. However, in our study the fixed dose approach severely over-predicted dose by a much higher percentage (27 %) versus 18 % in Shaw et al. [14] study. Percent of severely over-predicted patients provides a surrogate measure of safety. According to Shaw et al. [14] this may be because patients initiating warfarin who are under-dosed and remain sub-therapeutic with respect to INR, are often receiving concomitant bridging with parenteral anticoagulation.
The discordance from other studies in terms of MAE, lower percentages within ideal dose, and higher percentages of under-predicted dose using the pharmacogenetic dosing methods, may be attributed to the difference in race and to the higher mean therapeutic dose in our study cohort. The pharmacogenetic algorithms are especially unlikely to predict unusually high doses, because most do not include genetic variants associated with warfarin resistance [23]. In addition, the warfarin dosing table predicts doses within a range of 0.5-7 mg/day only, whereas 44.4 % of our patients had doses ≥7 mg [2]. Gage algorithm was developed in a cohort of 1015 individuals, 83 % of whom were white, in comparison with a cohort of 5700 individuals, 55 % of whom were white and 30 % were Asians in the IWPC study. Previous studies validating those two algorithms comprised mainly white individuals or Asians. [1, 14, 15, 18, 22] So, may be the two pharmacogenetic algorithms are less accurate in Egyptians due to these differences in ethnic background and the possible cultural differences from our study population.
Moreover, we assumed that, the reason behind this may be attributed to the need for incorporating other clinical factors that can affect warfarin dose, like comorbidities and vitamin K consumption [9, 24–29]. Although additional clinical factors could improve the performance of pharmacogenetic algorithms, this is difficult to apply for improvement of the warfarin dosing table. This may be attributed to the fact that the warfarin dosing table does not give guidelines on how both clinical and genetic factors should be combined to determine appropriate doses [18]. Also, genes other than CYP2C9 and VKORC1 may affect warfarin dose, such as CYP4F2 and ApoE, although their effects on warfarin dose require confirmation [21, 30–32]. Our assumptions are supported by Shahin et al. [33] study on Egyptian patients which reported that, pulmonary embolism was associated with higher warfarin dose, and that Calu rs339097 variant was also associated with higher warfarin dose requirement.
All dosing models provided similar accuracy in the intermediate dose group, although the non-pharmacogenetic methods produced higher percentage of predicted doses within ideal dose. The IWPC gave evidence that; the intermediate dose group is the group, which pharmacogenetic dosing has shown little improvement from the clinical dosing approach [10]. Marin-Leblanc et al. [15] study reported that, in the intermediate dose group, fixed dose approach produced a slightly greater percentage of patients within ideal dose compared to both IWPC and Gage algorithms.
Our data suggests that, the most reliable dosing method for predicting warfarin doses within ideal dose in patients requiring ≤3 mg/day are the Gage and IWPC pharmacogenetic algorithms. In the high dose group (≥7 mg/day), none of the dosing models predicted warfarin doses within ideal dose. This is in discordance with Roper et al. [12] study which, showed that in the high dose group, the Gage and IWPC algorithms predicted 41 % and 29 % within ideal dose, respectively. Also, Marin-Leblanc et al. [15], found that, in the high dose group, the Gage and IWPC algorithms predicted 18.2 % and 13.6 % respectively within ideal dose. However, both Roper et al. [12], and Marin-Leblanc et al. [15], results concerning the low and intermediate dose groups are in agreement with our study. Moreover, IWPC model supported the greatest benefits for using a genotype-based algorithm, compared with using a clinical algorithm or a fixed dose approach, in low and high dose groups [10].
Although this is the first study to be conducted in Egypt to validate pharmacogenetic dosing methods, it still has its limitations. The first is the small sample size recruited. The second is the absence of the higher-level variant CYP2C9 genotype (CYP2C9 *2/*3). Both limitation could be attributed to the financial constraint as the study was totally self-funded.
Conclusion
The present study showed that, genotype-based dosing improved prediction of warfarin therapeutic dose beyond that available with the non-pharmacogenetic methods, especially in the low dose group. We recommend working on the development of an algorithm based on our patient population within a larger sample set, incorporating additional genetic and non-genetic factors, and conducting a comparative prospective clinical trial, between the new algorithm, the online version of Gage et al. [8] pharmacogenetic algorithm available at www.warafrindosing.org, and the current dosing strategy in Egypt (fixed dose approach). This will provide direct evidence of the benefits, disadvantages and costs associated with pharmacogenetic testing, and the best algorithm for initiation of warfarin dosing in Egyptian patients.
References
Takeuchi F, Kashida M, Okazaki O, Tanaka Y, Fukuda S, Kashima T, et al. Evaluation of pharmacogenetic algorithm for warfarin dose requirements in Japanese patients. Circ J. 2010;74:977–82.
Cavallari LH, Shin J, Minoli A, Perera MA. Role of pharmacogenomics in the management of traditional and novel oral anticoagulants. Pharmacotherapy. 2011;31(12):1192–207.
Gage BF, Lesko LJ. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb Thrombolysis. 2008;25:45–51.
Bristol-Meyers Squibb Company. Coumadin® tablets (warfarin sodium tablets, USP) crystalline; Coumadin® for injection (warfarin sodium for injection, USP). Available at: http://www.accessdata.fda.gov./drugsatfda_docs/label/2010/009218s108lbl.pdf. Accessed June 2012.
Hillman MA, Wilke RA, Yale SH, Vidaillet HJ, Caldwell MD, Glurich I, et al. A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clin Med Res. 2005;3:137–45.
Millican EA, Lenzini PA, Milligan PE, Grosso L, Eby C, Deych E, et al. Genetic-based dosing in orthopedic patients beginning warfarin therapy. Blood. 2007;110:1511–5.
Tham LS, Goh BC, Nafziger A, Guo JY, Wang LZ, Soong R, et al. A warfarin-dosing model in Asians that uses single-nucleotide polymorphisms in vitamin K epoxide reductase complex and cytochrome P450 2C9. Clin Pharmacol Ther. 2006;80:346–55.
Gage BF, Eby C, Johnson JA, Deych E, Reider MJ, Ridker PM, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–31.
Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR. McLeod HL use pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004;91(1):87–94.
The International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–64.
Gage BF. Warfarin dosing. Washington University, St Louis. www.warfarindosing.org/Source/Home.aspx. Accessed June 2012.
Roper N, Storer B, Bona R, Fang M. Validation and comparison of pharmacogenetics-based warfarin dosing algorithms for application of pharmacogenetic testing. J Mol Diagn. 2010;12:283–91.
Shin J, Cao D. Comparison of warfarin pharmacogenetic dosing algorithms in a racially diverse large cohort. Pharmacogenomics. 2011;12:125–34.
Shaw PB, Donovan JL, Tran MT, Lemon SC, Burgwinkle P, Gore J. Accuracy assessment of pharmacogenetically predictive warfarin dosing algorithms in patients of an academic medical center anticoagulation clinic. J Thromb Thrombolysis. 2010;30:220–5.
Marin-Leblanc M, Perreault S, Bahroun I, Lapointe M, Mongrain I, Provost S, et al. Validation of warfarin pharmacogenetic algorithms in clinical practice. Pharmacogenomics. 2012;13(1):21–9.
Wu AH, Wang P, Smith A, Haller C, Drake K, Linder M, et al. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equations. Pharmacogenomics. 2008;9(2):169–78.
Lubitz SA, Scott SA, Rothlauf EB, Agarwal A, Peter I, Doheny D, et al. Comparative performance of gene-based warfarin dosing algorithms in a multiethnic population. J Thromb Haemost. 2010;8(5):1018–26.
Finkelman BS, Gage BF, Johnson JA, Brensinger CM, Kimmel SE. Genetic warfarin dosing: tables versus algorithms. J Am Coll Cardiol. 2011;57(5):612–8.
Declaration of Helsinki. World Medical Association. Available from: http://www.wma.net/en/30publications/10policies/b3/. Accessed June 2012.
Sagrieya H, Berube C, Wen A, Ramakrishnan R, Mir A, Hamilton A, Altman RB. Extending and evaluating a warfarin dosing algorithm that includes CYP4F2 and pooled rare variants of CYP2C9. Pharmacogenet Genom. 2010;20:407–13.
Cavallari LH, Langaee TY, Momary KM, Shapiro NL, Nutescu EA, Coty WA, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–64.
Shin J, Kayser SR. Accuracy of the pharmacogenetic dosing table in the warfarin label in predicting initial therapeutic warfarin doses in a large, racially diverse cohort. Pharmacotherapy. 2011;31:863–70.
Schwartz JB, Kane L, Moore K. Wu AH Failure of pharmacogenetic-based dosing algorithms to identify older patients requiring low daily doses of warfarin. J Am Med Dir Assoc. 2011;12(9):633–8.
Garcia D, Regan S, Crowther M, Hughes R, Hylek E. Warfarin maintenance dosing patterns in clinical practice: implications for safer anticoagulation in the elderly populations. Chest. 2005;127:2049–56.
Hirsh J, Dalen JE, Anderson DR, Poller L, Bussey H, Ansell J, et al. Oral anticoagulants: mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest. 1998;114:445S–69S.
James A, Britt R, Raskino C, Thompson S. Factors affecting the maintenance dose of warfarin. J Clin Pathol. 1992;45:704–6.
Booth S, Centurelli M. Vitamin k: a practical guide to the dietary management of patients on warfarin. Nutr Rev. 1999;57:288–96.
Cropp J, Bussey H. A review of enzyme induction of warfarin metabolism with recommendations for patient management. Pharmacotherapy. 1997;17:917–28.
Absher R, Moore M, Parker M. Patient-specific factors predictive of warfarin dosage requirements. Ann Pharmacother. 2002;36:1512–7.
Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433.
Carlquist J, Horne B, Mower C, Park J, Hutinghouse J, McKinney J, et al. An evaluation of nine genetic variants related to metabolism and mechanism of action of warfarin as applied to stable dose prediction. J Thromb Thrombolysis. 2010;30:358–64.
Lee M, Chen C, Chou C, Lu L, Chuang H, Chen Y, et al. Genetic determinants of warfarin dosing in the Han-Chinese population. Pharmacogenomics. 2009;10:1905–13.
Shahin MHA, Khalifa SI, Gong Y, Hammad LN, Sallam MTH, El Shafey M, et al. Genetic and non genetic factors associated with warfarin dose requirements in Egyptian patient. Pharmacogenet Genom. 2011;21:130–5.
Acknowledgments
We express our deep appreciation and thankfulness to the Critical Care Medicine Department, Cairo University Hospitals and all its members for all the help and support.The authors thank Walid Salah at Analysis, Cairo, Egypt for his laboratory assistance.
Funding
This study was completely self funded.
Conflicts of interest
None to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bazan, N.S., Sabry, N.A., Rizk, A. et al. Validation of pharmacogenetic algorithms and warfarin dosing table in Egyptian patients. Int J Clin Pharm 34, 837–844 (2012). https://doi.org/10.1007/s11096-012-9678-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11096-012-9678-3