Abstract
Purpose
Hesperidin, a glycoside flavonoid, is thought to act as an anti-cancer agent, since it has been found to exhibit both pro-apoptotic and anti-proliferative effects in several cancer cell types. The mechanisms underlying hesperidin-induced growth arrest and apoptosis are, however, not well understood. Here, we aimed to investigate the anti-proliferative and apoptotic effects of hesperidin on non-small cell lung cancer (NSCLC) cells and to investigate the mechanisms involved.
Methods
The anti-proliferative and apoptotic effects of hesperidin on two NSCLC-derived cell lines, A549 and NCI-H358, were determined using a WST-1 colorimetric assay, a LDH cytotoxicity assay, a Cell Death Detection assay, an AnnexinV-FITC assay, a caspase-3 assay and a JC-1 assay, respectively, all in a time- and dose-dependent manner. As a control, non-cancerous MRC-5 lung fibroblasts were included. Changes in whole genome gene expression profiles were assessed using an Illumina Human HT-12v4 beadchip microarray platform, and subsequent data analyses were performed using an Illumina Genome Studio and Ingenuity Pathway Analyser (IPA).
Results
We found that after hesperidin treatment, A549 and NCI-H358 cells exhibited decreasing cell proliferation and increasing caspase-3 and other apoptosis-related activities, in conjunction with decreasing mitochondrial membrane potential activities, in a dose- and time-dependent manner. Through a GO analysis, by which changes in gene expression profiles were compared, we found that the FGF and NF-κB signal transduction pathways were most significantly affected in the hesperidin treated NCI-H358 and A549 NSCLC cells.
Conclusions
Our results indicate that hesperidin elicits an in vitro growth inhibitory effect on NSCLC cells by modulating immune response-related pathways that affect apoptosis. When confirmed in vivo, hesperidin may serve as a novel anti-proliferative agent for non-small cell lung cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Worldwide, lung cancer is the leading cause of cancer death among both women and men, with an estimated number of more than 1000,000 deaths each year [1]. Based on their tendencies to metastasize and their responses to existing therapies, lung cancers can be divided into two major classes, i.e., non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) [2]. NSCLC accounts for ~80 % of all lung cancer cases, with adenocarcinoma being the primary subtype [3]. NSCLC is less responsive to chemotherapy than SCLC and, as a consequence, NSCLC is the main focus of current research through which several new therapeutic drugs are being investigated [4–6]. The overall prognosis of lung cancer is poor, which is primarily due to the fact that the diagnosis is frequently made late and/or at an advanced stage [7]. Although 70 % of the newly diagnosed patients present with locally advanced or metastatic lung cancers that require systemic treatment, currently available systemic chemotherapies and molecular targeted therapies have their limitations due to dose-dependent toxicities and the acquisition of drug resistancies [6–8]. In order to develop novel effective treatment strategies for lung cancer, several recent studies have been focusing on dietary agents including nutraceuticals, which are non-toxic, cost-effective and physiologically available [9–11].
Flavonoids are polyphenolic secondary metabolites that are abundantly present in vegetables, fruits and beverages such as tea and wine [12–13]. In particular, citrus flavonoids have been found to exhibit anti-carcinogenic and anti-inflammatory properties [14–16] and, therefore, these flavonoids may serve as potential chemotherapeutic agents [14, 17, 18]. In addition, they have been shown to exhibit low or no cytotoxicities towards normal cells [16]. Citrus flavonoids have also been found to exert synergistic effects with various known chemotherapeutic agents [17]. Hesperedin is a flavanone glycoside that is found abundantly in oranges and lemons [19–20] and has a wide range of properties including anti-inflammatory, analgesic, anti-fungal, anti-viral and anti-cancer properties [21–26]. Hesperidin has also been found to inhibit cell cycle progression in human pancreatic cells [27] and to contribute to the intracellular anti-oxidant defense system by acting as an agent against superoxide, singlet oxygen and hydroxyl radicals [28].
The effects of hesperidin on the prevention and treatment of diseases, including cancer, have recently been a focus of attention due to its pharmacological properties [29]. Although hesperidin is thought to exhibit anti-proliferative effects against many cancer types, its exact mode of action is not well understood [30–31]. Hence, we set out to investigate whether hesperidin has anti-proliferative and/or apoptotic effects in NSCLC cells, and to examine its genome-wide effects on (changes in) gene expression.
2 Materials and methods
2.1 Cells and culture conditions
The NCI-H358 bronchioalveolar carcinoma and MRC-5 lung fibroblast cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The A549 adenocarcinoma cell line was kindly provided by Dr.Yusuf Baran (Department of Molecular Biology and Genetics, IYTE). The NCI-H358 cell line was cultured in RPMI-1640 medium supplemented with 1 % penicillin/streptomycin and 10 % fetal bovine serum (FBS) at 37 °C in 5 % CO2. The A549 and MRC-5 cell lines were cultured in MEM-α medium supplemented with 1 % penicillin/streptomycin and 10 % FBS at 37 °C in 5 % CO2.
2.2 Reagents and assay kits
All cell culture reagents were purchased from GIBCO Life Technologies (Thermo Fisher Scientific, USA). Hesperidin was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The WST-1, Cell Death Detection ELISA PLUS and High Pure RNA Isolation kits were purchased from Roche Life Sciences (Germany). The CytoTox 96R Non-Radioactive Cytotoxicity Assay kit was purchased from Promega Corporation (Madison, WI, USA). The FITC Annexin V Apoptosis Detection kit was purchased from BD Biosciences (Franklin Lakes, NJ, USA). The Caspase-3 colorimetric kit was purchased from BioVision Research Products (Milpitas, CA, USA). The JC-1 Mitochondrial Membrane Potential Detection kit was purchased from Cayman Chemical (Ann Arbor, MI, USA). Human whole genome gene expression profiling was performed using Illumina Human HT-12v4 beadchips and microarray reagents (San Diego, CA, USA).
2.3 Cell growth and cytotoxicity assays
A549, NCI-H358 and MRC-5 cells were seeded at a density of 1 × 104/ml (0,1 ml/well) in 96-well microtiter plates and allowed to adhere and grow for 24 h. Cells in log phase were used for subsequent drug treatment. Hesperidin was dissolved in DMSO as a 100 mM stock solution and stored as aliquots in tightly sealed vials at −20 °C. Working solutions were prepared by serial dilution of the stock solution with culture medium. After the initial 24 h incubation period, cells were treated with increasing concentrations of hesperidin (5–50 μM) for 24, 48 and 72 h. A WST-1 colorimetric cell proliferation assay (Roche Life Sciences, Germany) was performed to assess its effects on cell viability. After each incubation period, cells were treated with 10 μl 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt (WST-1) for 4 h. Subsequently, color development was measured at 450 nm using a Multiscan ELISA reader (Thermo Fisher Scientific, Germany). Cells not treated with hesperidin were used as negative controls.
The cytotoxic effect of hesperidin was colorimetrically determined using a CytoTox 96R Non-Radioactive Cytotoxicity Assay kit (Promega; Madison, WI, USA). To this end, cells were treated as described above for the WST-1 assay and the culture media (10 μl) were transferred to a 96-well microtiter plate. Subsquently, the levels of lactate dehydrogenase (LDH) were determined by adding a freshly prepared substrate mix, incubation in the dark at room temperature for 30 min, addition of 50 μl STOP Solution and measurement of the optical density at 490 nm using a microplate reader (Bio-Rad; Hercules, CA, USA). The measurements were repeated in three independent experiments.
2.4 Apoptotic nucleosomal DNA release assay
The Cell Death Detection ELISA PLUS assay (CDDE; Roche Life Sciences, Germany) is a photometric enzyme-immunoassay for the quantitative in vitro determination of DNA in cytoplasmic histone-associated DNA fragments (mono- and oligo-nucleosomal), which is a measure of apoptosis. Color development of the samples was taken as an enrichment factor of DNA fragments in the cell supernatant relative to untreated cells. Briefly, 1 × 104/ml (0,1 ml/well) A549 and NCI-H358 cells (treated with 5–50 μM hesperidin for 48 h and 72 h, or negative controls) were centrifuged at 200×g for 10 min. After removal of the supernatants, cell pellets were resuspended in 200 μl lysis buffer and incubated for 30 min at room temperature. The resulting cell lysates were centrifuged at 200×g for 10 min after which 20 μl of the supernatants was transferred to streptavidin-coated microtiter plates. After a two hour incubation at room temperature, the solutions were removed and each well was rinsed three times with incubation buffer. Next, 100 μl ABTS solution (Roche Life Sciences, Germany) was added to each well and incubated until color development was sufficient for photometric analysis. Finally, 100 μl ABTS stop solution was added to each well and color development was measured at 405 and 490 nm using an Elisa plate reader (Thermo Fisher Scientific, Finland).
2.5 Caspase-3 enzyme activity assay
Changes in caspase-3 enzyme activity serve as important indicators of apoptosis [32]. These changes were examined using a caspase-3 colorimetric assay (BioVision Research Products; San Francisco, CA, USA). Briefly, 5 × 105 A549 and MRC-5 cells (treated with 5–50 μM hesperidin for 48 h and 72 h, or negative controls) were collected by centrifugation, lysed by adding 50 μl chilled Cell Lysis Buffer and incubated on ice for 10 min before centrifugation. The resulting supernatants were transferred to new tubes. Next, reaction mixtures were prepared by adding 50 μl Reaction Buffer, 50 μl sample and 5 μl DEVD-pNA substrate, and incubated for 2 h. The reactions were read at 405 nm as described above.
2.6 Mitochondrial membrane potential assay
We also examined loss of mitochondrial membrane potential (MMP) in response to hesperidin treatment for 48 h in A549 and MRC-5 cells by JC-1 assay (Cayman Chemical, Ann Arbor, MI, USA). Briefly, cells (5 × 105 cells/2 ml) were collected by centrifugation at 1000 rpm for 10 min. Supernatants were removed, pellets were homogenized in 200 μl medium, and 20 μl JC-1 dye was added to the cells. Next, the cells were incubated at 37 °C in 5 % CO2 for 30 min, centrifuged and, subsequently, 200 μl assay buffer was added to the resulting pellets. All pellets were re-suspended in 320 μl assay buffer and 100 μl of each of them was added to a 96-well plate in triplicate. The plate was read using a fluorescence Elisa plate reader (see above). Green/red (485 nm/560 nm) values were calculated to determine changes in MMP.
2.7 Phosphatidylserine exposure and cell permeability assay
During apoptotic cell death, phosphatidylserine (PS), a phospholipid component of the inner-leaflet of cell membranes [32], becomes available at the cell surface. This early marker of apoptotic cell death can be detected by staining with the green fluorescent dye Annexin V-FITC (BD Pharmingen, Germany). Briefly, 1 × 105/ml A549 and NCI-H358 cells (treated with 5–50 μM hesperidin for 48 h, or negative controls) were centrifuged, washed with HBSS (GIBCO, Germany) and stained with 150 μl buffer containing 5 μl Annexin V-FITC (1 mg/ml) and 1,5 μM 7-AAD at 37 °C. After 10 min, 500 μl ice-cold buffer was added and the cells were harvested by scraping off on ice. After centrifugation, the resulting cells were re-suspended in 250 μl buffer and analysed immediately using flow cytometry.
2.8 RNA extraction and microarray hybridization
Total RNA was isolated from A549, NCI-H358 and MRC-5 cells in response to 50 μM hesperidin treatment for 48 h using a High Pure RNA Isolation Kit (Roche; Madison, WI, USA) according to the manufacturer’s instructions. RNA concentrations were determined using a NanoDrop (Thermo Fisher Scientific) spectrophotometer (260 nm/280 nm ratios) and only the samples with an A260/A280 ratio between 1,9 and 2,1 were considered suitable for use. Illumina Human HT-12v4 beadchip microarrays containing 47,000 probes were used to assess the global gene expression profiles for each sample (Illumina, San Diego, CA, USA). Five hundred nanogram total RNA was amplified, converted to cRNA, fragmented, and biotin-labeled using an Illumina TotalPrep RNA-amplification kit (Ambion; Foster City, CA, USA). Next, 1,5 μg of labeled cRNA was hybridized to each microarray according to the Illumina whole-genome gene expression direct hybridization assay protocol, after which the microarrays were scanned using an Illumina BeadArray Reader. The images were processed and converted into signal intensities using the Illumina GenomeStudio software package. The same software package was used to perform hybridization quality controls (QC).
2.9 Statistical analysis
Signal intensities corresponding to the gene expression levels of individual microarrays were corrected for background noise and imported into text files using the Illumina GenomeStudio. Hierarchical cluster analysis was performed on datasets to evaluate the “proximity” between the time points. For data visualization, hierarchical clusters were constructed using the statistically significant (p < 0,05) genes. By computing a p-value for each gene assessed, the statistical significance of the differential expression of genes was determined. Genes were considered differentially expressed when logarithmic gene expression ratios in three independent hybridization experiments showed > 1,5-fold difference in expression level, and when the p-values were < 0,05. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA, USA) tools, and gene ontology (GO) and pathway analyses were used to retrieve biological meanings of the differentially expressed genes between the data points.
3 Results
3.1 Hesperidin inhibits the proliferation of A549 and NCI-H358 cells
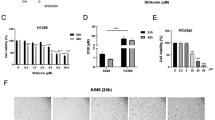
To determine the anti-proliferative effects of hesperidin on A549 and NCI-H358 non-small cell lung carcinoma (NSCLC) cells and MRC-5 normal lung fibroblasts, the respective cell lines were incubated with increasing concentrations (5–50 μM) of hesperidin for 24, 48 and 72 h and, next, subjected to a WST-1 colorimetric cell proliferation (viability) assay. By doing so, we observed significant (p < 0,05) time- and dose-dependent decreases in cell proliferation in the NSCLC cell lines, but not in the control fibroblast cell line nor in the non-treated control cells. The IC50 values of hesperidin for 24, 48 and 72 h were calculated from both cell proliferation and cytotoxicity (see below) plots and estimated to be 50 μM (Fig. 1a-c).
Effects of hesperidin on cell proliferation (viability) and cytotoxicity. Effects of hesperidin on proliferation and cytotoxicity of A549 (a–d), NCI-H358 (b–e) and MRC-5 (c–f.) cells. WST-1 proliferation and LDH cytotoxicity assays were performed using triplicate samples in three independent experiments. Statistical significance was determined using two-way analysis of variance, and p < 0.05 was considered significant (* p < 0,05; ** p < 0,01).
3.2 Hesperidin exerts a cytotoxic effect on A549 and NCI-H358 cells
To determine the cytotoxic effects of hesperidin on A549 and NCI-H358 NSCLC cells and MRC-5 normal lung fibroblasts, the respective cell lines were incubated with increasing concentrations (5–50 μM) of hesperidin for 24, 48, 72 h and, next, subjected to a LDH cytotoxicity assay (CytoTox 96R Non-Radioactive Cytotoxicity Assay). Through this assay, we observed significant (p < 0,05) time- and dose-dependent increases in cytotoxicity compared to the control fibroblast cell line and the non-treated control cells (Fig. 1d-f).
3.3 Hesperidin increases the apoptotic nucleosomal enrichment factor
To quantify apoptotic cell death colorimetrically, A549 and NCI-H358 NSCLC cells were incubated with 50 μM (see above) hesperidin for 48 and 72 h, and changes in nucleosomal enrichment factor (EF) were analyzed using a Cell Death Detection ELISA PLUS assay (CDDE). We observed 5,15-fold and 5,85-fold increases in EF after 48 and 72 h incubation with 50 μM hesperidin in A549 cells, respectively, as compared to the non-treated control cells (Fig. 2a; p < 0,05). In analogy, we observed 6,55-fold and 7,06-fold increases in EF in response to 48 and 72 h incubation with 50 μM hesperidin, respectively, in NCI-H358 cells compared to the non-treated control cells (Fig. 2b; p < 0,05).
Effects of hesperidin on apoptosis related nucleosomal enrichment factors. Effects of hesperidin on apoptosis-related nucleosomal enrichment factor of A549 (a) and NCI-H358 (b) cells. Cell Death Detection Plus ELISA assays were performed using triplicate samples in at least two independent experiments. Statistical significance was determined using two-way analysis of variance, and p < 0.05 was considered significant (* p < 0,05; ** p < 0,01)
3.4 Hesperidin increases apoptotic caspase-3 activity
To determine apoptotic effects of hesperidin on A549 NSCLC cells and MRC-5 normal lung fibroblasts, the respective cell lines were incubated with increasing concentrations of hesperidin for 48 h and changes in caspase-3 enzyme activities were measured using a colorimetric assay kit (see matarials and methods). We found a 1,19-fold increase in caspase-3 activity after a 48 h incubation with 50 μM hesperidin in A549 cells, as compared to non-treated control cells (Fig. 3; p < 0,05). Under the same conditions, we found no change in caspase-3 activity in MRC-5 fibroblasts when compared to A549 NSCLC cells (Fig. 3). These results indicate that a dose-dependent hesperidin-induced apoptosis of NSCLC cells may be related to caspase-3 activity.
3.5 Hesperidin induces loss of mitochondrial membrane potential
To assess the loss of mitochondrial membrane potential (MMP), A549 NSCLC cells and MRC-5 normal lung fibroblasts were exposed to 50 μM hesperidin for 48 h, after which a JC-1 MMP assay was performed. This assay revealed a 1,14-fold increase in loss of MMP in response to 50 μM hesperidin for 48 h in A549 cells, as compared to non-treated control cells (Fig. 4; p < 0,05). Under the same conditions, no significant MMP change in the MRC-5 cells was observed when compared to A549 cells (Fig. 4).
Effects of hesperidin on membrane potential in A549 and MRC-5 cells. Loss of mitochondrial membrane potential (MMP) in response to increasing concentrations of hesperidin in A549 and MRC-5 cells. The results are represented as the means of two independent experiments. p < 0.05 was considered significant
3.6 Hesperidin causes translocation of PS from the inner to the outer leaflet
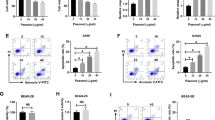
FITC AnnexinV/PI double staining was performed in A549 and NCI-H358 NSCLC cells after exposure to 50 μM hesperidin for 48 h. The results of this assay indicate that a 48 h incubation of A549 and NCI-H358 cells with 50 μM hesperidin increases apoptotic cell death compared to the non-treated control group (Fig. 5a-d; p < 0,05).
Effects of hesperidin on apoptosis. Effects of hesperidin (0 to 50 μM) on apoptosis in A549 (a–b) and NCI-H358 (c–d) cells. FITC AnnexinV/PI double staining was used on cells treated with increasing concentrations of hesperidin. Early apoptotic cells labeled with Annexin-V but not PI (in lower right quadrant, Q4) and apoptotic cells labelled with Annexin-V and PI (in upper right quadrant, Q2) are depicted in the flow cytometric graphics. Necrotic cells are in quadrant Q1 The results presented in b and d are means of two independent experiments. p < 0.05 was considered significant (* p < 0,05; ** p < 0,01)
3.7 Hesperidin induces changes in gene expression profiles
To reveal the cellular signaling pathways in NSCLC cells that are affected by hesperidin treatment, we performed Illumina Human HT-12v4 beadchip microarray-based whole-genome expression profiling on A549 and NC-H358 NSCLC cells and MRC-5 normal lung fibroblasts after 48 h hesperidin treatment (50 μM). As an initial step, we analyzed the transcripts that were most differentially expressed (i.e., most significant p-values and fold-changes) and clustered the transcripts according to their patterns of differential expression across the samples. Then, we annotated biofunctions and performed downstream effect analyses of hesperidin. To reveal biologically relevant networks beyond the canonical pathways, pathway analyses were performed for the gene sets using IPA. These networks indicate functional relationships between gene products based on known interactions reported in the literature (Fig. 6).
Functional analysis of genes selected through microarray-based expression profiling. The functional network was algorithmically constructed using Ingenuity Pathway Analysis (IPA) software on the basis of the functional and biological connectivity of genes in A549 (a), NCI-H358 (b) and MRC-5 (c) cells. The top-scoring genes regulated after 48 h of 50 μM hesperidin treatment, where Gpcr (a, c) and IFN-γ (b) are central nodes generated by the IPA software, are shown. Red and green represent up- and down-regulated genes, respectively, based on fold change as compared to non-treated samples
Through IPA analysis, three major goups of pathways emerged in hesperidin treated A549 cells, i.e., cell signaling, molecular transport, vitamin and mineral metabolism (score = 44), cellular assembly, organization and molecular transport (score = 35), and organ morphology and cellular development (score = 35) pathways (supplemental Table 1). In hesperidin treated NCI-H358 cells the major groups of pathways were cell-to-cell signaling and interaction, cell-mediated immune response and inflammatory response (score = 35), cellular response to therapeutics (score = 35), and organismal development, cell cycle, and cellular assembly and organization (score = 32) pathways (supplemental Table 2). In hesperidin treated MRC-5 cells the major groups of pathways were cellular signaling and nucleic acid metabolism (score = 36) and lipid metabolism and molecular transport networks (score = 34) pathways (supplemental Table 3).
Next, functional annotation analyses were performed comparing the hesperidin treated A549 and NCI-H358 NSCLC cell lines. We clustered functional annotations of putative hesperidin targets using lung tumor, non-small lung cancer cells, lung cancer and tumors, lung adenocarcinoma, and carcinoma in lung as queries. By doing so, we found that genes in apoptosis (p-value: 9,25E-13) and immune response (p-value: 3,77E-12) pathways were most significantly affected in the hesperidin treated NSCLC cell lines (supplemental Table 4).
Based on further detailed pathway analyses, we found that the FGF network genes FGF, PI3K, AKT, ERK and STAT were differentially expressed in hesperidin treated A549 and NCI-H358 cells (supplemental Fig.7). In addition, we found that the NF-κB pathway genes IL-1, IL-1/TLR, PKCβ, RANKL and BR3 were up-regulated, and the BMP 2/4, RAS and PI3K genes were down-regulated, in NCI-H358 cells (supplemental Fig. 8).
4 Discussion
Hesperidin is one of the most common flavanones and is abundantly present in citrus fruits. Recent studies indicate that hesperidin may exhibit cancer preventive and apoptotic effects [33]. The putative underlying mechanisms are, however, still unknown. In the colon cancer cell line SNU-C4 it has been shown that hesperidin can decrease cell viability by 65 % compared to untreated cells [29]. Subsequent assessment of the expression of apoptosis-regulating genes indicated that hesperidin decreased the expression of BCL2 mRNA and increased the expression of the BCL2-associated X (BAX) protein. The activity of the major apoptotic factor caspase-3 was found to increase significantly upon hesperidin treatment, i.e., hesperidin down-regulated the expression of pro-caspase-3 and upregulated the level of active caspase-3. These results suggest that hesperidin may induce apoptosis in human colon cancer cells through caspase-3 activation [29]. Similarly, Saiprasad et al. [35] showed that hesperidin can induce apoptosis through modulating BCL2/BAX levels to increase the activity of caspase-3 and caspase-9 in experimental colon cancer models. Furthermore, it has been found that hesperidin can induce cell death in hepatocellular carcinoma HepG2 cells in a caspase-independent manner by activating the ERK1/2 pathway [36]. Taken together, these results indicate that hesperidin may induce different cell death programs in different cancer cell types.
It has also been found that hesperidin can affect the proliferation of MCF-7 breast cancer cells transfected with green fluorescent protein (GFP)-coupled α-tubulin (MCF-7-GFP-Tubulin), androgen-independent PC-3 and DU-145 prostate cancer cells, and androgen-dependent LNCaP prostate cancer cells [34]. Specifically, hesperidin was found to inhibit the proliferation of MCF-7-GFP-Tubulin cells, most likely independent of an anti-mitotic mechanism. Hesperidin was also found to inhibit basal and testosterone-induced proliferation of LNCaP cells, whereas it did not significantly affect the proliferation of androgen-independent prostate cancer cells (PC-3 and DU-145). Based on these data, the authors concluded that further research on possible functional interactions between hesperidin and androgen receptors, including their downstream signaling pathways, is warranted [34].
Our data are in line with previous studies indicating that hesperidin may have growth inhibitory and apoptotic effects in cancer cells. Specifically, we found that hesperidin has a growth inhibitory effect and triggers the apoptotic pathway in two NSCLC cell lines (A549 and NCI-H358 cells) in a time- and dose-dependent manner. We also observed a very low cytotoxic effect of hesperidin in normal lung fibroblasts (MRC-5). Hesperidin was found to increase the caspase-3 enzyme level and to induce loss of mitochondrial membrane potential (MMP), which are important indicators of apoptosis, in a time- and dose-dependent manner. FITC AnnexinV/PI double staining confirmed the apoptotic effects of hesperidin on the NSCLC cells tested.
Citrus flavonoids may act in concert with known chemotherapeutic agents. It has e.g. been reported that hesperidin treatment of Ramos Burkitt’s lymphoma cells can abrogate constitutive and doxorubicin-induced NF-κB activation through inhibition of phosphorylation of its inhibitory subunit (IκB), and that hesperidin treatment can result in inhibition of proliferation and induction of apoptosis in a PPARγ-independent manner [31]. In another study, Ghorban et al. reported that hesperidin can induce the expression and transcriptional activity of PPARc, promote p53 accumulation and downregulate constitutive NF-κB activity in a PPARγ-dependent and PPARγ-independent manner in the pre-B cell line NALM-6 [30]. The growth-inhibitory effect of hesperidin was partially reduced when the NALM-6 cells were incubated with a PPARγ antagonist prior to the exposure to hesperidin. The findings of this latter study clearly demonstrate that hesperidin-mediated pro-apoptotic and anti-proliferative actions are regulated via both PPARγ-dependent and PPARγ-independent pathways in NALM-6 cells. Together, these data indicate that hesperidin has a potential to be developed into a therapeutic agent for hematologic malignancies.
In order to determine signaling networks and pathways regulated by hesperidin in NSCLC cells, we performed microarray-based expression profiling. By doing so, we found that inflammatory response pathways, including the FGF and NF-κB signal transduction pathways, were most significantly affected in the NCI-H358 and A549 NSCLC cells tested. Taken together, we conclude that hesperedin elicits a growth inhibitory effect on NSCLC cells by modulating immune response-related pathways that affect apoptosis. As such, hesperidin may serve as a novel anti-proliferative agent for non-small cell lung cancer.
Abbreviations
- EF:
-
Nucleosomal enrichment factor
- FBS:
-
Fetal bovine serum
- FGF:
-
Fibroblast growth factor
- FITC:
-
Fluorescein ısothiocyanate
- IPA:
-
Ingenuity pathway analysis
- LDH:
-
Lactate dehydrogenase
- MEM-α:
-
Eagle’s minimum essential medium
- NF-kB:
-
Nuclear factor kappa B
- NSCLC:
-
Non-small cell lung cancer
- PS:
-
Phosphatidylserine
- RPMI-1640:
-
Roswell Park Memorial Institute-1640
References
W. Pao, N. Girard, New driver mutations in non-small-cell lung cancer. Lancet Oncol. 12, 175–180 (2011)
W.D. Travis, Classification of lung cancer. Semin. Roentgenol. 46, 178–186 (2011)
Q. Wu, Y.F. Chen, J. Fu, Q.H. You, S.M. Wang, X. Huang, X.J. Feng, S.H. Zhang, Short hairpin RNA-mediated down-regulation of CENP-A attenuates the aggressive phenotype of lung adenocarcinoma cells. Cell. Oncol. 37, 399–407 (2014)
A. Koren, H. Motaln, T. Cufer, Lung cancer stem cells: a biological and clinical perspective. Cell. Oncol. 36, 265–275 (2013)
N. Peled, M.W. Wynes, N. Ikeda, T. Ohira, K. Yoshida, J. Qian, M. Ilouze, R. Brenner, Y. Kato, C. Mascaux, F.R. Hirsch, Insulin-like growth factor-1 receptor (IGF-1R) as a biomarker for resistance to the tyrosine kinase inhibitor gefitinib in non-small cell lung cancer. Cell. Oncol. 36, 277–288 (2013)
A. Maier, AL. Peille, V. Vuaroqueaux, M. Lahn. Anti-tumor activity of the TGF-β receptor kinase inhibitor galunisertib (LY2157299 monohydrate) in patient-derived tumor xenografts. Cell. Oncol. 2015 Jan 9. [Epub ahead of print] DOI 10.1007/s13402-014-0210-8
P. Ulivi, R. Silvestrini, Role of quantitative and qualitative characteristics of free circulating DNA in the management of patients with non-small cell lung cancer. Cell. Oncol. 36, 439–448 (2013)
G. Giaccone, Twenty-five years of treating advanced NSCLC: what have we achieved? Ann. Oncol. 15 Suppl 4: iv81–83 (2004)
J.W. Neal, L.V. Sequist, Exciting new targets in lung cancer therapy: ALK, IGF-1R, HDAC, and Hh. Curr. Treat. Options in Oncol. 11, 36–44 (2010)
R. Sangha, P.N. Lara, P.C. Mack, D.R. Gandara, Beyond antiepidermal growth factor receptors and antiangiogenesis strategies for nonsmall cell lung cancer: exploring a new frontier. Curr. Opin. Oncol. 21, 116–123 (2009)
S. Mateen, K. Raina, R. Agarwal, Chemopreventive and anti-cancer efficacy of silibinin against growth and progression of lung cancer. Nutr. Cancer J. 65, 3–11 (2013)
J. Nones, T.C.E. Spohr, F.C. Gomes, Hesperidin, a flavone glycoside, as mediator of neuronal survival. Neurochem. Res. 36, 1776–1784 (2011)
T. Tanaka, R. Takahashi, Flavonoids and asthma. Nutrients 10, 2128–2143 (2013)
J.A. Manthey, K. Grohmann, N. Guthrie, Biological properties of citrus flavonoids pertaining to cancer and inflammation. Curr. Med. Chem. 8, 135–153 (2001)
J. Yu, L. Wang, R.L. Walzem, E.G. Miller, L.M. Pike, B.S. Patil, Antioxidant activity of citrus limonoids, flavonoids, and coumarins. J. Agric. Food Chem. 53, 2009–2014 (2005)
S.L. Hwang, P.H. Shih, G.C. Yen, Neuroprotective effects of citrus flavonoids. J. Agric. Food Chem. 60, 877–885 (2012)
E. Meiyanto, A. Hermawan, Anindyajati: natural products for cancer-targeted therapy: citrus flavonoids as potent chemopreventive agents. Asian. Pac. J. Cancer Prev. 13, 427–1436 (2012)
J.A. Manthey, N. Guthrie, Antiproliferative activities of citrus flavonoids against six human cancer cell lines. J. Agric. Food Chem. 50, 5837–5843 (2002)
G. Saiprasad, P. Chitra, R. Manikandan, G. Sudhandiran, Hesperidin alleviates oxidative stress and downregulates the expressions of proliferative and inflammatory markers in azoxymethane-induced experimental colon carcinogenesis in mice. Inflamm. Res. 62, 425–440 (2013)
O.G. Benavente, J. Castillo, M. Alcaraz, V. Vicente, J.A. Del, A. Ortuno, Beneficial action of Citrus flavonoids on multiple cancer-related biological pathways. Curr. Cancer Drug Targets 7, 795–809 (2007)
J. Nones, T.C.E. Spohr, F.C. Gomes, Hesperidin, a flavone glycoside, as mediator of neuronal survival. Neurochem. Res. 36, 1776–1784 (2011)
V. Gaur, A. Kumar, Hesperidin pre-treatment attenuates NO-mediated cerebral ischemic reperfusion injury and memory dysfunction. Pharmacol. Rep. 62, 635–648 (2010)
S. Ou, Pharmacological action of hesperidin. Zhong. Yao. Cai. 25, 531–533 (2002)
O. Benavente-Garcia, J. Castillo, Update on uses and properties of citrus flavonoids: new findings in anticancer, cardiovascular, and anti-inflammatory activity. J. Agric. Food Chem. 56, 6185–6205 (2008)
A. Chanet, D. Milenkovic, C. Manach, A. Mazur, C. Morand, Citrus flavanones: what is their role in cardiovascular protection? J. Agric. Food Chem. 60, 8809–8822 (2012)
B.D. Sahu, M. Kuncha, G.J. Sindhura, R. Sistla, Hesperidin attenuates cisplatin-induced acute renal injury by decreasing oxidative stress, inflammation and DNA damage. Phytomedicine 20, 453–460 (2013)
J.R. Patil, K.N. Chidambara Murthy, G.K. Jayaprakasha, M.B. Chetti, B.S. Patil, Bioactive compounds from Mexican lime [Citrus aurantifolia] juice induce apoptosis in human pancreatic cells. J. Agric. Food Chem 57, 10933–10942 (2009)
M. Galleano, O. Pechanova, C.G. Fraga, Hypertension, nitric oxide, oxidants, and dietary plant polyphenols. Curr. Pharm. Biotechnol. 11, 837–848 (2010)
H.J. Park, M.J. Kim, E. Ha, J.H. Chung, Apoptotic effect of hesperidin through caspase3 activation in human colon cancer cells, SNU-C4. Phytomedicine 15, 147–151 (2008)
A. Ghorban, M. Nazari, M. Jeddi-Tehrani, H. Zand, The citrus flavonoid hesperidin induces p53 and inhibits NF-κB activation in order to trigger apoptosis in NALM-6 cells: involvement of PPARγ-dependent mechanism. Eur. J. Nutr. 51, 39–46 (2012)
M. Nazari, A. Ghorbani, A. Hekmat-Doost, M. Jeddi-Tehrani, H. Zand, Inactivation of nuclear factor-κB by citrus flavanone hesperidin contributes to apoptosis and chemo-sensitizing effect in Ramos cells. Eur. J. Pharmacol. 650, 526–533 (2011)
K. Mohankumar, S. Pajaniradje, S. Sridharan, V.K. Singh, L. Ronsard, A.C. Banerjea, B.C. Selvanesan, M.S. Coumar, L. Periyasamy, R. Rajagopalan, Apoptosis induction by an analog of curcumin (BDMC-A) in human laryngeal carcinoma cells through intrinsic and extrinsic pathways. Cell. Oncol. 37, 439–454 (2014)
S. Aranganathan, N. Nalini, Efficacy of the potential chemopreventive agent, hesperetin [citrus flavanone], on 1,2-dimethylhydrazine induced colon carcinogenesis. Food Chem. Toxicol. 47, 2594–2600 (2009)
C.J. Lee, L. Wilson, M.A. Jordan, V. Nguyen, J. Tang, G. Smiyun, Hesperidin suppressed proliferations of both human breast cancer and androgen-dependent prostate cancer cells. Phytother. Res. Suppl 1, S15–S19 (2010)
G. Saiprasad, P. Chitra, R. Manikandan, G. Sudhandiran, Hesperidin induces apoptosis and triggers autophagic markers through inhibition of Aurora-A mediated phosphoinositide-3-kinase/Akt/mammalian target of rapamycin and glycogen synthase kinase-3 beta signalling cascades in experimental colon carcinogenesis. Eur. J. Cancer 50, 2489–2507 (2014)
S. Yumnam, HS. Park, MK. Kim, A. Nagappan, GE. et al, Hesperidin induces paraptosis like cell death in hepatoblatoma, HepG2 cells: involvement of ERK1/2 MAPK. PLoS One 30;9(6):e101321 (2014)
Acknowledgments
This work was funded by Istanbul University Scientific Research Project number 9205. We would like to thank Mr. David Chapman for editing the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.

Supplemental Fig. 7
FGF signaling network. FGF network of 50 μM hesperidin-stimulated genes in A549 (a), NCI-H358(b) and MRC-5(c) cells. Ingenuity pathways analysis (IPA) software was used to identify genes involved in the FGF signaling that were differentially expressed in A549, NCI-H358 and MRC-5 cells. Genes labeled in red and green were identified as up- and down-regulated, respectively, whereas other genes were identified on the basis of the network analysis. (GIF 32 kb)
Fig. 7
High Resolution Image (TIFF 1736 kb)

Supplemental Fig. 8
NF-κB signaling network. NF-κB network of 50 μM hesperidin-stimulated genes in NCI-H358 cells. Ingenuity pathways analysis (IPA) software was used to identify genes involved in the NF-κB signaling that were differentially expressed in NCI-H358 cells. Genes labeled in red and green were identified as up- and down-regulated, respectively, whereas other genes were identified on the basis of the network analysis. (JPEG 131 kb)
ESM 1
(DOCX 14 kb)
ESM 2
(DOCX 14 kb)
ESM 3
(DOC 31 kb)
ESM 4
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Birsu Cincin, Z., Unlu, M., Kiran, B. et al. Anti-proliferative, apoptotic and signal transduction effects of hesperidin in non-small cell lung cancer cells. Cell Oncol. 38, 195–204 (2015). https://doi.org/10.1007/s13402-015-0222-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-015-0222-z